Application of a Novel Multiscale Global Graph Convolutional Neural Network to Improve the Accuracy of Forest Type Classification Using Aerial Photographs
Abstract
1. Introduction
2. Materials and Methods
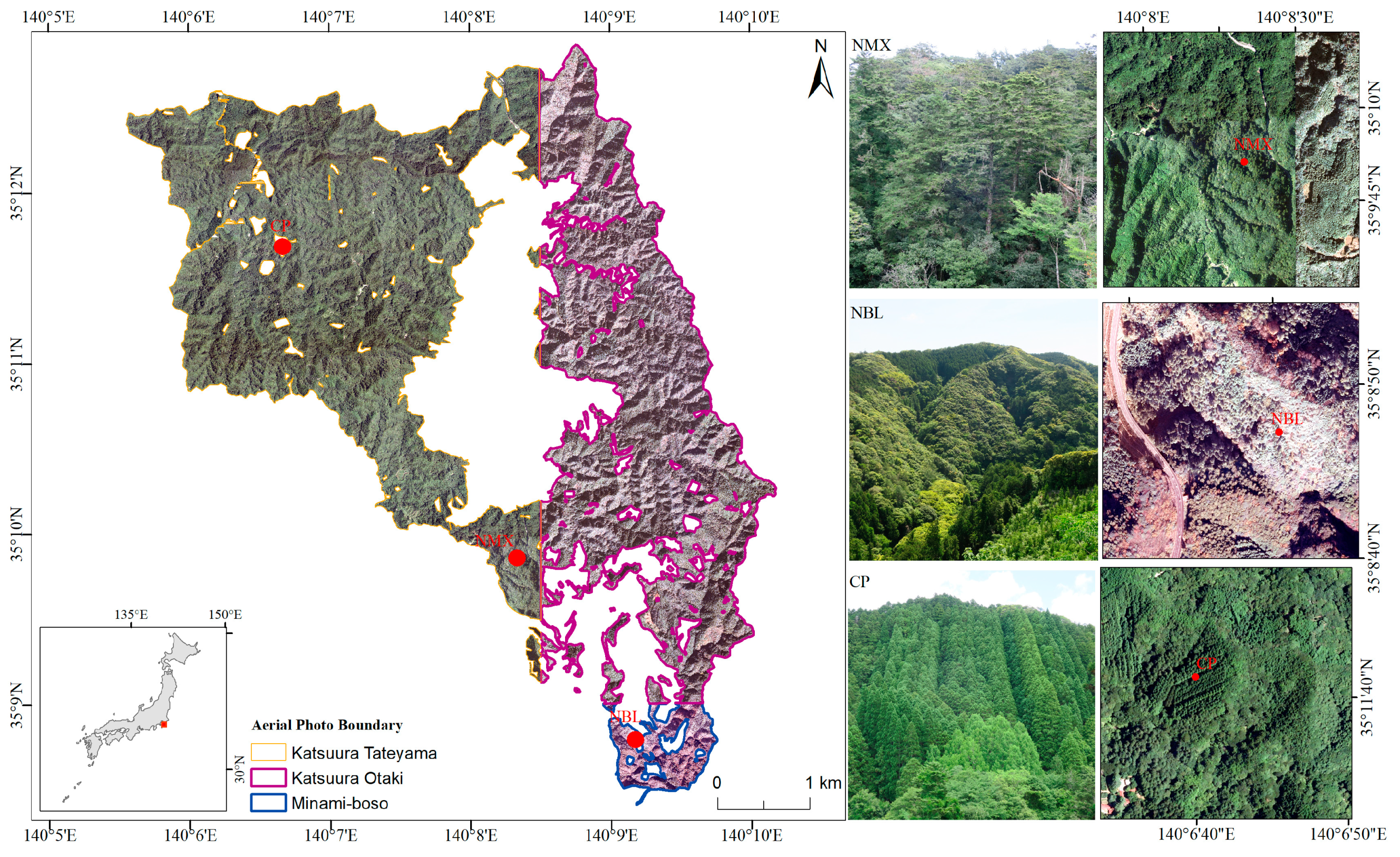
2.1. Study Area
2.2. Data Sources and Preprocessing
2.3. Model Architecture
2.3.1. Basic Overview
2.3.2. Encoding Module
2.3.3. Multiscale Graph Convolution Network (MSGCN) Module
2.3.4. Decoding Module
2.3.5. Loss Function
2.3.6. Comparison with SOTA Models and Experimental Settings
2.4. Data Analysis
2.4.1. Accuracy and Complexity Evaluation
2.4.2. Classification Difference Analysis
3. Results
3.1. Classification Accuracy Indices of Different Models
3.2. Area and Digital Number for Each Forest Type Predicted by the Various Models
3.3. Mapping and Spatial Distributions of Forest Types by the Different Models
4. Discussion
4.1. Classification Accuracy
4.2. Area and Digital Number for Each Forest Type Predicted by the Different Models
4.3. Mapping and Spatial Distributions of Forest Types for the Different Models
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thompson, I.D.; Baker, J.A.; Ter-Mikaelian, M. A review of the long-term effects of post-harvest silviculture on vertebrate wildlife, and predictive models, with an emphasis on boreal forests in Ontario, Canada. For. Ecol. Manag. 2003, 177, 441–469. [Google Scholar] [CrossRef]
- Wagner, F.H.; Sanchez, A.; Tarabalka, Y.; Lotte, R.G.; Ferreira, M.P.; Aidar, M.P.M.; Gloor, E.; Phillips, O.L.; Aragão, L.E.O.C. Using the U-net convolutional network to map forest types and disturbance in the Atlantic rainforest with very high resolution images. Remote Sens. Ecol. Conserv. 2019, 5, 360–375. [Google Scholar] [CrossRef]
- Kislov, D.E.; Korznikov, K.A.; Altman, J.; Vozmishcheva, A.S.; Krestov, P.V. Extending deep learning approaches for forest disturbance segmentation on very high-resolution satellite images. Remote Sens. Ecol. Conserv. 2021, 7, 355–368. [Google Scholar] [CrossRef]
- Muhammad, K.; Ahmad, J.; Baik, S.W. Early fire detection using convolutional neural networks during surveillance for effective disaster management. Neurocomputing 2018, 288, 30–42. [Google Scholar] [CrossRef]
- Zhao, F.; Sun, R.; Zhong, L.; Meng, R.; Huang, C.; Zeng, X.; Wang, M.; Li, Y.; Wang, Z. Monthly mapping of forest harvesting using dense time series Sentinel-1 SAR imagery and deep learning. Remote Sens. Environ. 2022, 269, 112822. [Google Scholar] [CrossRef]
- Pandit, S.; Tsuyuki, S.; Dube, T. Landscape-scale aboveground biomass estimation in buffer zone community forests of Central Nepal: Coupling in situ measurements with Landsat 8 Satellite Data. Remote Sens. 2018, 10, 1848. [Google Scholar] [CrossRef]
- Jayathunga, S.; Owari, T.; Tsuyuki, S. The use of fixed–wing UAV photogrammetry with LiDAR DTM to estimate merchantable volume and carbon stock in living biomass over a mixed conifer–broadleaf forest. Int. J. Appl. Earth Obs. Geoinf. 2018, 73, 767–777. [Google Scholar] [CrossRef]
- Reichstein, M.; Camps-Valls, G.; Stevens, B.; Jung, M.; Denzler, J.; Carvalhais, N. Prabhat Deep learning and process understanding for data-driven Earth system science. Nature 2019, 566, 195–204. [Google Scholar] [CrossRef]
- Yang, R.; Wang, L.; Tian, Q.; Xu, N.; Yang, Y. Estimation of the conifer-broadleaf ratio in mixed forests based on time-series data. Remote Sens. 2021, 13, 4426. [Google Scholar] [CrossRef]
- Ohsawa, T.; Saito, Y.; Sawada, H.; Ide, Y. Impact of altitude and topography on the genetic diversity of Quercus serrata populations in the Chichibu Mountains, central Japan. Flora Morphol. Distrib. Funct. Ecol. Plants 2008, 203, 187–196. [Google Scholar] [CrossRef]
- Pfeifer, M.; Lefebvre, V.; Peres, C.A.; Banks-Leite, C.; Wearn, O.R.; Marsh, C.J.; Butchart, S.H.M.; Arroyo-Rodríguez, V.; Barlow, J.; Cerezo, A.; et al. Creation of forest edges has a global impact on forest vertebrates. Nature 2017, 551, 187–191. [Google Scholar] [CrossRef]
