On Spectral-Spatial Classification of Hyperspectral Images Using Image Denoising and Enhancement Techniques, Wavelet Transforms and Controlled Data Set Partitioning
Abstract
:1. Introduction
2. Related Work
3. Methods and Materials
3.1. Addressing the Challenges in HSI Image Processing
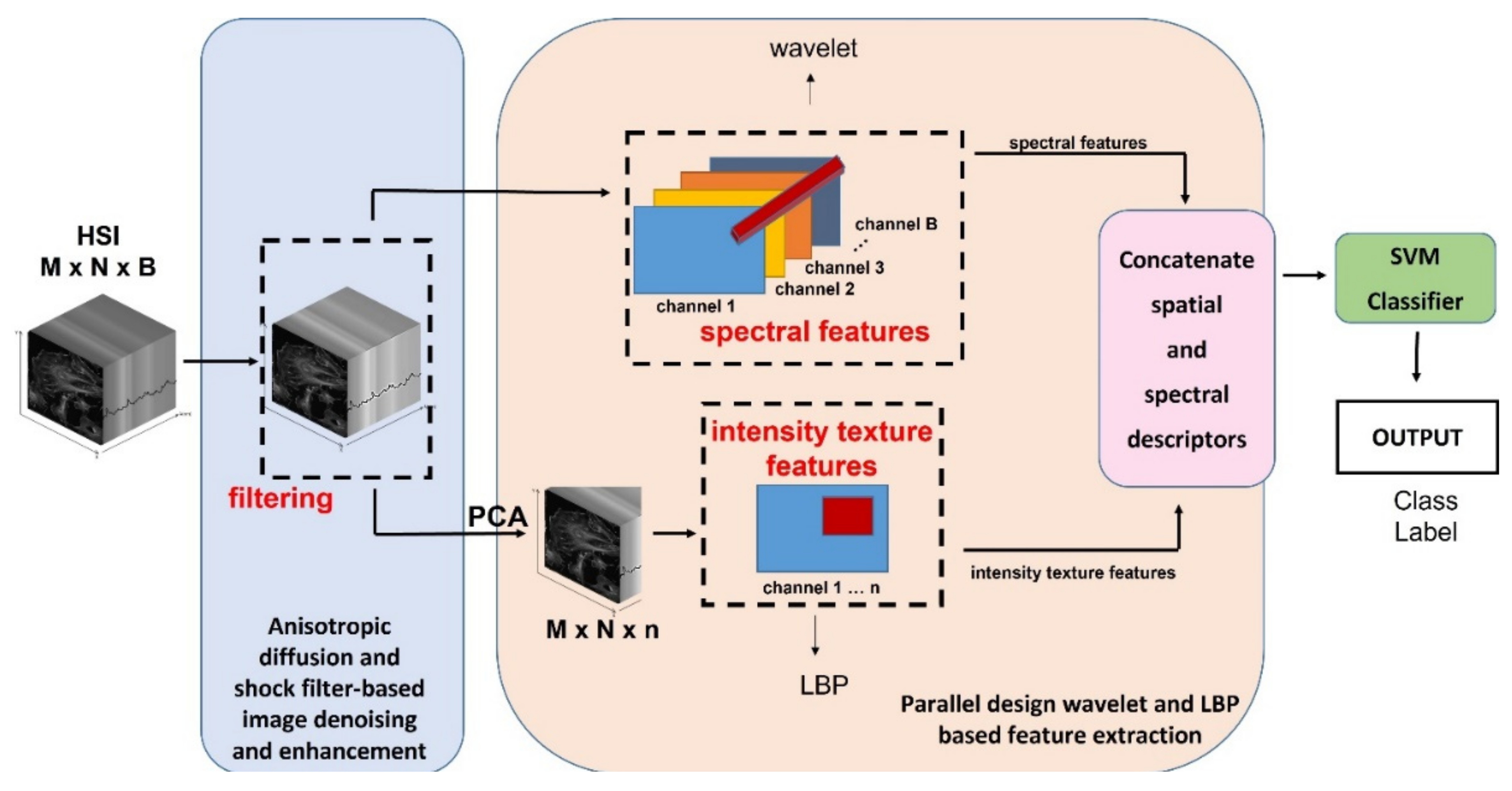
3.2. The Proposed Processing Method
3.2.1. The Spatial-Spectral Adaptive Shock Filtering Model
3.2.2. The Parallel Approach Based on Wavelet and LBP for Feature Extraction
3.2.3. The Concatenation of the Spatial and Spectral Descriptors
3.2.4. Controlled Sampling for Classification Accuracy Estimation
| Input: HSI data set U, the sampling rate ratio (the number of samples/percentage per class, the reference map, the desired distance between the pixels, the window representing the populated neighborhood |
| Output: training data set X_train |
| For each class c in U: |
| Determine all the partitions X in class C |
| For each partition x in X: |
| Get the centroid for each region in the partition |
| Get the spatial coordinates of the corresponding centroid |
| Calculate the number of training samples tot in the partition: |
| no_of_desired_samples = tot * ratio |
| While visited register is empty: |
| Add the samples situated at an established distance from the centroid and from the previous added sample |
| After each insertion of a sample in visited register, we verify: |
| if visited <= no_of_desired_samples; if not, continue with the region growing algorithm based on a window dimension representing the populated neighborhood |
| End while |
| End for Combine all the visited registers from the corresponding regions for the training samples End for |
| Combine all the samples from the partitions to obtain the final X_train training data set. |
4. Results
4.1. Hyperspectral Datasets
4.2. Ensuring the Independence between Data Training and Data Testing
4.3. The Experimental Configuration
4.4. Obtained Experimental Results
4.4.1. Random Sampling vs. Controlled Sampling Evaluation
4.4.2. Spatial vs. Spectral Information for Feature Extraction
4.4.3. Parallel Spatial-Spectral Feature Extraction and Classification
4.4.4. Contribution of the Data Filtering for End-to-End Classification
4.4.5. Classification Results on the Salinas Datasets
4.4.6. Classification Results on the Pavia University Datasets
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HSI | Hyperspectral image |
| TV | Total variation |
| CSSWHTV | Combined spatial and spectral weighted hyperspectral total variation |
| LBP | Local binary pattern |
| PCA | Principal component analysis |
| CA | Canonical analysis |
| SVM | Support vector machine |
| PDE | Partial differential equation |
| DWT | Discrete wavelet transform |
| OA | Overall accuracy |
| cA | Low pass (L) coefficients of the wavelet |
| cD | High pass (H) coefficients of the wavelet |
References
