Development of a UAS-Based Multi-Sensor Deep Learning Model for Predicting Napa Cabbage Fresh Weight and Determining Optimal Harvest Time
Abstract
:1. Introduction
1.1. The Growing Demand for Precision Agriculture
1.2. The Importance of Accurate Napa Cabbage Fresh Weight Prediction in Modern Agriculture
1.3. Limitations of Traditional Methods and the Rise of UAS-Based Remote Sensing
1.4. The Power of AI in Agriculture
1.5. Objectives and Contributions
2. Materials and Methods
2.1. Study Area and Experimental Plot Design
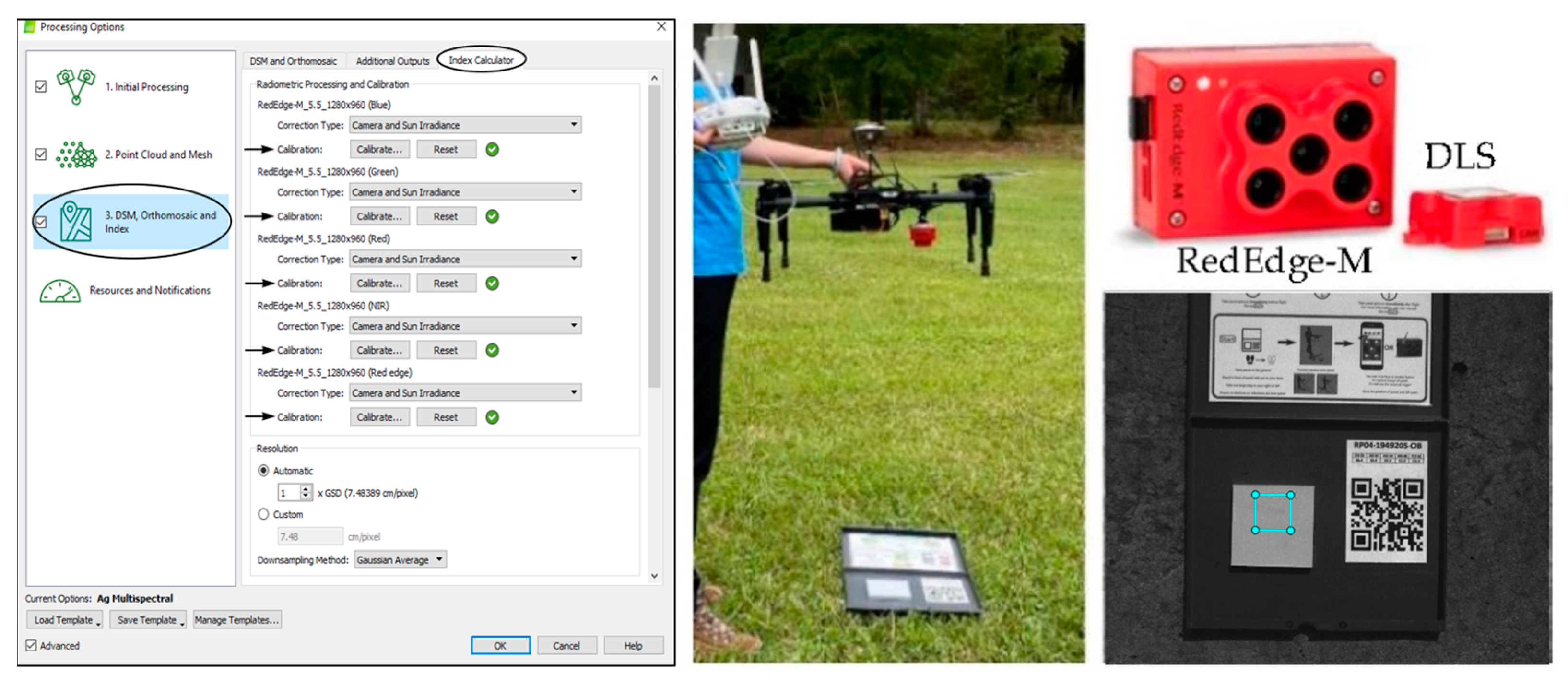
2.2. Unmanned Aerial System (UAS) and Sensors
2.3. Fall Napa Cabbage Growth Cycle and Data Collection Timeline
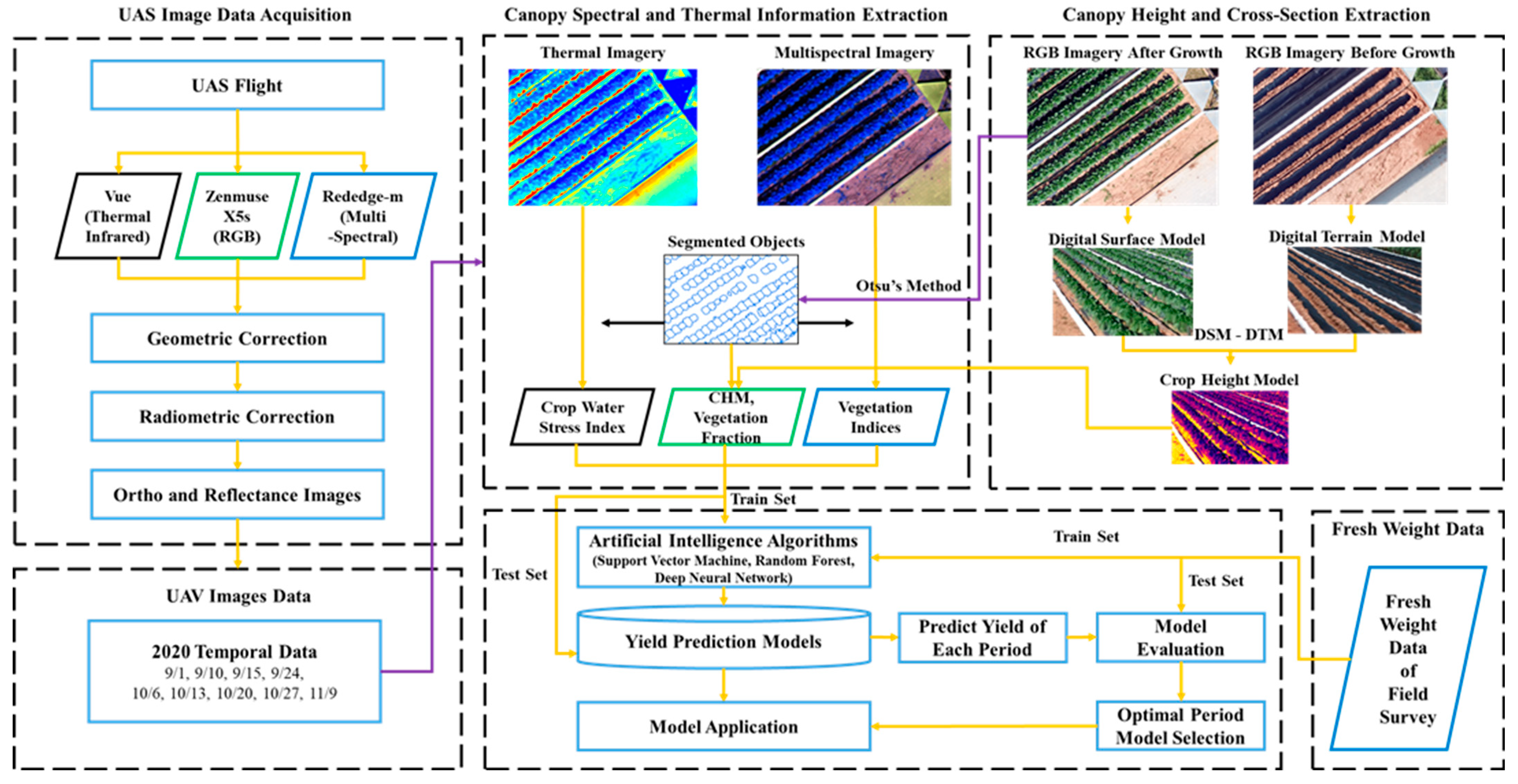
2.4. Study Flow Chart
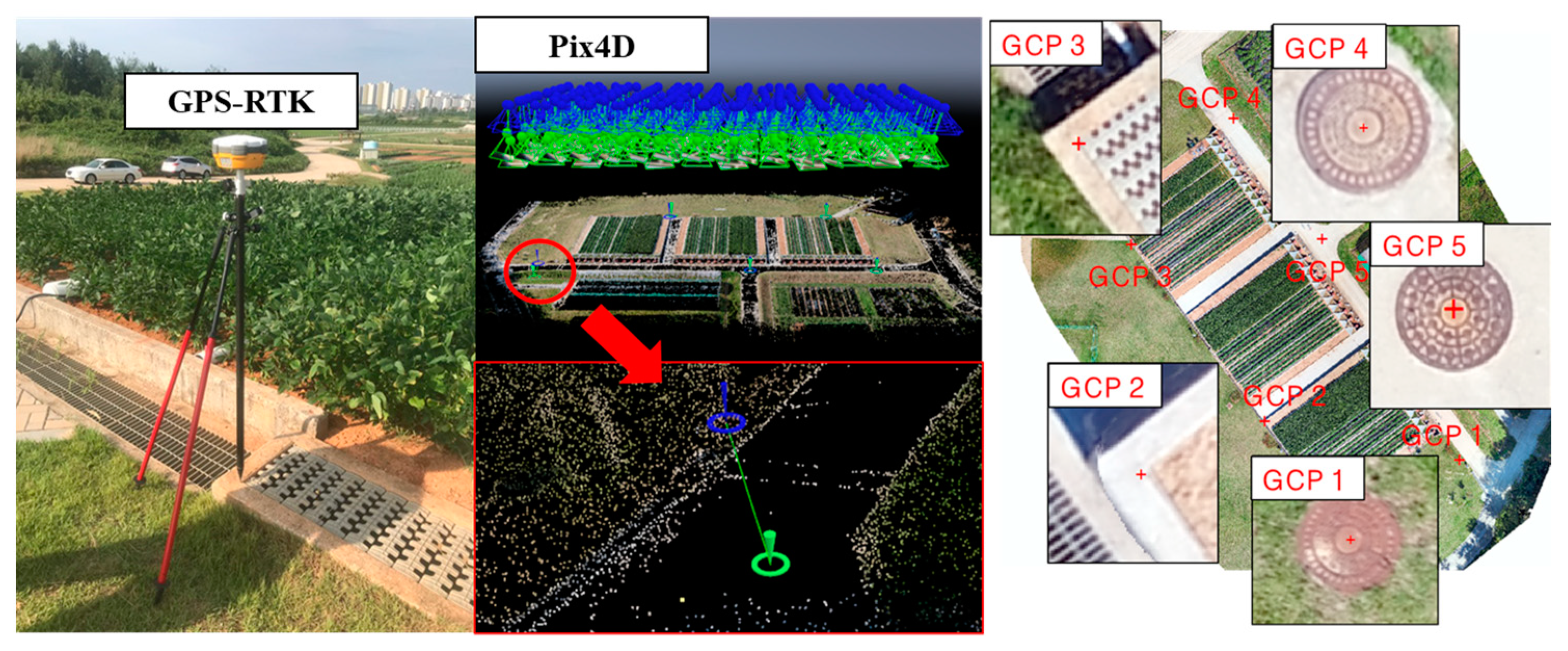
2.5. UAS Image Acquisition and Preprocessing
2.6. Field Survey
2.7. Individual Napa Cabbage Object Segmentation
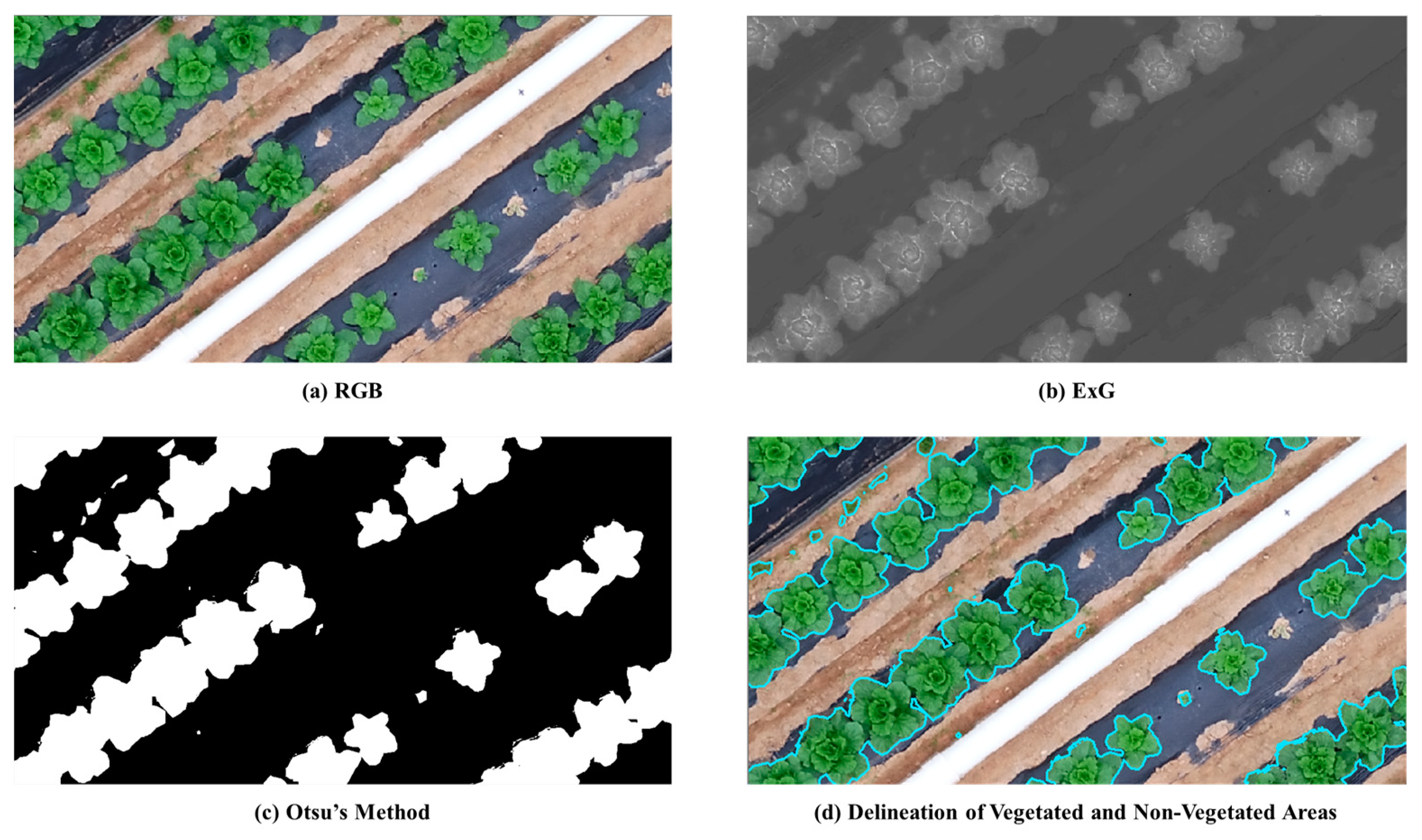
2.7.1. Vegetation Segmentation Using ExG and Otsu Methods
2.7.2. Otsu’s Method for Image Segmentation
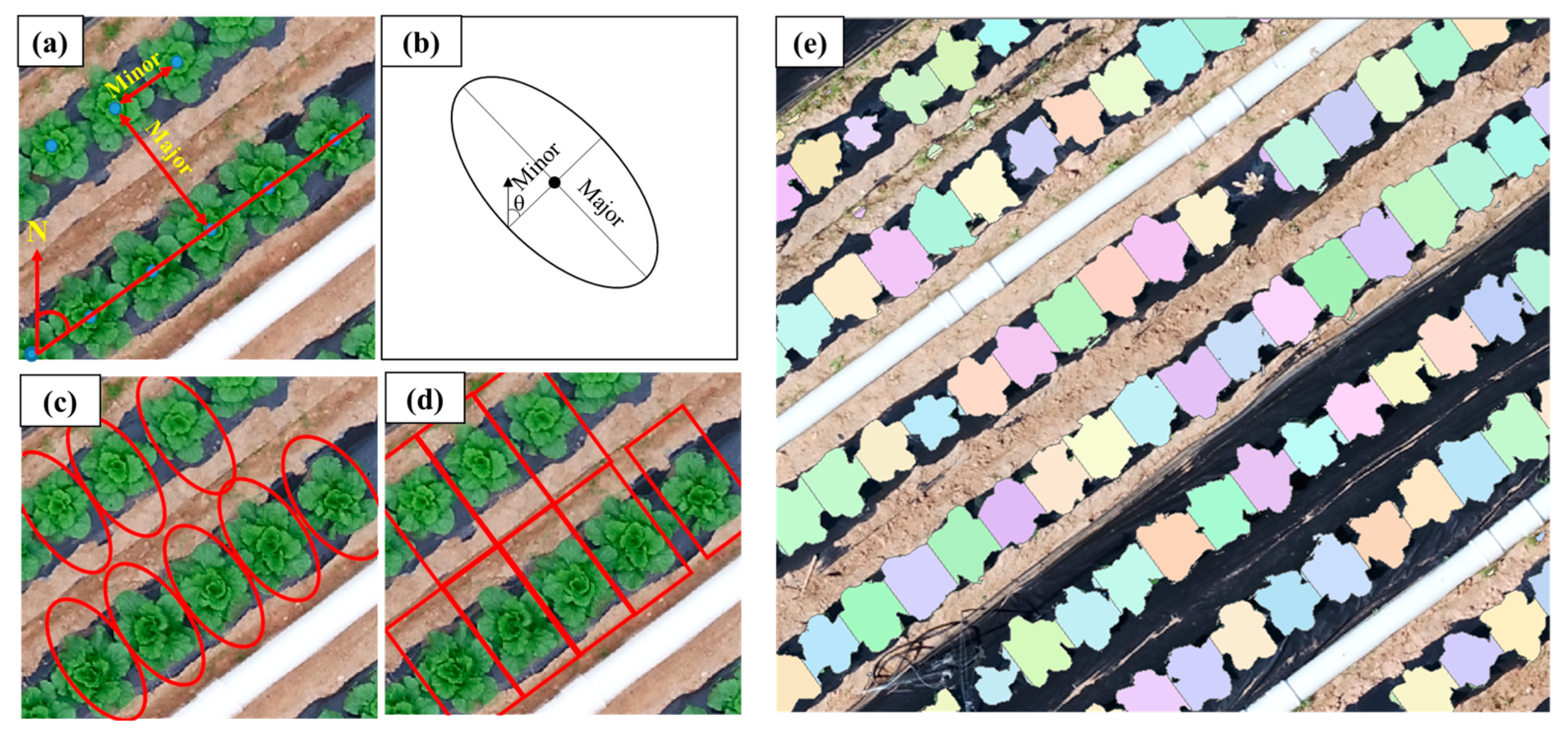
2.7.3. Process of Individual Napa Cabbage Object Segmentation
2.8. Definition of Independent Variables for AI Models
2.8.1. RGB-Based Independent Variables
2.8.2. Multispectral Sensor-Based Independent Variables
2.8.3. Thermal Infrared Sensor-Based Independent Variable
| Sensor Type | Vegetation Index | Equation | Reference |
|---|---|---|---|
| RGB | VF (Vegetation Fraction) | VF = (Area of each Napa cabbage object)/(Grid area) | - |
| CHM (Crop Height Model) | CHM = DSM_(growth stage) − DTM_(pre-planting) | [21] | |
| Multi Spectral | CIRE (Red Edge Chlorophyll Index) | CIRE = (RN/RRE) − 1 | [40] |
| VARI (Visible Atmospherically Resistant Index) | VARI = (RG − RR)/(RG + RR) | [41] | |
| CVI (Chlorophyll Vegetation Index) | CVI = (RN/RG) × (RR/RG) | [42] | |
| SR (Simple Ratio) | SR = (RN/RG) | [43] | |