- Bonan, G.B.; Pollard, D.; Thompson, S.L. Effects of boreal forest vegetation on global climate. Nature 1992, 359, 716–718. [Google Scholar] [CrossRef]
- Raft, A.; Oliier, H. Forest restoration, biodiversity and ecosystem functioning. BMC Ecol. 2011, 11, 29. [Google Scholar] [CrossRef]
- Rozendaal, D.M.A.; Requena Suarez, D.; De Sy, V.; Avitabile, V.; Carter, S.; Adou Yao, C.Y.; Alvarez-Davila, E.; Anderson-Teixeira, K.; Araujo-Murakami, A.; Arroyo, L.; et al. Aboveground forest biomass varies across continents, ecological zones and successional stages: Refined IPCC default values for tropical and subtropical forests. Environ. Res. Lett. 2022, 17, 014047. [Google Scholar] [CrossRef]
- Thurner, M.; Beer, C.; Santoro, M.; Carvalhais, N.; Wutzler, T.; Schepaschenko, D.; Shvidenko, A.; Kompter, E.; Ahrens, B.; Levick, S.R.; et al. Carbon stock and density of northern boreal and temperate forests. Glob. Ecol. Biogeogr. 2014, 23, 297–310. [Google Scholar] [CrossRef]
- Coppin, P.R.; Bauer, M.E. Digital Change Detection in Forest Ecosystems with Remote Sensing Imagery. Remote Sens. Rev. 1996, 13, 207–234. [Google Scholar] [CrossRef]
- Cowardin, L.M.; Myers, V.I. Remote Sensing for Identification and Classification of Wetland Vegetation. J. Wildl. Manag. 1974, 38, 308–314. [Google Scholar] [CrossRef]
- Schiefer, F.; Kattenborn, T.; Frick, A.; Frey, J.; Schall, P.; Koch, B.; Schmidtlein, S. Mapping forest tree species in high resolution UAV-based RGB-imagery by means of convolutional neural networks. ISPRS J. Photogramm. Remote Sens. 2020, 170, 205–215. [Google Scholar] [CrossRef]
- Kentsch, S.; Karatsiolis, S.; Kamilaris, A.; Tomhave, L.; Lopez Caceres, M.L. Identification of Tree Species in Japanese Forests based on Aerial Photography and Deep Learning. arXiv 2020. [Google Scholar] [CrossRef]
- Komárek, J. The perspective of unmanned aerial systems in forest management: Do we really need such details? Appl. Veg. Sci. 2020, 23, 718–721. [Google Scholar] [CrossRef]
- Ray, R.G. Aerial Photographs in Geologic Interpretation and Mapping; Professional Paper; US Government Printing Office: Washington, DC, USA, 1960. [CrossRef]
- Ozaki, K.; Ohsawa, M. Successional change of forest pattern along topographical gradients in warm-temperate mixed forests in Mt Kiyosumi, central Japan. Ecol. Res. 1995, 10, 223–234. [Google Scholar] [CrossRef]
- Chianucci, F.; Disperati, L.; Guzzi, D.; Bianchini, D.; Nardino, V.; Lastri, C.; Rindinella, A.; Corona, P. Estimation of canopy attributes in beech forests using true colour digital images from a small fixed-wing UAV. Int. J. Appl. Earth Obs. Geoinf. 2016, 47, 60–68. [Google Scholar] [CrossRef]
- Bagaram, M.B.; Giuliarelli, D.; Chirici, G.; Giannetti, F.; Barbati, A. UAV remote sensing for biodiversity monitoring: Are forest canopy gaps good covariates? Remote Sens. 2018, 10, 1397. [Google Scholar] [CrossRef]
- Sheykhmousa, M.; Mahdianpari, M.; Ghanbari, H.; Mohammadimanesh, F.; Ghamisi, P.; Homayouni, S. Support Vector Machine Versus Random Forest for Remote Sensing Image Classification: A Meta-Analysis and Systematic Review. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 6308–6325. [Google Scholar] [CrossRef]
- Heydari, S.S.; Mountrakis, G. Effect of classifier selection, reference sample size, reference class distribution and scene heterogeneity in per-pixel classification accuracy using 26 Landsat sites. Remote Sens. Environ. 2018, 204, 648–658. [Google Scholar] [CrossRef]
- Dostálová, A.; Wagner, W.; Milenković, M.; Hollaus, M. Annual seasonality in Sentinel-1 signal for forest mapping and forest type classification. Int. J. Remote Sens. 2018, 39, 7738–7760. [Google Scholar] [CrossRef]
- Liu, Y.; Gong, W.; Hu, X.; Gong, J. Forest type identification with random forest using Sentinel-1A, Sentinel-2A, multi-temporal Landsat-8 and DEM data. Remote Sens. 2018, 10, 946. [Google Scholar] [CrossRef]