- Chang, C.-I. Hyperspectral Imaging: Techniques for Spectral Detection and Classification; Springer Science & Business Media: New York, NY, USA, 2003; Volume 1. [Google Scholar]
- Liang, G.; Smith, R.T. Optical hyperspectral imaging in microscopy and spectroscopy—A review of data acquisition. J. Biophotonics 2015, 8, 441–456. [Google Scholar]
- Que, D.; Li, B. Medical images denoising based on total variation algorithm. In Proceedings of the International Conference on Environment Science and Biotechnology ICESB, Maldives, Spain, 25–26 November 2011; Volume 23, p. 227. [Google Scholar]
- Signoroni, A.; Savardi, M.; Baronio, A.; Benini, S. Deep learning meets hyperspectral image analysis: A multidisciplinary review. J. Imaging 2019, 5, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teke, M.; Deveci, H.S.; Haliloglu, O.; Gürbüz, S.Z.; Sakarya, U. A short survey of hyperspectral remote sensing applications in agriculture. In Proceedings of the 2013 6th International Conference on Recent Advances in Space Technologies (RAST), Istanbul, Turkey, 12–14 June 2013; pp. 171–176. [Google Scholar]
- Adão, T.; Hruška, J.; Pádua, L.; Bessa, J.; Peres, E.; Morais, R.; Sousa, J.J. Hyperspectral Imaging: A Review on UAV-Based Sensors, Data Processing and Applications for Agriculture and Forestry. Remote Sens. 2017, 9, 1110. [Google Scholar] [CrossRef] [Green Version]
- Paoletti, M.; Haut, J.; Plaza, J.; Plaza, A. Deep learning classifiers for hyperspectral imaging: A review. ISPRS J. Photogramm. Remote Sens. 2019, 158, 279–317. [Google Scholar] [CrossRef]
- Fei, B. Hyperspectral imaging in medical applications. In Data Handling in Science and Technology; Elsevier: Amsterdam, The Netherlands, 2020; Volume 32, pp. 523–565. [Google Scholar]
- Khan, M.J.; Khan, H.S.; Yousaf, A.; Khurshid, K.; Abbas, A. Modern Trends in Hyperspectral Image Analysis: A Review. IEEE Access 2018, 6, 14118–14129. [Google Scholar] [CrossRef]
- Rasti, B.; Scheunders, P.; Ghamisi, P.; Licciardi, G.; Chanussot, J. Noise Reduction in Hyperspectral Imagery: Overview and Application. Remote Sens. 2018, 10, 482. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Chova, L.; Tuia, D.; Moser, G.; Camps-Valls, G. Multimodal Classification of Remote Sensing Images: A Review and Future Directions. Proc. IEEE 2015, 103, 1560–1584. [Google Scholar] [CrossRef]
- Bowker, D.E. Spectral Reflectances of Natural Targets for Use in Remote Sensing Studies; NASA: Washington, DC, USA, 1985; Volume 1139. [Google Scholar]
- Hughes, G.F. On the mean accuracy of statistical pattern recognizers. IEEE Trans. Inf. Theory 1968, 14, 55–63. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Cui, J.; Wang, W.; Lin, C. A Study for Texture Feature Extraction of High-Resolution Satellite Images Based on a Direction Measure and Gray Level Co-Occurrence Matrix Fusion Algorithm. Sensors 2017, 17, 1474. [Google Scholar] [CrossRef] [Green Version]
