Focus on the Crop Not the Weed: Canola Identification for Precision Weed Management Using Deep Learning
Abstract
:1. Introduction
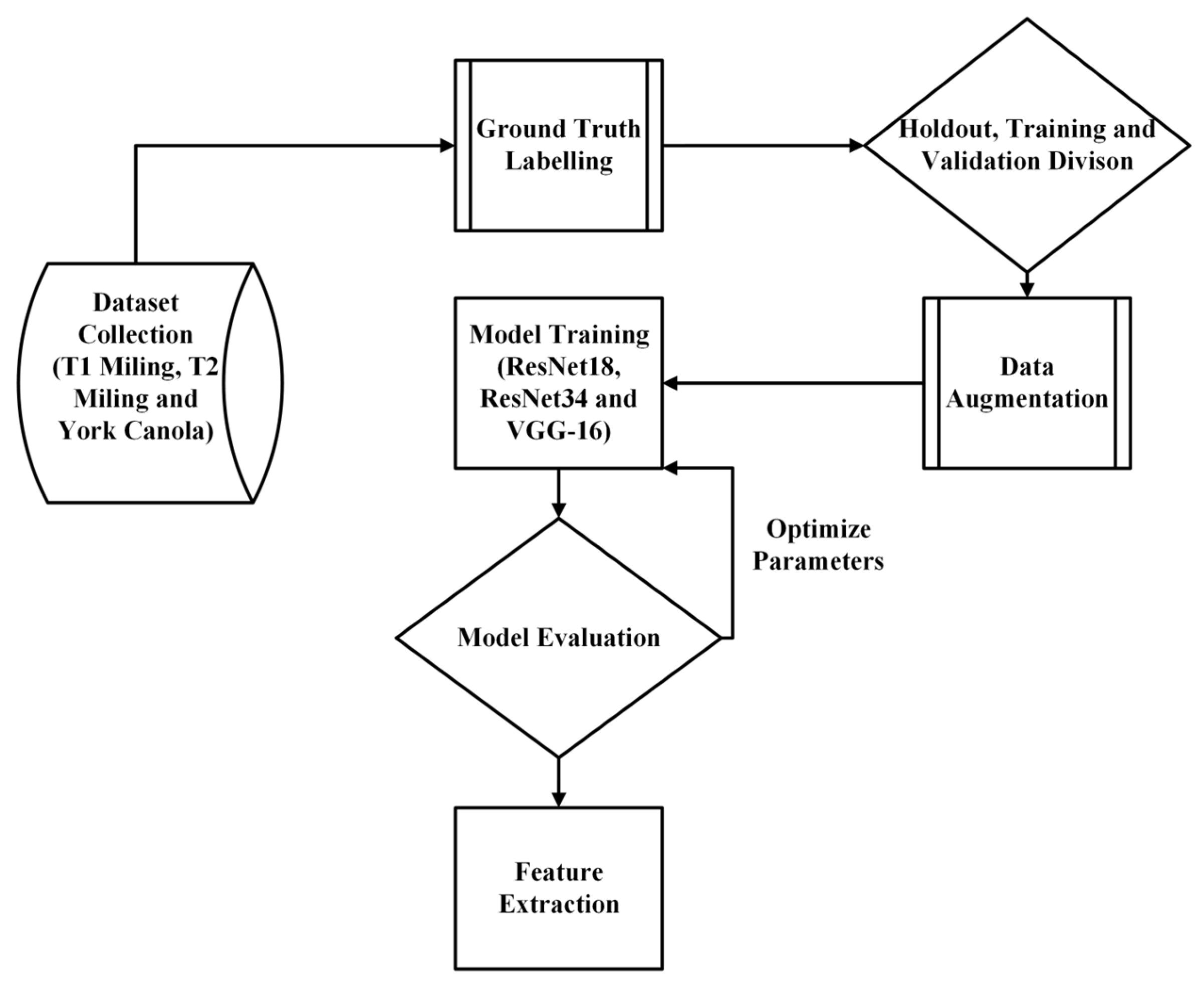
2. Materials and Methods
2.1. Miling Dataset Collection
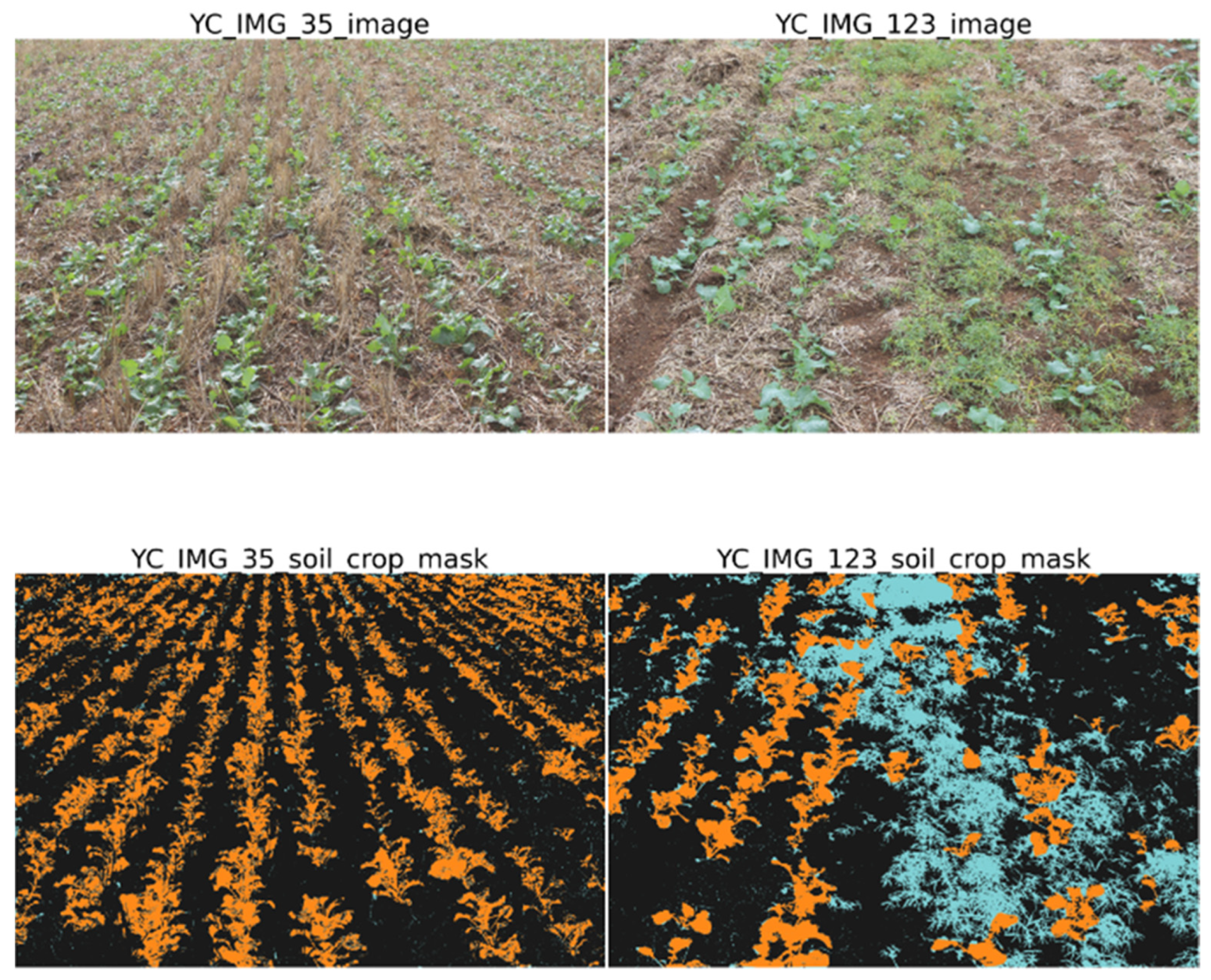
2.2. York Dataset Collection
2.3. Bounding-Box Image Labelling
2.4. Segmentation Mask Generation
2.5. Mask Cutting
2.6. Partition of Dataset into Training, Validation, and Holdout
2.7. Building the Dataloader and Hyperparameters
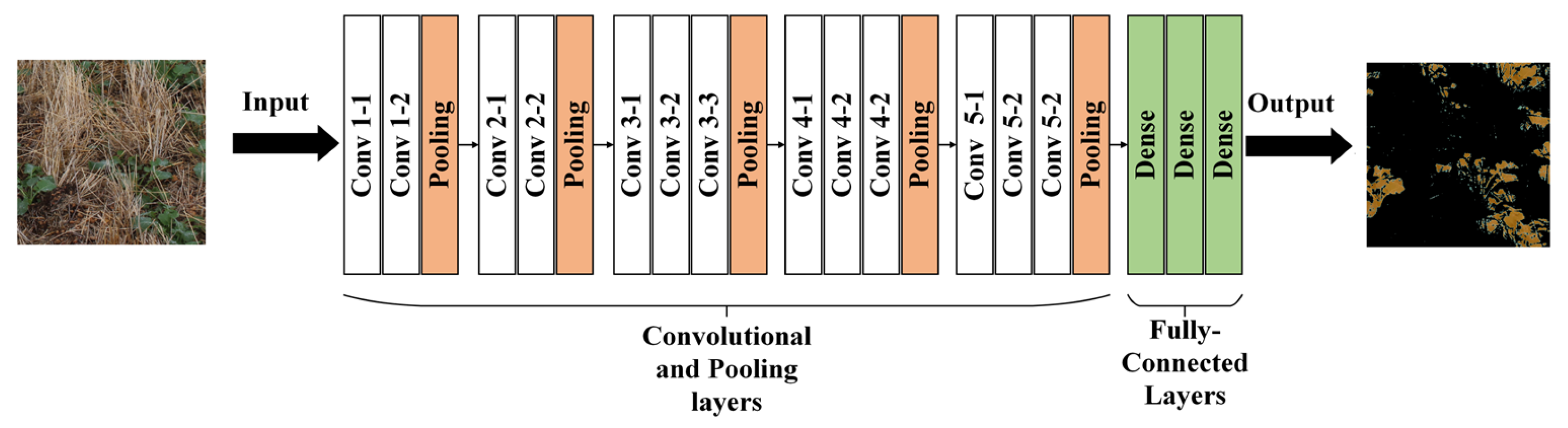
2.8. U-Net Model Architecture
2.9. Evaluation Metrics
2.10. Training
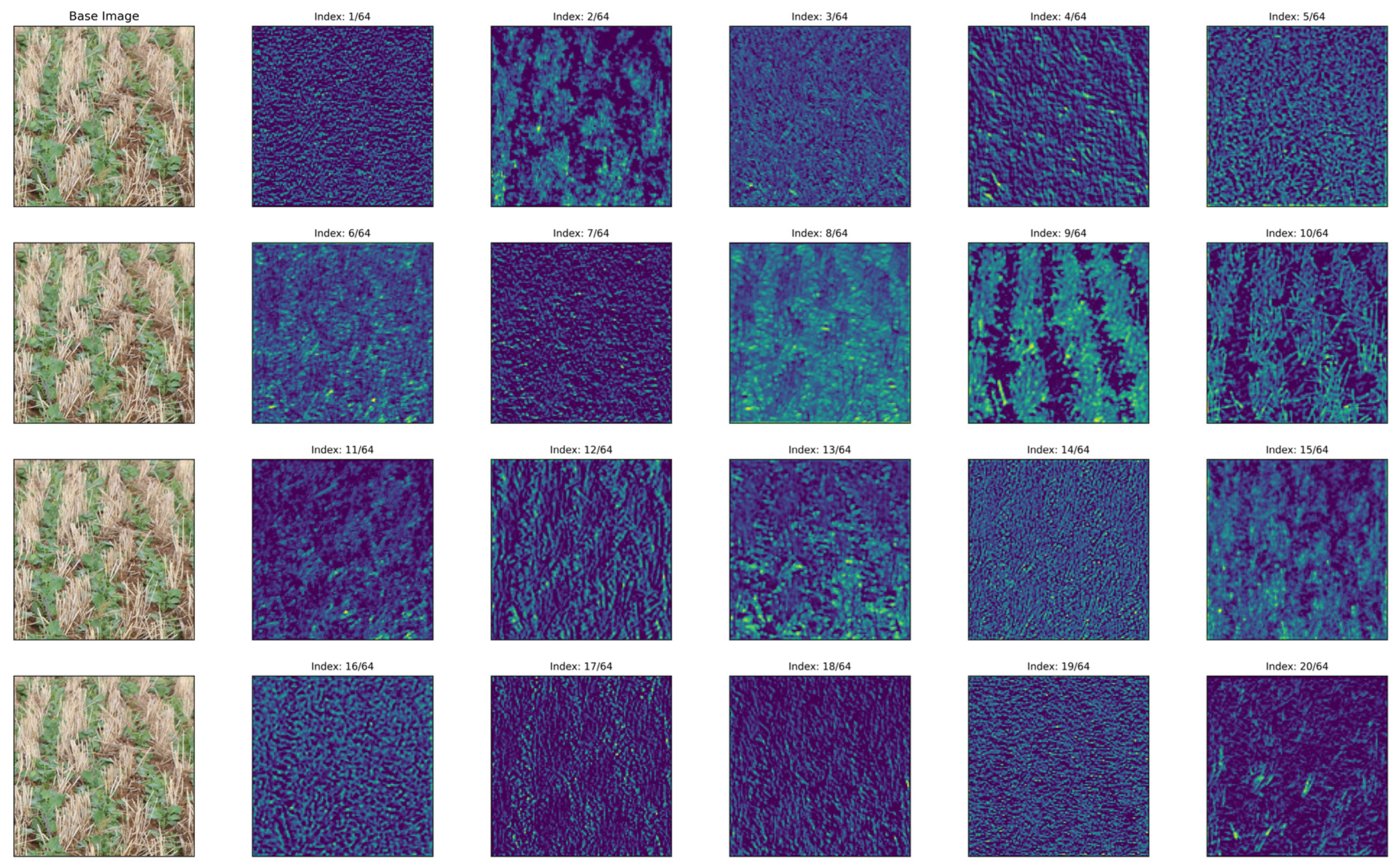
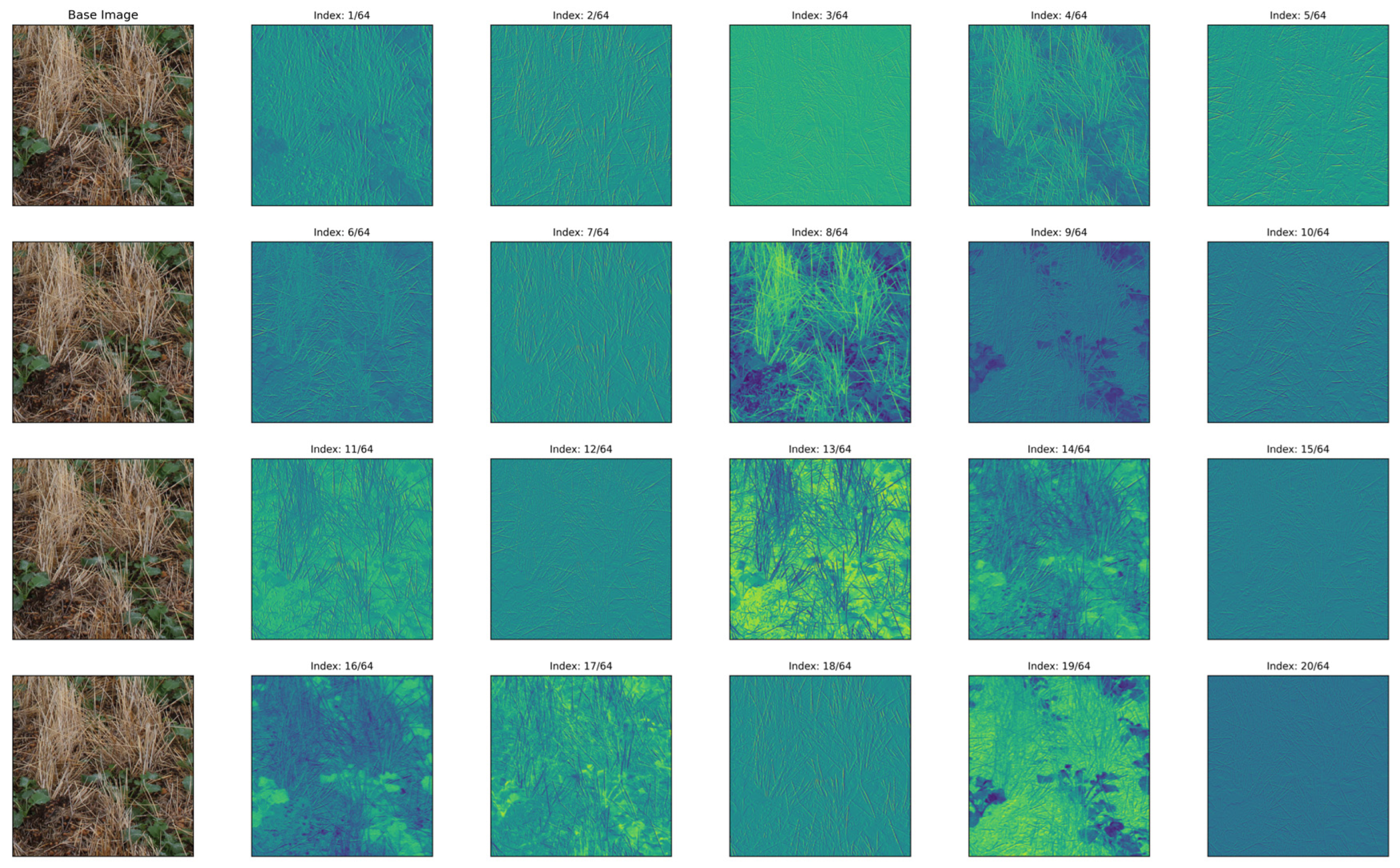
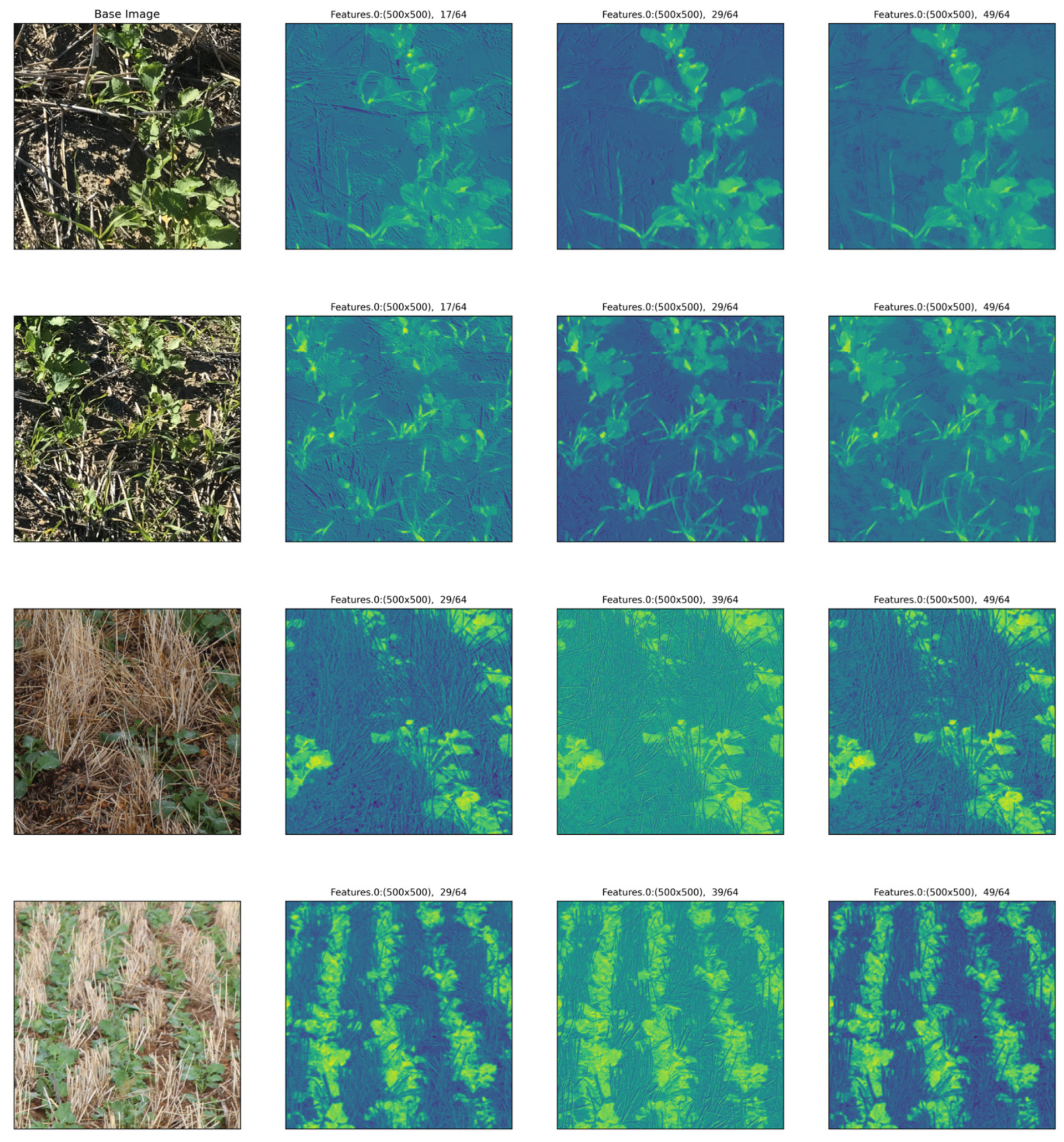
2.11. Feature Extraction Analysis
2.12. Displaying Results
3. Results
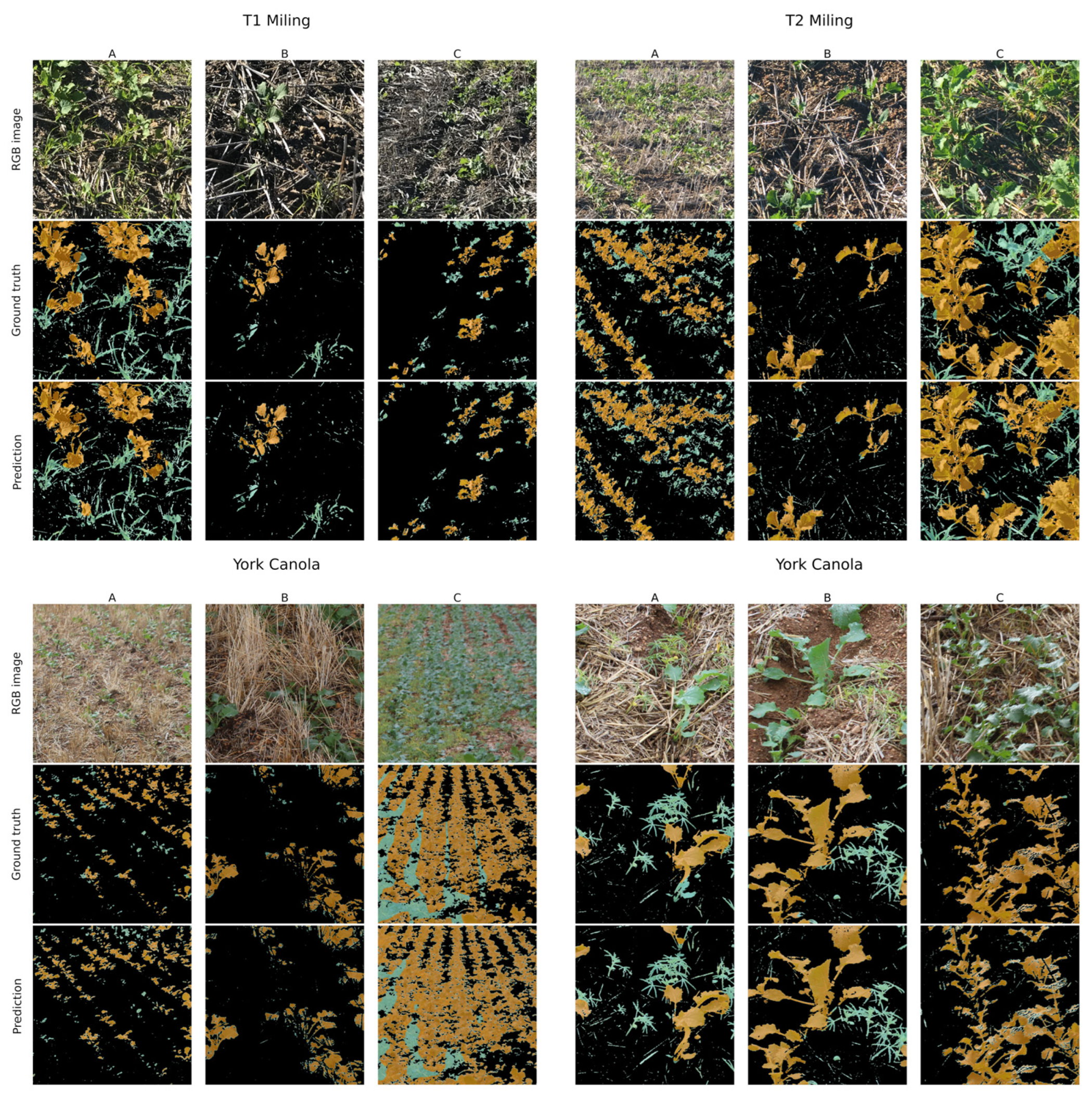
3.1. Segmentation Performance across Datasets
3.2. Variation in Architecture Prediction Performance
3.3. Feature Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shahbandeh, M. Consumption of Vegetable Oils Worldwide from 2013/14 to 2022/2023, by Oil Type. Available online: https://statista.com/statistics/263937/vegetable-oils-global-consumption/ (accessed on 22 October 2023).