- Griffiths, P.; Kuemmerle, T.; Baumann, M.; Radeloff, V.C.; Abrudan, I.V.; Lieskovsky, J.; Munteanu, C.; Ostapowicz, K.; Hostert, P. Forest disturbances, forest recovery, and changes in forest types across the carpathian ecoregion from 1985 to 2010 based on landsat image composites. Remote Sens. Environ. 2014, 151, 72–88. [Google Scholar] [CrossRef]
- Lapini, A.; Pettinato, S.; Santi, E.; Paloscia, S.; Fontanelli, G.; Garzelli, A. Comparison of machine learning methods applied to SAR images for forest classification in mediterranean areas. Remote Sens. 2020, 12, 369. [Google Scholar] [CrossRef]
- Pasquarella, V.J.; Holden, C.E.; Woodcock, C.E. Improved mapping of forest type using spectral-temporal Landsat features. Remote Sens. Environ. 2018, 210, 193–207. [Google Scholar] [CrossRef]
- Cheng, K.; Su, Y.; Guan, H.; Tao, S.; Ren, Y.; Hu, T. Mapping China’s planted forests using high resolution imagery and massive amounts of crowdsourced samples. ISPRS J. Photogramm. Remote Sens. 2023, 196, 356–371. [Google Scholar] [CrossRef]
- Kuppusamy, P.; Ieee, M. Retinal Blood Vessel Segmentation using Random Forest with Gabor and Canny Edge Features. In Proceedings of the 2022 International Conference on Smart Technologies and Systems for Next Generation Computing (ICSTSN), Villupuram, India, 5–26 March 2022; pp. 1–4. [Google Scholar] [CrossRef]
- Yoo, C.; Han, D.; Im, J.; Bechtel, B. Comparison between convolutional neural networks and random forest for local climate zone classification in mega urban areas using Landsat images. ISPRS J. Photogramm. Remote Sens. 2019, 157, 155–170. [Google Scholar] [CrossRef]
- Kattenborn, T.; Leitloff, J.; Schiefer, F.; Hinz, S. Review on Convolutional Neural Networks (CNN) in vegetation remote sensing. ISPRS J. Photogramm. Remote Sens. 2021, 173, 24–49. [Google Scholar] [CrossRef]
- Zhou, Z.; Siddiquee, M.R.; Tajbakhsh, N.; Liang, J. UNet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Trans. Med. Imaging 2020, 39, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.P.; de Almeida, D.R.A.; de Almeida Papa, D.; Minervino, J.B.S.; Veras, H.F.P.; Formighieri, A.; Santos, C.A.N.; Ferreira, M.A.D.; Figueiredo, E.O.; Ferreira, E.J.L. Individual tree detection and species classification of Amazonian palms using UAV images and deep learning. For. Ecol. Manag. 2020, 475, 118397. [Google Scholar] [CrossRef]
- Pyo, J.C.; Han, K.J.; Cho, Y.; Kim, D.; Jin, D. Generalization of U-Net Semantic Segmentation for Forest Change Detection in South Korea Using Airborne Imagery. Forests 2022, 13, 2170. [Google Scholar] [CrossRef]
- Fu, C.; Song, X.; Xie, Y.; Wang, C.; Luo, J.; Fang, Y.; Cao, B.; Qiu, Z. Research on the Spatiotemporal Evolution of Mangrove Forests in the Hainan Island from 1991 to 2021 Based on SVM and Res-UNet Algorithms. Remote Sens. 2022, 14, 5554. [Google Scholar] [CrossRef]
- Li, L.; Mu, X.; Chianucci, F.; Qi, J.; Jiang, J.; Zhou, J.; Chen, L.; Huang, H.; Yan, G.; Liu, S. Ultrahigh-resolution boreal forest canopy mapping: Combining UAV imagery and photogrammetric point clouds in a deep-learning-based approach. Int. J. Appl. Earth Obs. Geoinf. 2022, 107, 102686. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the 18th International Conference on Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Munich, Germany, 5–9 October 2015; Volume 9351, pp. 234–241. [Google Scholar] [CrossRef]
- Liu, Y.; Zhong, Y.; Qin, Q. Scene classification based on multiscale convolutional neural network. IEEE Trans. Geosci. Remote Sens. 2018, 56, 7109–7121. [Google Scholar] [CrossRef]
- Zhou, W.; Jin, J.; Lei, J.; Yu, L. CIMFNet: Cross-Layer Interaction and Multiscale Fusion Network for Semantic Segmentation of High-Resolution Remote Sensing Images. IEEE J. Sel. Top. Signal Process. 2022, 16, 666–676. [Google Scholar] [CrossRef]
- Zhao, W.; Du, S. Learning multiscale and deep representations for classifying remotely sensed imagery. ISPRS J. Photogramm. Remote Sens. 2016, 113, 155–165. [Google Scholar] [CrossRef]
- Hu, F.; Xia, G.S.; Hu, J.; Zhang, L. Transferring deep convolutional neural networks for the scene classification of high-resolution remote sensing imagery. Remote Sens. 2015, 7, 14680–14707. [Google Scholar] [CrossRef]