| GNDVI (Green Normalized Difference Vegetation Index) | GNDVI = (RN − RG)/(RN + RG) | [44] | |
| CIGreen (Green Chlorophyll Index) | CIGreen = (RN/RG) − 1 | [40] | |
| GEMI (Global Environmental Monitoring Index) | GEMI = n × [(1 − 0.25n) × (RR − 0.125)]/(1 − RR) n = [(RN2 − RR2) + 1.5 × RN + 0.5 × RR]/(RN + RR + 0.5) | [45] | |
| NDVI (Normalized Difference Vegetation Index) | NDVI = (RN − RR)/(RN + RR) | [46] | |
| Thermal Infrared | CWSI (Crop Water Stress Index) | CWSI = (T − Tc)/(Th − Tc) | [39] |
2.9. Construction of Datasets and AI Models
2.10. Accuracy Evaluation
2.10.1. Model Accuracy Evaluation
2.10.2. Feature Importance
3. Results
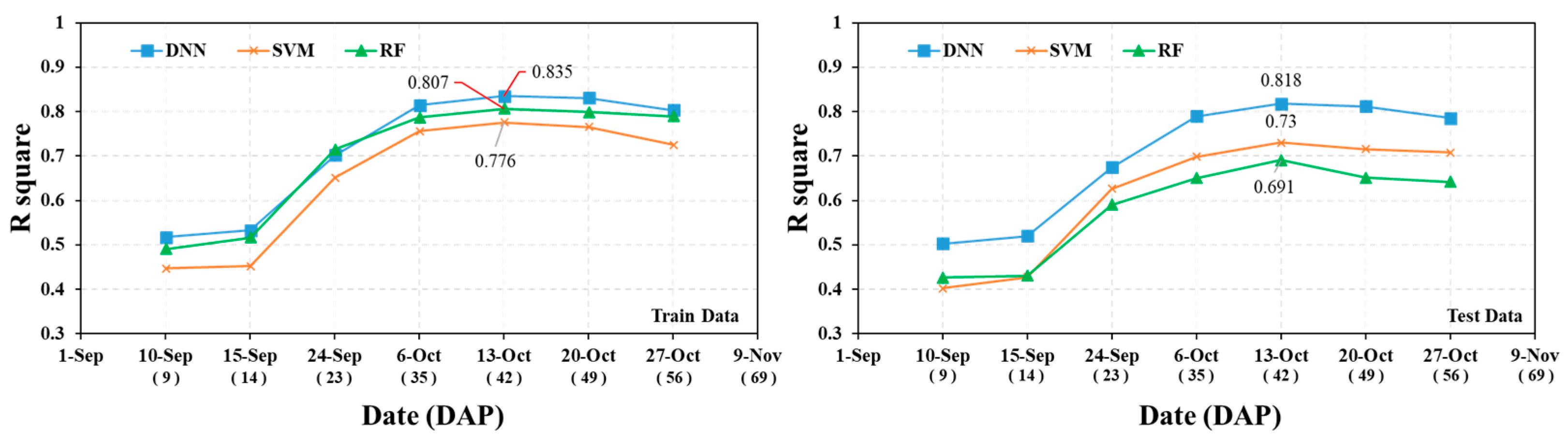
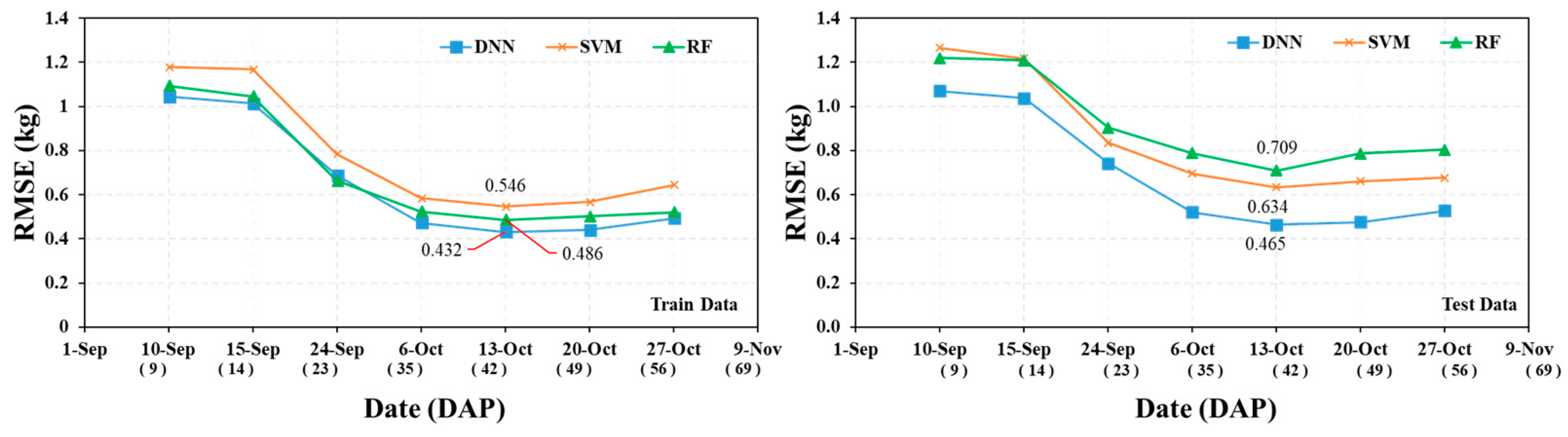
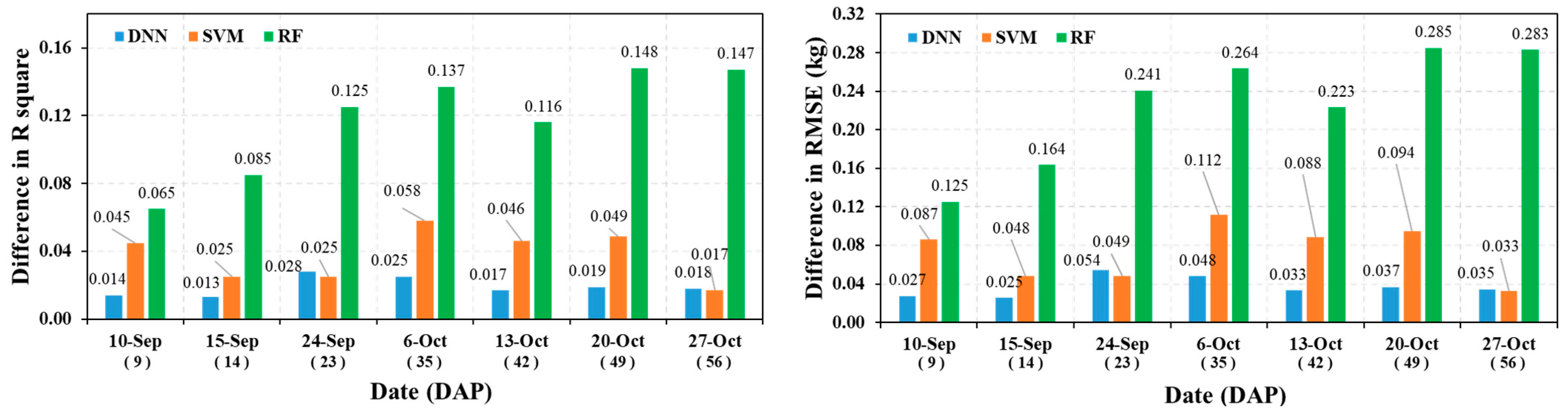
3.1. Model Accuracy Evaluation by Survey Period
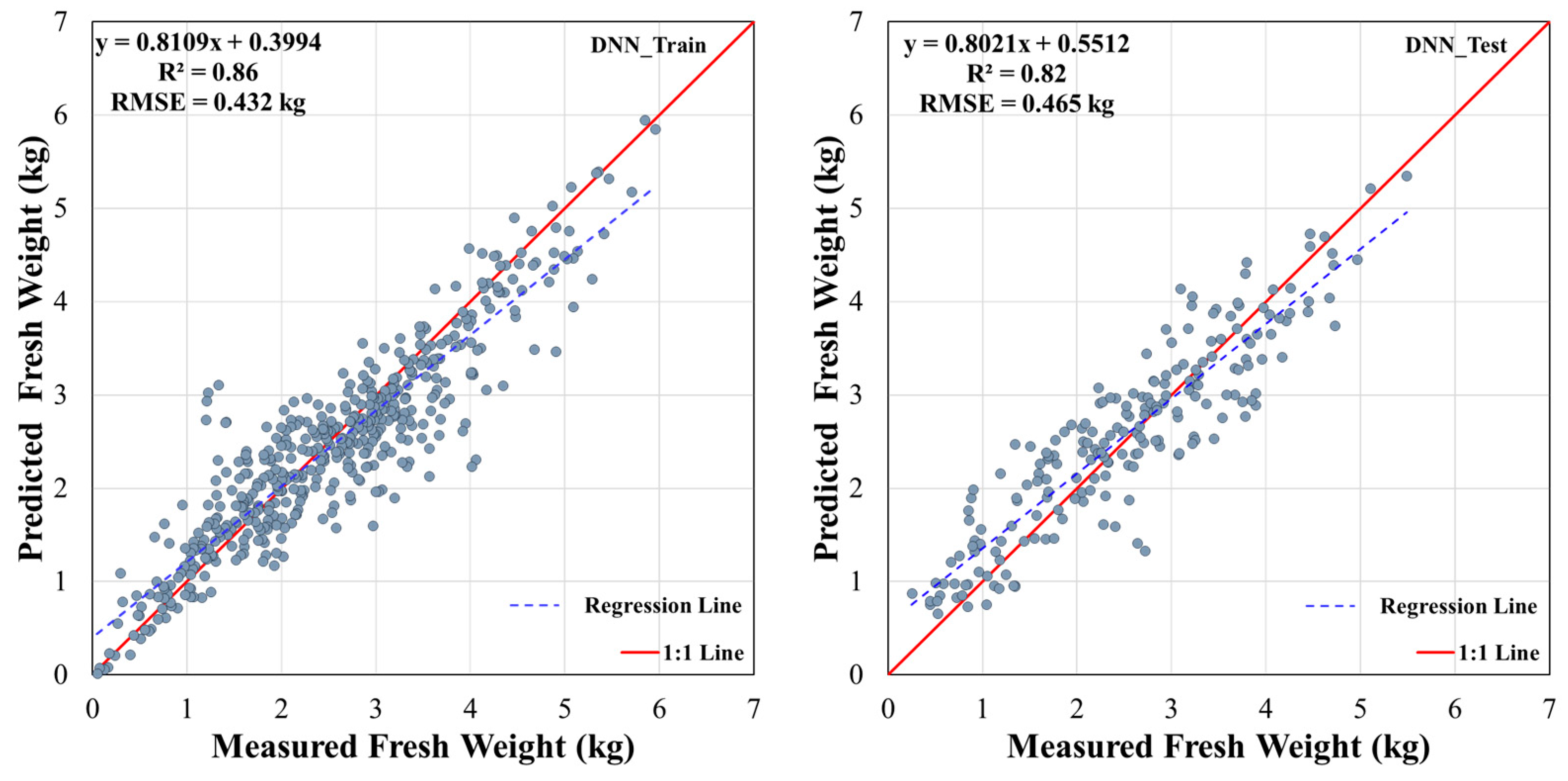
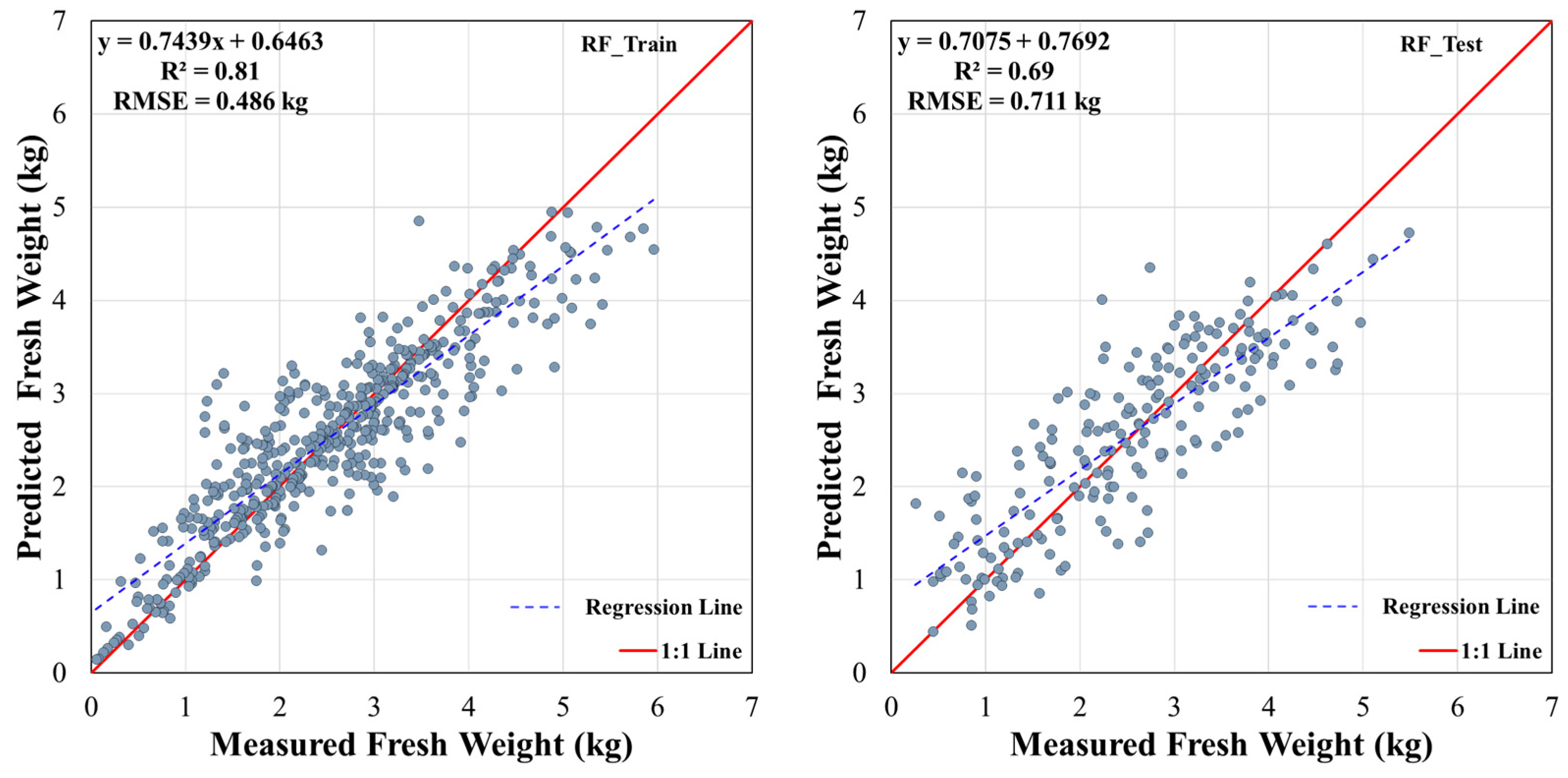
3.2. Model Overfitting Analysis
3.3. Fresh Weight Prediction Performance of DNN, SVM, and RF Models
3.4. Bias Analysis for Overall and Fresh Weight Sections of DNN, SVM, and RF Models
- DNN: The highest bias (underestimation of 0.365 kg) is observed in the 4–5 kg range.
- RF: The highest bias (underestimation of 0.541 kg) occurs for Napa cabbages exceeding 5 kg.
- SVM: Similar to the RF model, the highest bias (underestimation of 0.88 kg) is also observed in the >5 kg range.
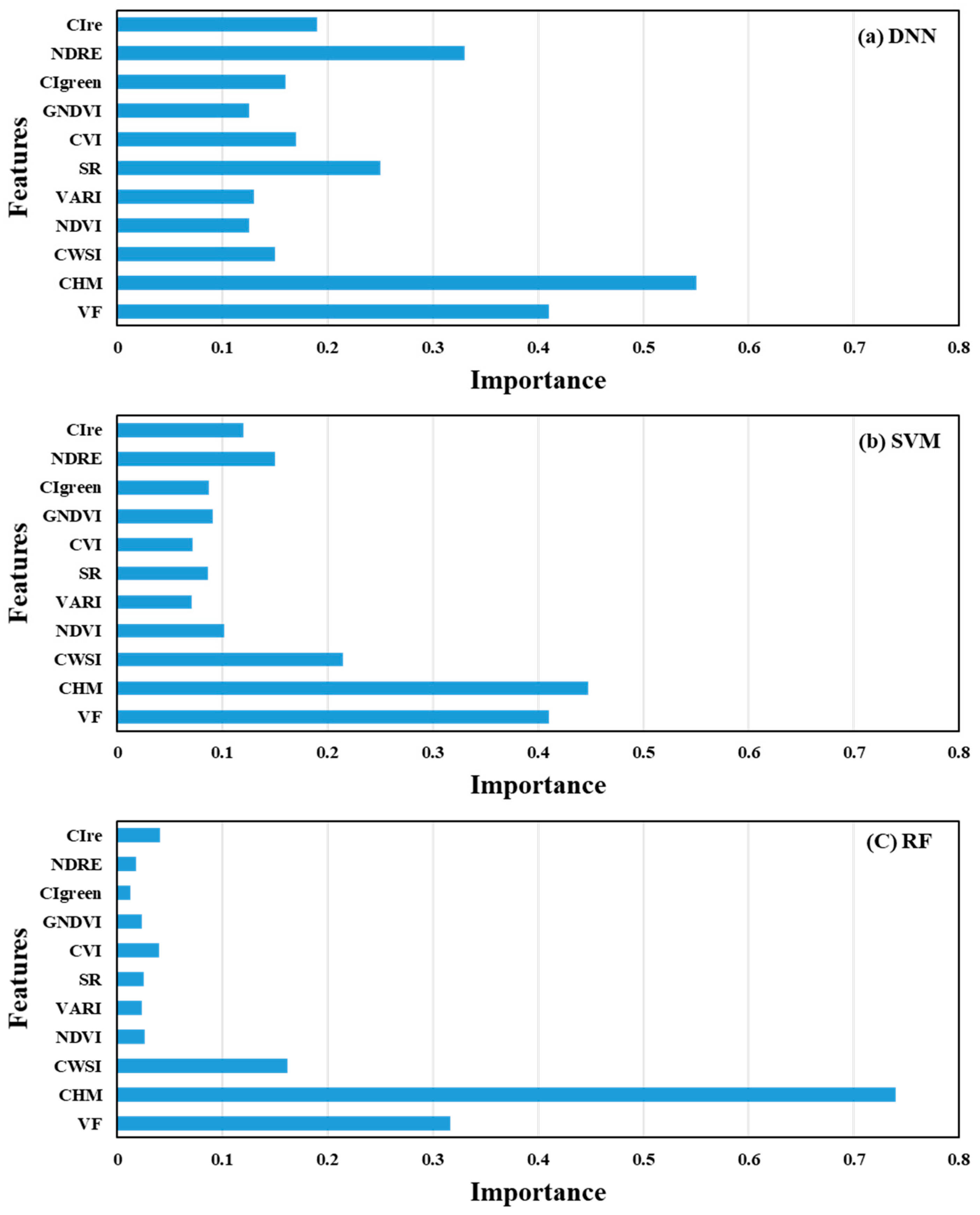
3.5. Evaluation of Feature Importance in DNN, SVM, and RF Models Using Permutation Feature Importance (PFI)
3.6. Spatial Analysis of Measured and Predicted Napa Cabbage Fresh Weight Using the DNN Model
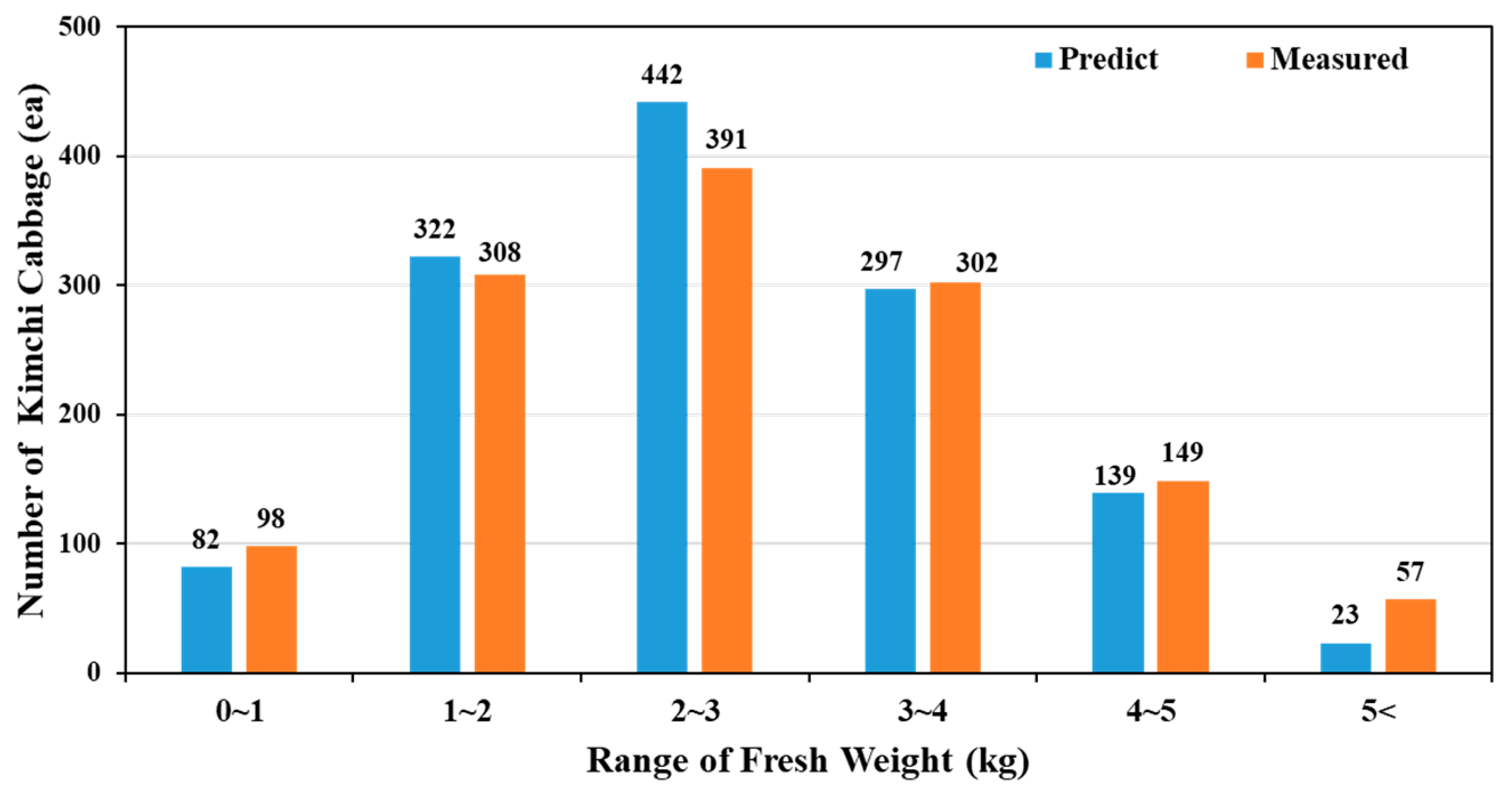
3.6.1. Comparison of Spatial Distributions and Weight Category Frequencies
3.6.2. Overall Model Performance and Spatial Variability