- Asaduzzaman, M.; Pratley, J.E.; Luckett, D.; Lemerle, D.; Wu, H. Weed management in canola (Brassica napus L.): A review of current constraints and future strategies for Australia. Arch. Agron. Soil Sci. 2020, 66, 427–444. [Google Scholar] [CrossRef]
- Lemerle, D.; Luckett, D.J.; Lockley, P.; Koetz, E.; Wu, H. Competitive ability of Australian canola (Brassica napus) genotypes for weed management. Crop Pasture Sci. 2014, 65, 1300–1310. [Google Scholar] [CrossRef]
- Oerke, E.-C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Mennan, H.; Jabran, K.; Zandstra, B.H.; Pala, F. Non-chemical weed management in vegetables by using cover crops: A review. Agronomy 2020, 10, 257. [Google Scholar] [CrossRef]
- Kuehne, G.; Llewellyn, R.; Pannell, D.J.; Wilkinson, R.; Dolling, P.; Ouzman, J.; Ewing, M. Predicting farmer uptake of new agricultural practices: A tool for research, extension and policy. Agric. Syst. 2017, 156, 115–125. [Google Scholar] [CrossRef]
- Lemerle, D.; Blackshaw, R.; Smith, A.B.; Potter, T.; Marcroft, S. Comparative survey of weeds surviving in triazine-tolerant and conventional canola crops in south-eastern Australia. Plant Prot. Q. 2001, 16, 37–40. [Google Scholar]
- Deirdre, L.; Rodney, M. Influence of wild radish on yield and quality of canola. Weed Sci. 2002, 50, 344–349. [Google Scholar]