- Liu, Q.; Hang, R.; Song, H.; Li, Z. Learning multiscale deep features for high-resolution satellite image scene classification. IEEE Trans. Geosci. Remote Sens. 2018, 56, 117–126. [Google Scholar] [CrossRef]
- Wang, Q.; Member, S.; Liu, S.; Chanussot, J. Scene Classification with Recurrent Attention of VHR Remote Sensing Images. IEEE Trans. Geosci. Remote Sens. 2019, 57, 1155–1167. [Google Scholar] [CrossRef]
- Bi, Q.; Qin, K.; Li, Z.; Zhang, H.; Xu, K.; Xia, G.S. A Multiple-Instance Densely-Connected ConvNet for Aerial Scene Classification. IEEE Trans. Image Process. 2020, 29, 4911–4926. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 20, 2018; Springer: Cham, Witzerland, 2018; Volume 11045 LNCS, pp. 3–11. [Google Scholar] [CrossRef]
- Deng, Y.; Hou, Y.; Yan, J.; Zeng, D. ELU-Net: An Efficient and Lightweight U-Net for Medical Image Segmentation. IEEE Access 2022, 10, 35932–35941. [Google Scholar] [CrossRef]
- Cao, W.; Zheng, J.; Xiang, D.; Ding, S.; Sun, H.; Yang, X.; Liu, Z.; Dai, Y. Edge and neighborhood guidance network for 2D medical image segmentation. Biomed. Signal Process. Control 2021, 69, 102856. [Google Scholar] [CrossRef]
- Yan, Y.; Ren, J.; Liu, Q.; Zhao, H.; Sun, H.; Zabalza, J. PCA-domain Fused Singular Spectral Analysis for fast and Noise-Robust Spectral-Spatial Feature Mining in Hyperspectral Classification. IEEE Geosci. Remote Sens. Lett. 2021. [Google Scholar] [CrossRef]
- Bazi, Y.; Bashmal, L.; Al Rahhal, M.M.; Dayil, R.A.; Ajlan, N. Al Vision transformers for remote sensing image classification. Remote Sens. 2021, 13, 516. [Google Scholar] [CrossRef]
- Liang, J.; Deng, Y.; Zeng, D. A Deep Neural Network Combined CNN and GCN for Remote Sensing Scene Classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 4325–4338. [Google Scholar] [CrossRef]
- Xiong, Z.; Cai, J. Multi-scale Graph Convolutional Networks with Self-Attention. arXiv 2021. [CrossRef]
- Khan, N.; Chaudhuri, U.; Banerjee, B.; Chaudhuri, S. Graph convolutional network for multi-label VHR remote sensing scene recognition. Neurocomputing 2019, 357, 36–46. [Google Scholar] [CrossRef]
- Yuan, J.; Qiu, Y.; Wang, L.; Liu, Y. Non-Intrusive Load Decomposition Based on Graph Convolutional Network. In Proceedings of the 2022 IEEE 5th International Electrical and Energy Conference (CIEEC), Nangjing, China, 27–29 May 2022; pp. 1941–1944. [Google Scholar] [CrossRef]
- Liu, Q.; Xiao, L.; Huang, N.; Tang, J.; Member, S. Composite Neighbor-Aware Convolutional Metric Networks for Hyperspectral Image Classification. IEEE Trans. Neural Netw. Learn. Syst. 2022, 1–15. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, Y.; Zhao, D.; Chen, J. Graph-FCN for Image Semantic Segmentation. Comput. Vis. Pattern Recognit. 2019, 11554, 97–105. [Google Scholar] [CrossRef]
- Liu, Q.; Xiao, L.; Yang, J.; Wei, Z. Multilevel Superpixel Structured Graph U-Nets for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2022, 60, 5516115. [Google Scholar] [CrossRef]
- Liu, Q.; Xiao, L.; Yang, J.; Wei, Z. CNN-Enhanced Graph Convolutional Network with Pixel- and Superpixel-Level Feature Fusion for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2021, 59, 8657–8671. [Google Scholar] [CrossRef]
- Ding, Y.; Zhang, Z.; Zhao, X.; Hong, D.; Cai, W.; Yu, C.; Yang, N.; Cai, W. Multi-feature fusion: Graph neural network and CNN combining for hyperspectral image classification. Neurocomputing 2022, 501, 246–257. [Google Scholar] [CrossRef]
- Wang, S.H.; Govindaraj, V.V.; Górriz, J.M.; Zhang, X.; Zhang, Y.D. COVID-19 classification by FGCNet with deep feature fusion from graph convolutional network and convolutional neural network. Inf. Fusion 2021, 67, 208–229. [Google Scholar] [CrossRef]
- Peng, F.; Lu, W.; Tan, W.; Qi, K.; Zhang, X.; Zhu, Q. Multi-Output Network Combining GNN and CNN for Remote Sensing Scene Classification. Remote Sens. 2022, 14, 1478. [Google Scholar] [CrossRef]
- Knight, J. From timber to tourism: Recommoditizing the Japanese forest. Dev. Chang. 2000, 31, 341–359. [Google Scholar] [CrossRef]