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lobell, D.B.; Schlenker, W.; Costa-Roberts, J. Climate Trends and Global Crop Production since 1980. Science 2011, 333, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Walsh, K.J.; McBride, J.L.; Klotzbach, P.J.; Balachandran, S.; Camargo, S.J.; Holland, G.; Knutson, T.R.; Kossin, J.P.; Lee, T.; Sobel, A. Tropical Cyclones and Climate Change. Wiley Interdiscip. Rev. Clim. Change 2016, 7, 65–89. [Google Scholar] [CrossRef]
- Ummenhofer, C.C.; Meehl, G.A. Extreme Weather and Climate Events with Ecological Relevance: A Review. Philos. Trans. R. Soc. B. 2017, 372, 20160135. [Google Scholar] [CrossRef] [PubMed]
- Sishodia, R.P.; Ray, R.L.; Singh, S.K. Applications of Remote Sensing in Precision Agriculture: A Review. Remote Sens. 2020, 12, 3136. [Google Scholar] [CrossRef]
- Kwaghtyo, D.K.; Eke, C.I. Smart Farming Prediction Models for Precision Agriculture: A Comprehensive Survey. Artif. Intell. Rev. 2023, 56, 5729–5772. [Google Scholar] [CrossRef]
- Kim, I.; Jeong, S.; Jeong, G. An Analysis of the Impact of Changes in Kimchi Imports on the Korean Kimchi Industry. Korean J. Org. Agric. 2022, 30, 151–170. [Google Scholar] [CrossRef]
- Eum, H.L.; Kim, B.; Yang, Y.J.; Hong, S.J. Quality Evaluation and Optimization of Storage Temperature with Eight Cultivars of Kimchi Cabbage Produced in Summer at Highland Areas. Hortic. Sci. Technol. 2013, 31, 211–218. [Google Scholar] [CrossRef]
- Choi, E.J.; Jeong, M.C.; Ku, K.H. Effect of Seasonal Cabbage Cultivar (Brassica rapa L. Ssp. Pekinesis) on the Quality Characteristics of Salted-Kimchi Cabbages during Storage Period. Korean J. Food Preserv. 2015, 22, 303–313. [Google Scholar] [CrossRef]
- Radoglou-Grammatikis, P.; Sarigiannidis, P.; Lagkas, T.; Moscholios, I. A Compilation of UAV Applications for Precision Agriculture. Comput. Netw. 2020, 172, 107148. [Google Scholar] [CrossRef]
- Tripathi, M.K.; Maktedar, D.D. A Role of Computer Vision in Fruits and Vegetables among various Horticulture Products of Agriculture Fields: A Survey. Inf. Process. Agric. 2020, 7, 183–203. [Google Scholar] [CrossRef]