- Gomez, C.; Drost, A.; Roger, J.-M. Analysis of the uncertainties affecting predictions of clay contents from VNIR/SWIR hyperspectral data. Remote Sens. Environ. 2015, 156, 58–70. [Google Scholar] [CrossRef] [Green Version]
- Engesl, J.M.; Chakravarthy, B.L.; Rothwell, D.; Chavan, A. SEEQ™ MCT wearable sensor performance correlated to skin irritation and temperature. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Milan, Italy, 25–29 August 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 2030–2033. [Google Scholar]
- Jia, S.; Jiang, S.; Lin, Z.; Li, N.; Xu, M.; Yu, S. A survey: Deep learning for hyperspectral image classification with few labeled samples. Neurocomputing 2021, 448, 179–204. [Google Scholar] [CrossRef]
- Mohan, B.K.; Porwal, A. Hyperspectral image processing and analysis. Curr. Sci. 2015, 108, 833–841. [Google Scholar]
- Taskin, G.; Kaya, H.; Bruzzone, L. Feature Selection Based on High Dimensional Model Representation for Hyperspectral Images. IEEE Trans. Image Process. 2017, 26, 2918–2928. [Google Scholar] [CrossRef] [PubMed]
- Miclea, A.; Borda, M.; Terebes, R.; Meza, S. Hyperspectral Image Enhancement using Diffusion and Shock Filtering Techniques. In Proceedings of the 2019 E-Health and Bioengineering Conference (EHB), Iasi, Romania, 21–23 November 2019; pp. 1–6. [Google Scholar]
- Miclea, A.; Terebes, R. A Novel Local Binary Patterns and Wavelet Transform-based Approach for Hyperspectral Image Classification. Acta Teh. Napoc. 2021, 61, 29–34. [Google Scholar]
- Cernadas, E.; Delgado, M.F.; González-Rufino, E.; Carrión, P. Influence of normalization and color space to color texture classification. Pattern Recognit. 2017, 61, 120–138. [Google Scholar] [CrossRef]
- Willett, R.; Krishnamurthy, K.; Raginsky, M. Multiscale photon-limited spectral image reconstruction. SIAM J. Imaging Sci. 2010, 3, 619–645. [Google Scholar]
- Shen, H.; Yuan, Q.; Zhang, L. Hyperspectral image denoising employing a spectral-spatial adaptive total variation model. IEEE Trans. Geosci. Remote Sens. 2012, 50, 3660–3677. [Google Scholar]
- Rudin, L.I.; Osher, S.; Fatemi, E. Nonlinear total variation based noise removal algorithms. Phys. D Nonlinear Phenom. 1992, 60, 259–268. [Google Scholar] [CrossRef]
- Othman, H.; Qian, S. Noise reduction of hyperspectral imagery using hybrid spatial-spectral derivative-domain wavelet shrinkage. IEEE Trans. Geosci. Remote Sens. 2006, 44, 397–408. [Google Scholar] [CrossRef]
- Cheng, J.; Zhang, H.; Zhang, L.; Shen, H.; Yuan, Q. Hyperspectral image denoising with a combined spatial and spectral weighted hyperspectral total variation model. Can. J. Remote Sens. 2016, 42, 53–72. [Google Scholar]
- Tu, B.; Li, N.; Fang, L.; He, D.; Ghamisi, P. Hyperspectral Image Classification with Multi-Scale Feature Extraction. Remote Sens. 2019, 11, 534. [Google Scholar] [CrossRef] [Green Version]