- Kosztra, B.; Büttner, G.; Hazeu, G.; Arnold, S. Updated CLC Illustrated Nomenclature Guidelines; European Environment Agency: Wien, Austria, 2017; Available online: https://land.copernicus.eu/user-corner/technical-library/corine-land-cover-nomenclature-guidelines/docs/pdf/CLC2018_Nomenclature_illustrated_guide_20190510.pdf (accessed on 8 December 2022).
- de la Cuesta, I.R.; Blanco, J.A.; Imbert, J.B.; Peralta, J.; Rodríguez-Pérez, J. Changes in Long-Term Light Properties of a Mixed Conifer—Broadleaf Forest in Southwestern Europe Ignacio. Forests 2021, 12, 1485. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Spectral and chemical analysis of tropical forests: Scaling from leaf to canopy levels. Remote Sens. Environ. 2008, 112, 3958–3970. [Google Scholar] [CrossRef]
- Zhang, C.; Ma, L.; Chen, J.; Rao, Y.; Zhou, Y.; Chen, X. Assessing the impact of endmember variability on linear Spectral Mixture Analysis (LSMA): A theoretical and simulation analysis. Remote Sens. Environ. 2019, 235, 111471. [Google Scholar] [CrossRef]
- Wang, Q.; Ding, X.; Tong, X.; Atkinson, P.M. Spatio-temporal spectral unmixing of time-series images. Remote Sens. Environ. 2021, 259, 112407. [Google Scholar] [CrossRef]
- Knyazikhin, Y.; Schull, M.A.; Stenberg, P.; Mõttus, M.; Rautiainen, M.; Yang, Y.; Marshak, A.; Carmona, P.L.; Kaufmann, R.K.; Lewis, P.; et al. Hyperspectral remote sensing of foliar nitrogen content. Proc. Natl. Acad. Sci. USA 2013, 110, E185–E192. [Google Scholar] [CrossRef]
- Oreti, L.; Giuliarelli, D.; Tomao, A.; Barbati, A. Object oriented classification for mapping mixed and pure forest stands using very-high resolution imagery. Remote Sens. 2021, 13, 2508. [Google Scholar] [CrossRef]
- Kattenborn, T.; Eichel, J.; Wiser, S.; Burrows, L.; Fassnacht, F.E.; Schmidtlein, S. Convolutional Neural Networks accurately predict cover fractions of plant species and communities in Unmanned Aerial Vehicle imagery. Remote Sens. Ecol. Conserv. 2020, 6, 472–486. [Google Scholar] [CrossRef]
- Jayathunga, S.; Owari, T.; Tsuyuki, S. Analysis of forest structural complexity using airborne LiDAR data and aerial photography in a mixed conifer–broadleaf forest in northern Japan. J. For. Res. 2018, 29, 479–493. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Hornero, A.; Beck, P.S.A.; Kattenborn, T.; Kempeneers, P.; Hernández-Clemente, R. Chlorophyll content estimation in an open-canopy conifer forest with Sentinel-2A and hyperspectral imagery in the context of forest decline. Remote Sens. Environ. 2019, 223, 320–335. [Google Scholar] [CrossRef]
- Peng, C.; Zhang, X.; Yu, G.; Luo, G.; Sun, J. Large kernel matters—Improve semantic segmentation by global convolutional network. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2017, Honolulu, HI, USA, 21–26 July 2016; pp. 1743–1751. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2018, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar] [CrossRef]
- Ouyang, S.; Li, Y. Combining deep semantic segmentation network and graph convolutional neural network for semantic segmentation of remote sensing imagery. Remote Sens. 2021, 13, 119. [Google Scholar] [CrossRef]
- Li, L.; Tang, S.; Deng, L.; Zhang, Y.; Tian, Q. Image caption with global-local attention. In Proceedings of the 31st AAAI Conference on Artificial Intelligence AAAI 2017, San Francisco, CA, USA, 4–9 February 2017; Volume 31, pp. 4133–4139. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, X.; Ji, S. Semantic image segmentation for sea ice parameters recognition using deep convolutional neural networks. Int. J. Appl. Earth Obs. Geoinf. 2022, 112, 102885. [Google Scholar] [CrossRef]
- The University of Tokyo Forests, Graduate School of Agricultural and Life Sciences. Education and Research Plan (2021–2030) of the University of Tokyo Forests: Part 2 Standing Technical Committee Plans; The University of Tokyo Forests: Tokyo, Japan, 2022; Volume 64, pp. 33–49. [Google Scholar] [CrossRef]