- De Ocampo, A.L.P.; Montalbo, F.J.P. A Multi-Vision Monitoring Framework for Simultaneous Real-Time Unmanned Aerial Monitoring of Farmer Activity and Crop Health. Smart Agric. Technol. 2024, 8, 100466. [Google Scholar] [CrossRef]
- Mezera, J.; Lukas, V.; Horniaček, I.; Smutný, V.; Elbl, J. Comparison of Proximal and Remote Sensing for the Diagnosis of Crop Status in Site-Specific Crop Management. Sensors 2021, 22, 19. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Srivastava, P.; Lamba, J.; Ortiz, B.V.; Way, T.R.; Sangha, L.; Takhellambam, B.S.; Morata, G.; Molinari, R. Within-Field Variability in Nutrients for Site-Specific Agricultural Management in Irrigated Cornfield. J. ASABE 2022, 65, 865–880. [Google Scholar] [CrossRef]
- Flores, R.; Lázaro, E.; Ramos, E.; Provost, K.; Habib, M. Demand Management in the Supply Chain: A Focus on Agribusiness. In Advances in Manufacturing, Production Management and Process Control: Proceedings of the AHFE 2020 Virtual Conferences on Human Aspects of Advanced Manufacturing, Advanced Production Management and Process Control, and Additive Manufacturing, Modeling Systems and 3D Prototyping, San Diego, CA, USA, 16–20 July 2020; Springer: Cham, Switzerland, 2020; pp. 333–340. [Google Scholar]
- Karthikeyan, L.; Chawla, I.; Mishra, A.K. A Review of Remote Sensing Applications in Agriculture for Food Security: Crop Growth and Yield, Irrigation, and Crop Losses. J. Hydrol. 2020, 586, 124905. [Google Scholar] [CrossRef]
- Shi, Y.; Ji, S.; Shao, X.; Tang, H.; Wu, W.; Yang, P.; Zhang, Y.; Ryosuke, S. Framework of SAGI Agriculture Remote Sensing and its Perspectives in Supporting National Food Security. J. Integr. Agric. 2014, 13, 1443–1450. [Google Scholar] [CrossRef]
- Saranya, T.; Deisy, C.; Sridevi, S.; Anbananthen, K.S.M. A Comparative Study of Deep Learning and Internet of Things for Precision Agriculture. Eng. Appl. Artif. Intell. 2023, 122, 106034. [Google Scholar] [CrossRef]