- Beckie, H.J.; Warwick, S.I.; Nair, H.; Séguin-Swartz, G. Gene flow in commercial fields of herbicide-resistant canola (Brassica napus). Ecol. Appl. 2003, 13, 1276–1294. [Google Scholar] [CrossRef]
- Dill, G.M. Glyphosate-resistant crops: History, status and future. Pest Manag. Sci. 2005, 61, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Gaines, T.A.; Duke, S.O.; Morran, S.; Rigon, C.A.G.; Tranel, P.J.; Küpper, A.; Dayan, F.E. Mechanisms of evolved herbicide resistance. J. Biol. Chem. 2020, 295, 10307–10330. [Google Scholar] [CrossRef]
- Lemerle, D.; Luckett, D.J.; Wu, H.; Widderick, M.J. Agronomic interventions for weed management in canola (Brassica napus L.)—A review. Crop Prot. 2017, 95, 69–73. [Google Scholar]
- Heap, I.; Duke, S.O. Overview of glyphosate-resistant weeds worldwide. Pest Manag. Sci. 2018, 74, 1040–1049. [Google Scholar] [CrossRef] [PubMed]
- Neve, P.; Sadler, J.; Powles, S.B. Multiple herbicide resistance in a glyphosate-resistant rigid ryegrass (Lolium rigidum) population. Weed Sci. 2004, 52, 920–928. [Google Scholar] [CrossRef]
- Ashworth, M.B.; Walsh, M.J.; Flower, K.C.; Powles, S.B. Identification of the first glyphosate-resistant wild radish (Raphanus raphanistrum L.) populations. Pest Manag. Sci. 2014, 70, 1432–1436. [Google Scholar] [CrossRef] [PubMed]