- Fadnavis, S. Image Interpolation Techniques in Digital Image Processing: An Overview. Int. J. Eng. Res. Appl. 2014, 4, 70–73. [Google Scholar]
- Ohsato, S.; Negisi, K. Miscellaneous Information, the University of Tokyo Forests; The Tokyo University Forests: Tokyo, Japan, 1994; Volume 32, pp. 9–35. (In Japanese) [Google Scholar] [CrossRef]
- Gu, Z.; Cheng, J.; Fu, H.; Zhou, K.; Hao, H.; Zhao, Y.; Zhang, T.; Gao, S.; Liu, J. CE-Net: Context Encoder Network for 2D Medical Image Segmentation. IEEE Trans. Med. Imaging 2019, 38, 2281–2292. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Guo, Y.; Liu, H.; Lei, Y.; Wen, G. Global context reasoning for semantic segmentation of 3D point clouds. In Proceedings of the 2020 IEEE Winter Conference on Applications of Computer Vision (WACV), Snowmass, CO, USA, 1–5 March 2020; pp. 2920–2929. [Google Scholar] [CrossRef]
- Li, X.; Sun, X.; Meng, Y.; Liang, J.; Wu, F.; Li, J. Dice Loss for Data-imbalanced NLP Tasks. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Online, 5–10 July 2020; pp. 465–476. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-Net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar] [CrossRef]
- Chen, H.; Liu, X.; Jia, Z.; Liu, Z.; Shi, K.; Cai, K. A combination strategy of random forest and back propagation network for variable selection in spectral calibration. Chemom. Intell. Lab. Syst. 2018, 182, 101–108. [Google Scholar] [CrossRef]
- Shao, Z.; Zhou, W.; Deng, X.; Zhang, M.; Cheng, Q. Multilabel Remote Sensing Image Retrieval Based on Fully Convolutional Network. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 318–328. [Google Scholar] [CrossRef]
- Deng, P.; Xu, K.; Huang, H. When CNNs Meet Vision Transformer: A Joint Framework for Remote Sensing Scene Classification. IEEE Geosci. Remote Sens. Lett. 2022, 19, 8020305. [Google Scholar] [CrossRef]
- Sangeetha, V.; Prasad, K.J.R. Deep Residual Learning for Image Recognition Kaiming. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2006, 45, 1951–1954. [Google Scholar] [CrossRef]
- Loshchilov, I.; Hutter, F. Decoupled weight decay regularization. In Proceedings of the 7th International Conference on Learning Representations (ICLR 2019), New Orleans, LA, USA, 6–9 May 2019. [Google Scholar]
- Culjak, I.; Abram, D.; Pribanic, T.; Dzapo, H.; Cifrek, M. A brief introduction to OpenCV. In Proceedings of the 2012 Proceedings of the 35th International Convention MIPRO, Opatija, Croatia, 21–25 May 2012; pp. 1725–1730. [Google Scholar]
- Barupal, D.K.; Fiehn, O. Scikit-learn: Machine Learning in Python. Environ. Health Perspect. 2019, 127, 2825–2830. [Google Scholar] [CrossRef]
- Acharjya, P.P.; Das, R.; Ghoshal, D. Study and Comparison of Different Edge Detectors for Image Segmentation. Glob. J. Comput. Sci. Technol. Graph. Vis. 2012, 12, 29–32. [Google Scholar]
- Basu, M.; Member, S. Gaussian-Based Edge-Detection Methods—A Survey. IEEE Trans. Syst. Man Cybern. Part C (Appl. Rev.) 2002, 32, 252–260. [Google Scholar] [CrossRef]
- Adrian, J.; Sagan, V.; Maimaitijiang, M. Sentinel SAR-optical fusion for crop type mapping using deep learning and Google Earth Engine. ISPRS J. Photogramm. Remote Sens. 2021, 175, 215–235. [Google Scholar] [CrossRef]
- Carbonneau, P.E.; Dugdale, S.J.; Breckon, T.P.; Dietrich, J.T.; Fonstad, M.A.; Miyamoto, H.; Woodget, A.S. Adopting deep learning methods for airborne RGB fluvial scene classification. Remote Sens. Environ. 2020, 251, 112107. [Google Scholar] [CrossRef]
- Molchanov, P.; Tyree, S.; Karras, T.; Aila, T.; Kautz, J. Pruning convolutional neural networks for resource efficient inference. In Proceedings of the 5th International Conference on Learning Representations ICLR 2017—ICLR 2017 Conference Track, Toulon, France, 24–26 April 2017; pp. 1–17. [Google Scholar] [CrossRef]
- Markoulidakis, I.; Rallis, I.; Georgoulas, I.; Kopsiaftis, G.; Doulamis, A.; Doulamis, N. Multiclass Confusion Matrix Reduction Method and Its Application on Net Promoter Score Classification Problem. Technologies 2021, 9, 81. [Google Scholar] [CrossRef]