- Benos, L.; Tagarakis, A.C.; Dolias, G.; Berruto, R.; Kateris, D.; Bochtis, D. Machine Learning in Agriculture: A Comprehensive Updated Review. Sensors 2021, 21, 3758. [Google Scholar] [CrossRef]
- Lee, D.; Shin, H.; Park, J. Developing a P-NDVI Map for Highland Kimchi Cabbage using Spectral Information from UAVs and a Field Spectral Radiometer. Agronomy 2020, 10, 1798. [Google Scholar] [CrossRef]
- Zhang, C.; Kovacs, J.M. The Application of Small Unmanned Aerial Systems for Precision Agriculture: A Review. Precis. Agric. 2012, 13, 693–712. [Google Scholar] [CrossRef]
- Bendig, J.; Bolten, A.; Bennertz, S.; Broscheit, J.; Eichfuss, S.; Bareth, G. Estimating Biomass of Barley using Crop Surface Models (CSMs) Derived from UAV-Based RGB Imaging. Remote Sens. 2014, 6, 10395–10412. [Google Scholar] [CrossRef]
- Go, S.; Lee, D.; Na, S.; Park, J. Analysis of Growth Characteristics of Kimchi Cabbage using Drone-Based Cabbage Surface Model Image. Agriculture 2022, 12, 216. [Google Scholar] [CrossRef]
- Hassan, M.A.; Yang, M.; Rasheed, A.; Yang, G.; Reynolds, M.; Xia, X.; Xiao, Y.; He, Z. A Rapid Monitoring of NDVI Across the Wheat Growth Cycle for Grain Yield Prediction using a Multi-Spectral UAV Platform. Plant Sci. 2019, 282, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Messina, G.; Modica, G. Applications of UAV Thermal Imagery in Precision Agriculture: State of the Art and Future Research Outlook. Remote Sens. 2020, 12, 1491. [Google Scholar] [CrossRef]
- Shuai, L.; Li, Z.; Chen, Z.; Luo, D.; Mu, J. A Research Review on Deep Learning Combined with Hyperspectral Imaging in Multiscale Agricultural Sensing. Comput. Electron. Agric. 2024, 217, 108577. [Google Scholar] [CrossRef]
- Lee, D.; Kim, H.; Park, J. UAV, a Farm Map, and Machine Learning Technology Convergence Classification Method of a Corn Cultivation Area. Agronomy 2021, 11, 1554. [Google Scholar] [CrossRef]