- Elham, K.G. Hyperspectral image classification using a spectral–spatial random walker method. Int. J. Remote Sens. 2019, 40, 3948–3967. [Google Scholar]
- Wei, L.; Chen, C.; Su, H.; Qian, D. Local Binary Patterns and Extreme Learning Machine for Hyperspectral Imagery Classification. IEEE Trans. Geosci. Remote Sens. 2015, 53, 3681–3693. [Google Scholar]
- Demel, M.; Ecker, G.; Janecek, A.; Gansterer, W. On the relationship between feature selection and classification accuracy. In Proceedings of the Machine Learning Research, New Challenges for Feature Selection in Data Mining and Knowledge Discovery, Antwerp, Belgium, 15 September 2008; Volume 4, pp. 90–105. [Google Scholar]
- Shan, J.; Rodarmel, C. Principal component analysis for hyperspectral image classification. Surveying and Land Information System. Surv. Land Inf. Sci. 2002, 62, 115. [Google Scholar]
- Samat, A.; Persello, C.; Gamba, P.; Liu, S.; Abuduwaili, J.; Li, E. Supervised and Semi-Supervised Multi-View Canonical Correlation Analysis Ensemble for Heterogeneous Domain Adaptation in Remote Sensing Image Classification. Remote Sens. 2017, 9, 337. [Google Scholar] [CrossRef] [Green Version]
- Fang, B.; Li, Y.; Zhang, H.; Chan, J.C.-W. Hyperspectral Images Classification Based on Dense Convolutional Networks with Spectral-Wise Attention Mechanism. Remote Sens. 2019, 11, 159. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Bioucas-Dias, M.; Plaza, A. Semisupervised Hyperspectral Image Segmentation Using Multinomial Logistic Regression with Active Learning. IEEE Trans. Geosci. Remote Sens. 2010, 48, 4085–4098. [Google Scholar] [CrossRef] [Green Version]
- Su, J.; Vargas, D.V.; Sakurai, K. One Pixel Attack for Fooling Deep Neural Networks. IEEE Trans. Evol. Comput. 2019, 23, 828–841. [Google Scholar] [CrossRef] [Green Version]
- Pop, S.; Lavialle, O.; Donias, M.; Terebes, R.; Borda, M.; Guillon, S.; Keskes, N. A PDE-Based Approach to Three-Dimensional Seismic Data Fusion, Geoscience and Remote Sensing. IEEE Trans. Geosci. Remote Sens. 2008, 46, 1385–1393. [Google Scholar] [CrossRef]
- Ojala, T.; Pietikainen, M.; Maenpaa, T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 971–987. [Google Scholar] [CrossRef]
- Feng, S.; Yuki, P.; Duarte, M. Wavelet-Based Semantic Features for Hyperspectral Signature Discrimination. arXiv 2016, arXiv:1602.03903. [Google Scholar]
- Ghaderpour, E.; Pagiatakis, S.D.; Hassan, Q.K. A Survey on Change Detection and Time Series Analysis with Applications. Appl. Sci. 2021, 11, 6141. [Google Scholar] [CrossRef]
- Qian, Y.; Wen, L.; Bai, X.; Liang, J.; Zhou, J. On the sampling strategy for evaluation of spectral-spatial methods in hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 862–88042. [Google Scholar]
- Hyperspectral Remote Sensing Scenes. Available online: http://www.ehu.eus/ccwintco/index.php/Hyperspectral_Remote_Sensing_Scenes (accessed on 16 March 2022).
