- Aamir, M.; Li, Z.; Bazai, S.; Wagan, R.A.; Bhatti, U.A.; Nizamani, M.M.; Akram, S. Spatiotemporal Change of Air-Quality Patterns in Hubei Province—A Pre- to Post-COVID-19 Analysis Using Path Analysis and Regression. Atmosphere 2021, 12, 1338. [Google Scholar] [CrossRef]
- Wilebore, B.; Coomes, D. Combining spatial data with survey data improves predictions of boundaries between settlements. Appl. Geogr. 2016, 77, 1–7. [Google Scholar] [CrossRef]
- Perez, L.; Wang, J. The effectiveness of data augmentation in image classification using deep learning. arXiv 2017. [Google Scholar] [CrossRef]
- Karatas, G.; Demir, O.; Sahingoz, O.K. Increasing the Performance of Machine Learning-Based IDSs on an Imbalanced and Up-to-Date Dataset. IEEE Access 2020, 8, 32150–32162. [Google Scholar] [CrossRef]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.W.; Wu, J. UNet 3+: A Full-Scale Connected UNet for Medical Image Segmentation. In Proceedings of the ICASSP 2020—2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1055–1059. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, H.; Li, J.; Wang, L.; Fang, Y.; Sun, J. Supervised graph regularization based cross media retrieval with intra and inter-class correlation. J. Vis. Commun. Image Represent. 2019, 58, 1–11. [Google Scholar] [CrossRef]
- Kosaka, N.; Akiyama, T.; Tsai, B.; Kojima, T. Forest type classification using data fusion of multispectral and panchromatic high-resolution satellite imageries. Int. Geosci. Remote Sens. Symp. 2005, 4, 2980–2983. [Google Scholar] [CrossRef]
- Johnson, B.; Tateishi, R.; Xie, Z. Using geographically weighted variables for image classification. Remote Sens. Lett. 2011, 3, 491–499. [Google Scholar] [CrossRef]
- Mellor, A.; Boukir, S.; Haywood, A.; Jones, S. Exploring issues of training data imbalance and mislabelling on random forest performance for large area land cover classification using the ensemble margin. ISPRS J. Photogramm. Remote Sens. 2015, 105, 155–168. [Google Scholar] [CrossRef]
- Schlerf, M.; Atzberger, C. Vegetation structure retrieval in beech and spruce forests using spectrodirectional satellite data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 8–17. [Google Scholar] [CrossRef]
- Grabska, E.; Hostert, P.; Pflugmacher, D.; Ostapowicz, K. Forest Stand Species Mapping Using the Sentinel-2 Time Series. Remote Sens. 2019, 11, 1197. [Google Scholar] [CrossRef]
- McIlrath, L.D. A CCD/CMOS Focal-Plane Array Edge Detection Processor Implementing the Multi-Scale Veto Algorithm. IEEE J. Solid-State Circuits 1996, 31, 1239–1247. [Google Scholar] [CrossRef][Green Version]
- Wu, J.; Zhou, W.; Luo, T.; Yu, L.; Lei, J. Multiscale multilevel context and multimodal fusion for RGB-D salient object detection. Signal Process. 2021, 178, 63–65. [Google Scholar] [CrossRef]
- Li, J.; Hong, D.; Gao, L.; Yao, J.; Zheng, K.; Zhang, B.; Chanussot, J. Deep Learning in Multimodal Remote Sensing Data Fusion: A Comprehensive Review. Int. J. Appl. Earth Obs. Geoinf. 2022, 112, 102926. [Google Scholar] [CrossRef]
- Jin, H.; Mountrakis, G. Fusion of optical, radar and waveform LiDAR observations for land cover classification. ISPRS J. Photogramm. Remote Sens. 2022, 187, 171–190. [Google Scholar] [CrossRef]
- Hong, D.; Hu, J.; Yao, J.; Chanussot, J.; Zhu, X.X. Multimodal remote sensing benchmark datasets for land cover classification with a shared and specific feature learning model. ISPRS J. Photogramm. Remote Sens. 2021, 178, 68–80. [Google Scholar] [CrossRef]
- Hong, D.; Yokoya, N.; Ge, N.; Chanussot, J.; Zhu, X. Learnable manifold alignment (LeMA): A semi-supervised cross-modality learning framework for land cover and land use classification. ISPRS J. Photogramm. Remote Sens. 2019, 147, 193–205. [Google Scholar] [CrossRef]









| Models | OA | Kappa | IoU_NMX | IoU_NBL | IoU_CP | F1_NMX | F1_NBL | F1_CP | Pre_NMX | Pre_NBL | Pre_CP | Rec_NMX | Rec_NBL | Rec_CP | FLOPs (GMac) | Params(M) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RF | 0.5739 | 0.3373 | 0.0596 | 0.4605 | 0.1584 | 0.1124 | 0.6306 | 0.2734 | 0.1017 | 0.6964 | 0.2307 | 0.1257 | 0.5762 | 0.3357 | - | - |