- Shafi, U.; Mumtaz, R.; García-Nieto, J.; Hassan, S.A.; Zaidi, S.A.R.; Iqbal, N. Precision Agriculture Techniques and Practices: From Considerations to Applications. Sensors 2019, 19, 3796. [Google Scholar] [CrossRef]
- Gebbers, R.; Adamchuk, V.I. Precision Agriculture and Food Security. Science 2010, 327, 828–831. [Google Scholar] [CrossRef] [PubMed]
- Villette, S.; Maillot, T.; Guillemin, J.-P.; Douzals, J.-P. Assessment of nozzle control strategies in weed spot spraying to reduce herbicide use and avoid under-or over-application. Biosyst. Eng. 2022, 219, 68–84. [Google Scholar] [CrossRef]
- Vieira, B.C.; Luck, J.D.; Amundsen, K.L.; Werle, R.; Gaines, T.A.; Kruger, G.R. Herbicide drift exposure leads to reduced herbicide sensitivity in Amaranthus spp. Nature 2020, 10, 2146. [Google Scholar] [CrossRef]
- Egan, J.F.; Bohnenblust, E.; Goslee, S.; Mortensen, D.; Tooker, J. Herbicide drift can affect plant and arthropod communities. Agric. Ecosyst. Environ. 2014, 185, 77–87. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, M.; Wang, N. Precision agriculture—A worldwide overview. Comput. Electron. Agric. 2002, 36, 113–132. [Google Scholar] [CrossRef]