| Number of Bands after the PCA Band Reduction Process (n1) | Number of Neighbors (Param. P of LBP) (n2) | Spatial Features | Spectral Features | Size of the Feature Vector | |
|---|---|---|---|---|---|
| Number of LBP Histogram Bins (nbins) | Wavelet Level | ||||
| 10 | 8 | 9 | N = 1 (L) | 100 | 190 |
| N = 2 (LL) | 50 | 140 | |||
| Feature Extraction Method | Window Size | |||
|---|---|---|---|---|
| w3 | w5 | w11 | ||
| 75.35 ± 1.42 | 97.69 ± 1.81 | 98.19 ± 0.71 | ||
| 77.09 ± 1.67 | 97.30 ± 1.24 | 99.93 ± 0.55 | ||
| Methods and Corresponding Parameters | Window Size | ||
|---|---|---|---|
| w = 3 | w = 5 | ||
| 25.74 ± 1.21 | 26.27 ± 0.99 | ||
| 25.74 ± 1.29 | 26.27 ± 1.32 | ||
| N = 1 (cA) | 76.40 ± 1.52 | 75.68 ± 1.78 | |
| N = 2 (cA) | 76.85 ± 1.98 | 78.05 ± 1.78 | |
| N = 3 (cA + cD) | 80.52 ± 1.18 | 74.91 ± 1.27 | |
| Spectral Feature | Window Size | |||
|---|---|---|---|---|
| Spatial feature | Wavelet Level | High/Low Pass Coefficients | w3 | w5 |
| N = 1 | cA | 84.49 ± 1.54 | 83.62 ± 1.78 | |
| N = 2 | cA | 83.45 ± 1.32 | 82.42 ± 1.69 | |
| N = 3 | cA + cD | 81.59 ± 1.25 | 78.04 ± 2.12 | |
| N = 1 | cA | 84.98 ± 1.82 | 84.54 ± 1.89 | |
| N = 2 | cA | 84.10 ± 1.63 | 82.41 ± 1.33 | |
| N = 3 | cA + cD | 83.23 ± 1.47 | 72.24 ± 1.85 | |
| Filtering Method | Spatial Feature | Spectral Feature | Window Size | ||
|---|---|---|---|---|---|
| Wavelet Level | High Pass (cD)/Low Pass (cA) Coeff. | w3 | w5 | ||
| TV filtering | N = 1 | cA | 90.2 ± 1.3 | 87.83 ± 0.96 | |
| N = 2 | cA | 88.16 ± 0.92 | 86.69 ± 1.62 | ||
| N = 3 | cA + cD | 80.99 ± 1.45 | 82.95 ± 1.55 | ||
| N = 1 | cA | 90.36 ± 0.96 | 88.12 ± 1.02 | ||
| N = 2 | cA | 88.24 ± 1.78 | 86.45 ± 1.63 | ||
| N = 3 | cA + cD | 81.1 ± 1.65 | 83.15 ± 1.54 | ||
| CSSWHTV filtering | N = 1 | cA | 86.46 ± 1.26 | 84.56 ± 1.65 | |
| N = 2 | cA | 84.12 ± 1.24 | 82.83 ± 1.28 | ||
| N = 3 | cA + cD | 74.73 ± 1.96 | 83.03 ± 1.63 | ||
| N = 1 | cA | 86.46 ± 1.62 | 84.56 ± 1.21 | ||
| N = 2 | cA | 84.12 ± 0.76 | 82.83 ± 1.03 | ||
| N = 3 | cA + cD | 74.73 ± 1.26 | 83.03 ± 1.69 | ||
| proposed anisotropic diffusion and shock filter | N = 1 | cA | 93.85 ± 1.57 | 90.04 ± 0.99 | |