- Kwak, G.; Park, N. Unsupervised Domain Adaptation with Adversarial Self-Training for Crop Classification using Remote Sensing Images. Remote Sens. 2022, 14, 4639. [Google Scholar] [CrossRef]
- Stewart, E.L.; Wiesner-Hanks, T.; Kaczmar, N.; DeChant, C.; Wu, H.; Lipson, H.; Nelson, R.J.; Gore, M.A. Quantitative Phenotyping of Northern Leaf Blight in UAV Images using Deep Learning. Remote Sens. 2019, 11, 2209. [Google Scholar] [CrossRef]
- Zhu, W.; Sun, Z.; Yang, T.; Li, J.; Peng, J.; Zhu, K.; Li, S.; Gong, H.; Lyu, Y.; Li, B.; et al. Estimating Leaf Chlorophyll Content of Crops Via Optimal Unmanned Aerial Vehicle Hyperspectral Data at Multi-Scales. Comput. Electron. Agric. 2020, 178, 105786. [Google Scholar] [CrossRef]
- Lee, D.; Jeong, C.; Go, S.; Park, J. Evaluation of Applicability of RGB Image using Support Vector Machine Regression for Estimation of Leaf Chlorophyll Content of Onion and Garlic. Korean J. Remote Sens. 2021, 37, 1669–1683. [Google Scholar] [CrossRef]
- Maimaitijiang, M.; Sagan, V.; Sidike, P.; Hartling, S.; Esposito, F.; Fritschi, F.B. Soybean Yield Prediction from UAV using Multimodal Data Fusion and Deep Learning. Remote Sens. Environ. 2020, 237, 111599. [Google Scholar] [CrossRef]
- Kim, D.; Yun, H.S.; Jeong, S.; Kwon, Y.; Kim, S.; Lee, W.S.; Kim, H. Modeling and Testing of Growth Status for Chinese Cabbage and White Radish with UAV-Based RGB Imagery. Remote Sens. 2018, 10, 563. [Google Scholar] [CrossRef]
- Killeen, P.; Kiringa, I.; Yeap, T.; Branco, P. Corn Grain Yield Prediction using UAV-Based High Spatiotemporal Resolution Imagery, Machine Learning, and Spatial Cross-Validation. Remote Sens. 2024, 16, 683. [Google Scholar] [CrossRef]
- Zhang, T.; Yang, Z.; Xu, Z.; Li, J. Wheat Yellow Rust Severity Detection by Efficient DF-UNet and UAV Multispectral Imagery. IEEE Sens. J. 2022, 22, 9057–9068. [Google Scholar] [CrossRef]