- Hunter, J.E.; Gannon, T.W.; Richardson, R.J.; Yelverton, F.H.; Leon, R.G. Integration of remote-weed mapping and an autonomous spraying unmanned aerial vehicle for site-specific weed management. Pest Manag. Sci. 2020, 76, 1386–1392. [Google Scholar] [CrossRef] [PubMed]
- Andrade, R.; Ramires, T.G. Precision Agriculture: Herbicide Reduction with AI Models. In Proceedings of the 4th International Conference on Statistics: Theory and Applications (ICSTA’22), Prague, Czech Republic, 28–30 July 2022. [Google Scholar]
- Danilevicz, M.F.; Roberto Lujan, R.; Batley, J.; Bayer, P.E.; Bennamoun, M.; Edwards, D.; Ashworth, M.B. Segmentation of Sandplain Lupin Weeds from Morphologically Similar Narrow-Leafed Lupins in the Field. Remote Sens. 2023, 15, 1817. [Google Scholar] [CrossRef]
- Peña, J.M.; Torres-Sánchez, J.; de Castro, A.I.; Kelly, M.; López-Granados, F. Weed mapping in early-season maize fields using object-based analysis of unmanned aerial vehicle (UAV) images. PLoS ONE 2013, 8, e77151. [Google Scholar] [CrossRef] [PubMed]
- Sa, I.; Popović, M.; Khanna, R.; Chen, Z.; Lottes, P.; Liebisch, F.; Nieto, J.; Stachniss, C.; Walter, A.; Siegwart, R. WeedMap: A Large-Scale Semantic Weed Mapping Framework Using Aerial Multispectral Imaging and Deep Neural Network for Precision Farming. Remote Sens. 2018, 10, 1423. [Google Scholar] [CrossRef]