| N = 2 | cA | 91.38 ± 1.2 | 89.4 ± 1.85 | ||
| N = 3 | cA + cD | 81.64 ± 1.87 | 85.95 ± 1.27 | ||
| N = 1 | cA | 93.37 ± 1.25 | 90.73 ± 1.12 | ||
| N = 2 | cA | 91.53 ± 1.22 | 88.84 ± 1.28 | ||
| N = 3 | cA + cD | 83.25 ± 1.98 | 85.88 ± 1.22 | ||
| Indian Pines Class Name | Average Accuracy for Each Method | ||
|---|---|---|---|
| TV(%) | CSSWHTV (%) | Anisotropic Diffusion (%) | |
| Corn-notill | 87.54 | 81 | 90.09 |
| Corn-mintill | 87.91 | 76.37 | 91.76 |
| Grass-pasture | 95.9 | 92.43 | 95.9 |
| Grass-trees | 97.37 | 96.24 | 99.06 |
| Hay-windrowed | 98.27 | 99.14 | 99.71 |
| Soybean-notill | 70.91 | 67.84 | 70.18 |
| Soybean-mintill | 90.42 | 85.03 | 92.79 |
| Soybean-clean | 93.4 | 87.07 | 95.25 |
| Woods | 99.12 | 98.75 | 99.37 |
| Filtering Method | Spatial Features | Spectral Feature | Window Size | ||
|---|---|---|---|---|---|
| Wavelet Level | High Pass (cD)/Low Pass (cA) Coeff. | w3 | w5 | ||
| TV filtering | N = 1 | cA | 94.1 ± 1.45 | 93.59 ± 1.87 | |
| N = 2 | cA | 93.88 ± 1.85 | 93.45 ± 1.48 | ||
| N = 3 | cA + cD | 92.78 ± 1.89 | 95.7 ± 1.75 | ||
| N = 1 | cA | 94.85 ± 1.42 | 93.77 ± 1.49 | ||
| N = 2 | cA | 93.56 ± 1.85 | 93.41 ± 1.47 | ||
| N = 3 | cA + cD | 95.93 ± 1.45 | 92.67 ± 1.81 | ||
| CSSWHTV filtering | N = 1 | cA | 92.82 ± 1.98 | 92.67 ± 1.52 | |
| N = 2 | cA | 92.81 ± 1.84 | 92.67 ± 1.22 | ||
| N = 3 | cA + cD | 90.68 ± 1.14 | 92.13 ± 1.87 | ||
| N = 1 | cA | 92.99 ± 1.48 | 92.81 ± 1.47 | ||
| N = 2 | cA | 92.47 ± 1.85 | 92.12 ± 1.36 | ||
| N = 3 | cA + cD | 91.62 ± 1.58 | 92.69 ± 1.74 | ||
| proposed anisotropic diffusion and shock filter | N = 1 | cA | 95.41 ± 1.22 | 95.1 ± 1.22 | |
| N = 2 | cA | 95.49 ± 1.82 | 94.71 ± 1.56 | ||
| N = 3 | cA + cD | 94.36 ± 1.32 | 94.1 ± 1.75 | ||
| N = 1 | cA | 96.54 ± 1.87 | 94.91 ± 1.78 | ||
| N = 2 | cA | 95.88 ± 1.45 | 93.89 ± 1.36 | ||
| N = 3 | cA + cD | 93.78 ± 1.61 | 93.22 ± 1.55 | ||
| Filtering Method | Spatial Feature | Spectral Feature | Window Size | ||
|---|---|---|---|---|---|
| Wavelet Level | High Pass (cD)/Low Pass (cA) Coeff. | w3 | w5 | ||
| TV filtering | N = 1 | cA | 84.02 ± 1.52 | 83.86 ± 1.42 | |
| N = 2 | cA | 88.29 ± 1.77 | 83.25 ± 1.33 | ||