- Vélez, S.; Ariza-Sentís, M.; Valente, J. Mapping the Spatial Variability of Botrytis Bunch Rot Risk in Vineyards using UAV Multispectral Imagery. Eur. J. Agron. 2023, 142, 126691. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep Learning in Agriculture: A Survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Woebbecke, D.M.; Meyer, G.E.; Von Bargen, K.; Mortensen, D.A. Color Indices for Weed Identification Under various Soil, Residue, and Lighting Conditions. Trans. ASAE 1995, 38, 259–269. [Google Scholar] [CrossRef]
- Ostu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Jones, H.G. Use of Infrared Thermometry for Estimation of Stomatal Conductance as a Possible Aid to Irrigation Scheduling. Agric. For. Meteorol. 1999, 95, 139–149. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Viña, A.; Ciganda, V.; Rundquist, D.C.; Arkebauer, T.J. Remote Estimation of Canopy Chlorophyll Content in Crops. Geophys. Res. Lett. 2005, 32, L08403. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Stark, R.; Rundquist, D. Novel Algorithms for Remote Estimation of Vegetation Fraction. Remote Sens. Environ. 2002, 80, 76–87. [Google Scholar] [CrossRef]
- Vincini, M.; Frazzi, E.; D’Alessio, P. A Broad-Band Leaf Chlorophyll Vegetation Index at the Canopy Scale. Precis. Agric. 2008, 9, 303–319. [Google Scholar] [CrossRef]
- Jordan, C.F. Derivation of Leaf-area Index from Quality of Light on the Forest Floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a Green Channel in Remote Sensing of Global Vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Pinty, B.; Verstraete, M.M. GEMI: A Non-Linear Index to Monitor Global Vegetation from Satellites. Vegetatio 1992, 101, 15–20. [Google Scholar] [CrossRef]
- Rouse, J.; Haas, R.; Schell, J.; Deering, D. Monitoring Vegetation Systems in the Great Plains with ERTS; Goddard Space Flight Center 3d ERTS-1 Symp.; NASA: Washington, DC, USA, 1973; Volume 1, pp. 309–317. Available online: http://hdl.handle.net/2060/19740022614 (accessed on 2 July 2024).