- Huang, H.; Lan, Y.; Deng, J.; Yang, A.; Deng, X.; Zhang, L.; Wen, S. A semantic labeling approach for accurate weed mapping of high resolution UAV imagery. Sensors 2018, 18, 2113. [Google Scholar] [CrossRef] [PubMed]
- Gerhards, R.; Christensen, S. Real-time weed detection, decision making and patch spraying in maize, sugarbeet, winter wheat and winter barley. Weed Res. 2003, 43, 385–392. [Google Scholar] [CrossRef]
- Khan, S.; Tufail, M.; Khan, M.T.; Khan, Z.A.; Anwar, S. Deep learning-based identification system of weeds and crops in strawberry and pea fields for a precision agriculture sprayer. Precis. Agric. 2021, 22, 1711–1727. [Google Scholar] [CrossRef]
- Murad, N.Y.; Mahmood, T.; Forkan, A.R.M.; Morshed, A.; Jayaraman, P.P.; Siddiqui, M.S. Weed detection using deep learning: A systematic literature review. Sensors 2023, 23, 3670. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Zhang, W.; Wei, X. A review on weed detection using ground-based machine vision and image processing techniques. Comput. Electron. Agric. 2019, 158, 226–240. [Google Scholar] [CrossRef]
- Lottes, P.; Khanna, R.; Pfeifer, J.; Siegwart, R.; Stachniss, C. UAV-based crop and weed classification for smart farming. In Proceedings of the 2017 IEEE International Conference on Robotics and Automation (ICRA), Singapore, 29 May–3 June 2017; pp. 3024–3031. [Google Scholar]
- López-Granados, F. Weed detection for site-specific weed management: Mapping and real-time approaches. Weed Res. 2011, 51, 1–11. [Google Scholar] [CrossRef]
- Maes, W.H.; Steppe, K. Perspectives for remote sensing with unmanned aerial vehicles in precision agriculture. Trends Plant Sci. 2019, 24, 152–164. [Google Scholar] [CrossRef]
- Asad, M.H.; Bais, A. Weed detection in canola fields using maximum likelihood classification and deep convolutional neural network. Inf. Process. Agric. 2020, 7, 535–545. [Google Scholar] [CrossRef]
- Che’Ya, N.N.; Dunwoody, E.; Gupta, M. Assessment of weed classification using hyperspectral reflectance and optimal multispectral UAV imagery. Agronomy 2021, 11, 1435. [Google Scholar] [CrossRef]
- Raeva, P.L.; Šedina, J.; Dlesk, A. Monitoring of crop fields using multispectral and thermal imagery from UAV. Eur. J. Remote Sens. 2019, 52, 192–201. [Google Scholar] [CrossRef]