| SVM | 0.6344 | 0.0028 | 0.1245 | 0.2324 | 0.1497 | 0.2214 | 0.3772 | 0.2605 | 0.3416 | 0.2938 | 0.3659 | 0.1637 | 0.5264 | 0.2022 | - | - |
| U-Net | 0.8098 | 0.6706 | 0.344 | 0.6679 | 0.6776 | 0.5119 | 0.8009 | 0.8078 | 0.5151 | 0.7758 | 0.8504 | 0.5088 | 0.8277 | 0.7693 | 218.94 | 31.04 |
| U-Net++ | 0.8143 | 0.7263 | 0.3367 | 0.6741 | 0.6992 | 0.5037 | 0.8054 | 0.8229 | 0.5221 | 0.7919 | 0.8358 | 0.4867 | 0.8192 | 0.8105 | 153.00 | 47.18 |
| FCN | 0.6895 | 0.5209 | 0.0007 | 0.5531 | 0.4367 | 0.0014 | 0.7123 | 0.6079 | 0.3857 | 0.6149 | 0.6561 | 0.0007 | 0.8462 | 0.5664 | 102.19 | 3.93 |
| ViT | 0.7914 | 0.6942 | 0.3499 | 0.6454 | 0.6530 | 0.5184 | 0.7845 | 0.7900 | 0.5913 | 0.7475 | 0.8189 | 0.4615 | 0.8254 | 0.7632 | 22.66 | 23.28 |
| GCN | 0.8240 | 0.7473 | 0.5463 | 0.6651 | 0.6811 | 0.7066 | 0.7989 | 0.8103 | 0.7232 | 0.8154 | 0.7781 | 0.6907 | 0.7830 | 0.8453 | 57.66 | 9.18 |
| MSG-GCN | 0.8523 | 0.7808 | 0.4374 | 0.7341 | 0.7451 | 0.6086 | 0.8467 | 0.8539 | 0.6103 | 0.8475 | 0.8510 | 0.6069 | 0.8459 | 0.8569 | 104.99 | 88.10 |
| Classification | Ground Truth | MSG-GCN | U-Net++ | U-Net | RF | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number of Pixels | Percentage (%) | Number of Pixels | Percentage (%) | Number of Pixels | Percentage (%) | Number of Pixels | Percentage (%) | Number of Pixels | Percentage (%) | |
| BG | 11,654,816 | 100 | 11,624,871 | 99.74 | 11,651,030 | 99.97 | 11,650,029 | 99.96 | 11,645,984 | 99.92 |
| BG-NMX | 1138 | 0.01 | 343 | 0 | 55 | 0 | 665 | 0.01 | ||
| BG-NBL | 18,005 | 0.16 | 1597 | 0.01 | 2461 | 0.02 | 5039 | 0.04 | ||
| BG-CP | 10,802 | 0.09 | 1846 | 0.02 | 2271 | 0.02 | 3128 | 0.03 | ||
| NMX | 6,282,528 | 100 | 3,833,885 | 61.02 | 3,279,871 | 52.21 | 3,235,934 | 51.51 | 638,718 | 10.17 |
| NMX-BG | 3691 | 0.06 | 2741 | 0.04 | 5495 | 0.09 | 4416 | 0.07 | ||
| NMX-NBL | 2,342,218 | 37.28 | 2,742,520 | 43.65 | 2,633,321 | 41.91 | 4,320,615 | 68.77 | ||
| NMX-CP | 102,734 | 1.64 | 257,396 | 4.10 | 407,778 | 6.49 | 1,318,779 | 20.99 | ||
| NBL | 30,431,072 | 100 | 25,789,099 | 84.75 | 24,098,604 | 79.19 | 23,609,032 | 77.58 | 21,192,944 | 69.64 |
| NBL-BG | 37,705 | 0.12 | 30,617 | 0.10 | 51,540 | 0.17 | 42,704 | 0.14 | ||
| NBL-NMX | 2,351,894 | 7.73 | 3,308,442 | 10.87 | 2,935,850 | 9.65 | 2,920,111 | 9.60 | ||
| NBL-CP | 2,252,374 | 7.40 | 2,993,409 | 9.84 | 3,834,650 | 12.60 | 6,275,313 | 20.62 | ||
| CP | 16,643,296 | 100 | 14,163,402 | 85.1 | 13,911,171 | 83.58 | 14,153,704 | 85.04 | 3,838,865 | 23.07 |
| CP-BG | 13,638 | 0.08 | 9119 | 0.05 | 22,016 | 0.13 | 17,659 | 0.10 | ||
| CP-NMX | 129,378 | 0.78 | 150,747 | 0.91 | 188,197 | 1.13 | 1,521,940 | 9.15 | ||
| CP-NBL | 2,336,878 | 14.04 | 2,572,259 | 15.46 | 2,279,379 | 13.70 | 11,264,832 | 67.68 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pei, H.; Owari, T.; Tsuyuki, S.; Zhong, Y. Application of a Novel Multiscale Global Graph Convolutional Neural Network to Improve the Accuracy of Forest Type Classification Using Aerial Photographs. Remote Sens. 2023, 15, 1001. https://doi.org/10.3390/rs15041001
Pei H, Owari T, Tsuyuki S, Zhong Y. Application of a Novel Multiscale Global Graph Convolutional Neural Network to Improve the Accuracy of Forest Type Classification Using Aerial Photographs. Remote Sensing. 2023; 15(4):1001. https://doi.org/10.3390/rs15041001
Chicago/Turabian StylePei, Huiqing, Toshiaki Owari, Satoshi Tsuyuki, and Yunfang Zhong. 2023. "Application of a Novel Multiscale Global Graph Convolutional Neural Network to Improve the Accuracy of Forest Type Classification Using Aerial Photographs" Remote Sensing 15, no. 4: 1001. https://doi.org/10.3390/rs15041001
APA StylePei, H., Owari, T., Tsuyuki, S., & Zhong, Y. (2023). Application of a Novel Multiscale Global Graph Convolutional Neural Network to Improve the Accuracy of Forest Type Classification Using Aerial Photographs. Remote Sensing, 15(4), 1001. https://doi.org/10.3390/rs15041001