- Camps-Valls, G.; Bruzzone, L.; Rojo-Álvarez, J.L.; Melgani, F. Robust Support Vector Regression for Biophysical Variable Estimation from Remotely Sensed Images. IEEE Geosci. Remote Sens. Lett. 2006, 3, 339–343. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Otchere, D.A.; Mohammed, M.A.A.; Al-Hadrami, H.; Boakye, T.B. Enhancing Drilling Fluid Lost-circulation Prediction: Using Model Agnostic and Supervised Machine Learning. In Data Science and Machine Learning Applications in Subsurface Engineering; CRC Press: Boca Raton, FL, USA, 2022; pp. 6–32. [Google Scholar]
- Li, F.; Miao, Y.; Feng, G.; Yuan, F.; Yue, S.; Gao, X.; Liu, Y.; Liu, B.; Ustin, S.L.; Chen, X. Improving Estimation of Summer Maize Nitrogen Status with Red Edge-Based Spectral Vegetation Indices. Field Crops Res. 2014, 157, 111–123. [Google Scholar] [CrossRef]
- Schrader-Patton, C.; Grulke, N.E.; Anderson, P.D.; Chaitman, J.; Webb, J. Assessing Tree Water Balance After Forest Thinning Treatments using Thermal and Multispectral Imaging. Remote Sens. 2024, 16, 1005. [Google Scholar] [CrossRef]
- Yang, F.; Zhou, Y.; Du, J.; Wang, K.; Lv, L.; Long, W. Prediction of Fruit Characteristics of Grafted Plants of Camellia Oleifera by Deep Neural Networks. Plant Methods 2024, 20, 23. [Google Scholar] [CrossRef]
- Fei, S.; Hassan, M.A.; Xiao, Y.; Su, X.; Chen, Z.; Cheng, Q.; Duan, F.; Chen, R.; Ma, Y. UAV-Based Multi-Sensor Data Fusion and Machine Learning Algorithm for Yield Prediction in Wheat. Precis. Agric. 2023, 24, 187–212. [Google Scholar] [CrossRef]
- Kuradusenge, M.; Hitimana, E.; Hanyurwimfura, D.; Rukundo, P.; Mtonga, K.; Mukasine, A.; Uwitonze, C.; Ngabonziza, J.; Uwamahoro, A. Crop Yield Prediction using Machine Learning Models: Case of Irish Potato and Maize. Agriculture 2023, 13, 225. [Google Scholar] [CrossRef]

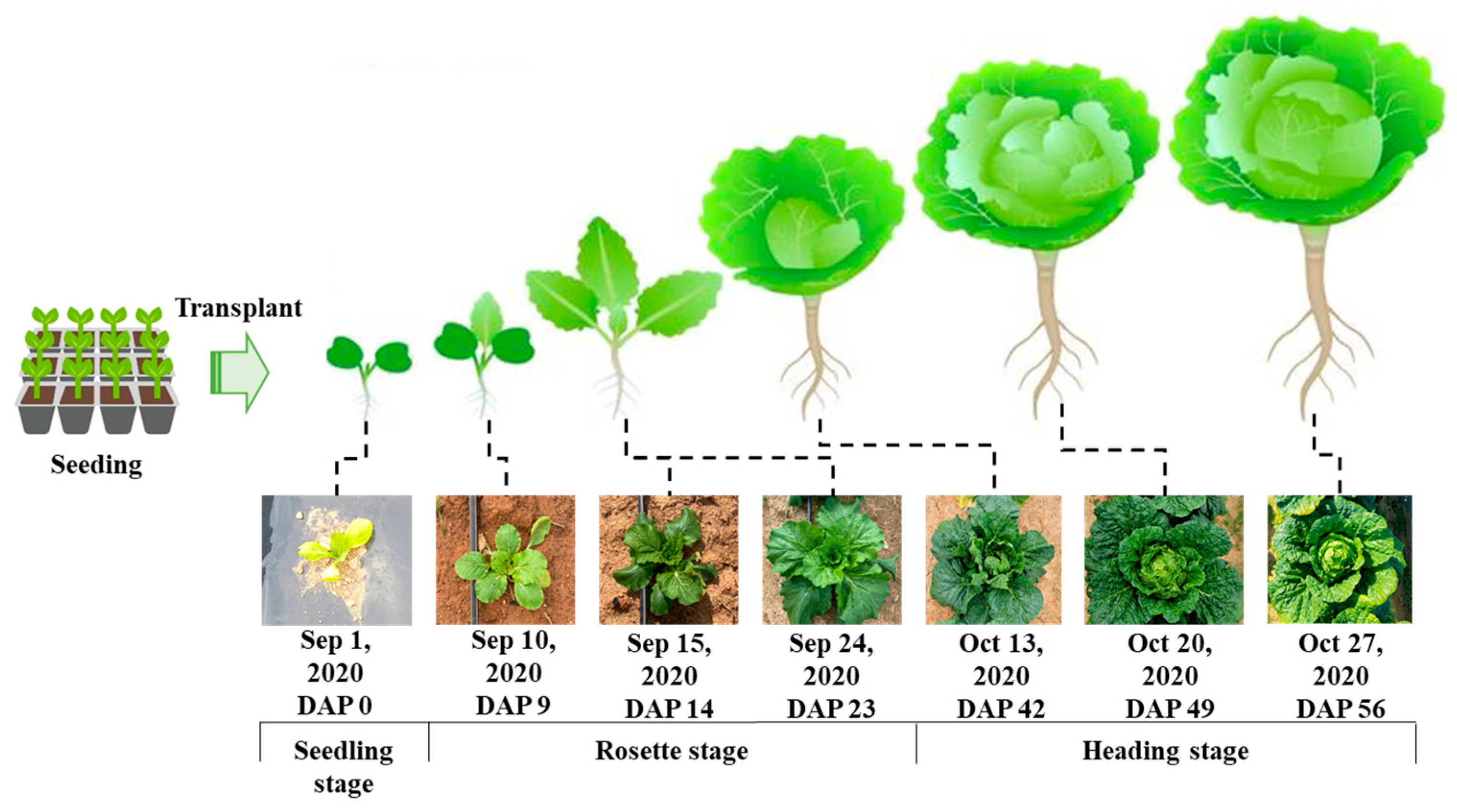




| Year | Date (mm/dd) |
|---|---|
| 2020 | 9/1, 9/10, 9/15, 9/24, 10/6, 10/13, 10/20, 10/27, 11/9 |
| Model | Data Set | Metrics | Date | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 10 Sep | 15 Sep | 24 Sep | 6 Oct | 13 Oct | 20 Oct | 27 Oct | |||
| DNN | Train | R2 | 0.52 | 0.53 | 0.70 | 0.81 | 0.84 * | 0.83 | 0.80 |
| Test | 0.50 | 0.52 | 0.67 | 0.79 | 0.82 | 0.81 | 0.79 | ||
| RF | Train | 0.44 | 0.46 | 0.66 | 0.73 | 0.75 | 0.75 | 0.74 | |
| Test | 0.43 | 0.43 | 0.59 | 0.65 | 0.69 | 0.65 | 0.64 | ||
| SVM | Train | 0.45 | 0.45 | 0.65 | 0.76 | 0.78 | 0.77 | 0.73 | |
| Test | 0.40 | 0.43 | 0.63 | 0.70 | 0.73 | 0.72 | 0.71 | ||
| DNN | Train | RMSE (kg) | 1.04 | 1.01 | 0.69 | 0.47 | 0.43 | 0.44 | 0.49 |
| Test | 1.07 | 1.04 | 0.74 | 0.52 | 0.47 | 0.48 | 0.53 | ||
| RF | Train | 1.09 | 1.05 | 0.66 | 0.52 | 0.49 | 0.50 | 0.52 | |
| Test | 1.22 | 1.21 | 0.90 | 0.79 | 0.71 | 0.79 | 0.80 | ||
| SVM | Train | 1.18 | 1.17 | 0.79 | 0.58 | 0.55 | 0.57 | 0.64 | |
| Test | 1.27 | 1.22 | 0.83 | 0.70 | 0.63 | 0.66 | 0.68 | ||
| Range of Fresh Wight (kg) | Models | ||
|---|---|---|---|
| DNN | RF | SVM | |
| <1 | −0.021 | −0.002 | −0.015 |
| 1~2 | −0.212 | −0.229 | −0.335 |
| 2~3 | 0.086 | −0.007 | −0.026 |
| 3~4 | 0.354 | 0.200 | 0.219 |
| 4~5 | 0.365 | 0.227 | 0.464 |
| >5 | 0.109 | 0.541 | 0.88 |
| All Range | 0.008 | 0.031 | 0.041 |
| Year | Number of Kimchi Cabbage | Measured Fresh Weight (kg) [A] | Predicted Fresh Weight (kg) [B] | Error Rate (%) [1 − B/A × 100] |
|---|---|---|---|---|
| 2020 | 1305 | 3507 | 3413 | 2.69 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, D.-H.; Park, J.-H. Development of a UAS-Based Multi-Sensor Deep Learning Model for Predicting Napa Cabbage Fresh Weight and Determining Optimal Harvest Time. Remote Sens. 2024, 16, 3455. https://doi.org/10.3390/rs16183455
Lee D-H, Park J-H. Development of a UAS-Based Multi-Sensor Deep Learning Model for Predicting Napa Cabbage Fresh Weight and Determining Optimal Harvest Time. Remote Sensing. 2024; 16(18):3455. https://doi.org/10.3390/rs16183455
Chicago/Turabian StyleLee, Dong-Ho, and Jong-Hwa Park. 2024. "Development of a UAS-Based Multi-Sensor Deep Learning Model for Predicting Napa Cabbage Fresh Weight and Determining Optimal Harvest Time" Remote Sensing 16, no. 18: 3455. https://doi.org/10.3390/rs16183455
APA StyleLee, D.-H., & Park, J.-H. (2024). Development of a UAS-Based Multi-Sensor Deep Learning Model for Predicting Napa Cabbage Fresh Weight and Determining Optimal Harvest Time. Remote Sensing, 16(18), 3455. https://doi.org/10.3390/rs16183455