| N = 3 | cA + cD | 82.93 ± 0.99 | 82.01 ± 1.02 | ||
| N = 1 | cA | 85.51 ± 1.47 | 82.25 ± 1.41 | ||
| N = 2 | cA | 87.62 ± 1.78 | 82.45 ± 1.30 | ||
| N = 3 | cA + cD | 82.85 ± 1.31 | 82.07 ± 1.52 | ||
| CSSWHTV filtering | N = 1 | cA | 89.86 ± 1.52 | 85.91 ± 1.28 | |
| N = 2 | cA | 88.4 ± 1.27 | 82.66 ± 1.34 | ||
| N = 3 | cA + cD | 83.73 ± 1.87 | 83.98 ± 1.85 | ||
| N = 1 | cA | 89.16 ± 1.74 | 84.88 ± 1.51 | ||
| N = 2 | cA | 87.25 ± 1.31 | 83.62 ± 1.78 | ||
| N = 3 | cA + cD | 83.96 ± 1.88 | 82.11 ± 1.39 | ||
| proposed anisotropic diffusion and shock filter | N = 1 | cA | 92.86 ± 1.32 | 91.41 ± 1.78 | |
| N = 2 | cA | 92.29 ± 1.99 | 90.98 ± 1.22 | ||
| N = 3 | cA + cD | 91.75 ± 1.18 | 89.89 ± 1.54 | ||
| N = 1 | cA | 92.12 ± 1.94 | 90.27 ± 1.83 | ||
| N = 2 | cA | 90.51 ± 1.15 | 89.15 ± 1.48 | ||
| N = 3 | cA + cD | 90.02 ± 1.54 | 87.32 ± 1.52 | ||
| Model Name | Indian Pines Dataset | Pavia University Dataset |
|---|---|---|
| CNN2D40 with disjoint sample selection (results taken from [7]) | 82.97 | 84.98 |
| CNN3D with disjoint sample selection (results taken from [7]) | 79.58 | 86.82 |
| Proposed end-to-end (results taken from Table 5 and Table 7) | 93.85 | 92.86 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miclea, A.V.; Terebes, R.M.; Meza, S.; Cislariu, M. On Spectral-Spatial Classification of Hyperspectral Images Using Image Denoising and Enhancement Techniques, Wavelet Transforms and Controlled Data Set Partitioning. Remote Sens. 2022, 14, 1475. https://doi.org/10.3390/rs14061475
Miclea AV, Terebes RM, Meza S, Cislariu M. On Spectral-Spatial Classification of Hyperspectral Images Using Image Denoising and Enhancement Techniques, Wavelet Transforms and Controlled Data Set Partitioning. Remote Sensing. 2022; 14(6):1475. https://doi.org/10.3390/rs14061475
Chicago/Turabian StyleMiclea, Andreia Valentina, Romulus Mircea Terebes, Serban Meza, and Mihaela Cislariu. 2022. "On Spectral-Spatial Classification of Hyperspectral Images Using Image Denoising and Enhancement Techniques, Wavelet Transforms and Controlled Data Set Partitioning" Remote Sensing 14, no. 6: 1475. https://doi.org/10.3390/rs14061475