- Qiao, M.; He, X.; Cheng, X.; Li, P.; Luo, H.; Zhang, L.; Tian, Z. Crop yield prediction from multi-spectral, multi-temporal remotely sensed imagery using recurrent 3D convolutional neural networks. Int. J. Appl. Earth Obs. Geoinf. 2021, 102, 102436. [Google Scholar] [CrossRef]
- Lottes, P.; Behley, J.; Milioto, A.; Stachniss, C. Fully Convolutional Networks with Sequential Information for Robust Crop and Weed Detection in Precision Farming. IEEE Robot. Autom. Lett. 2018, 3, 2870–2877. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, B.; Singh, V.; Neely, C.; Rajan, N.; Bagavathiannan, M. Detection of Italian Ryegrass in Wheat and Prediction of Competitive Interactions Using Remote-Sensing and Machine-Learning Techniques. Remote Sens. 2020, 12, 2977. [Google Scholar] [CrossRef]
- Dos Santos Ferreira, A.; Matte Freitas, D.; Gonçalves da Silva, G.; Pistori, H.; Theophilo Folhes, M. Weed detection in soybean crops using ConvNets. Comput. Electron. Agric. 2017, 143, 314–324. [Google Scholar] [CrossRef]
- Farooq, A.; Hu, J.; Jia, X. Analysis of Spectral Bands and Spatial Resolutions for Weed Classification Via Deep Convolutional Neural Network. IEEE Geosci. Remote Sens. Lett. 2019, 16, 183–187. [Google Scholar] [CrossRef]
- Sunil, G.C.; Zhang, Y.; Koparan, C.; Ahmed, M.R.; Howatt, K.; Sun, X. Weed and crop species classification using computer vision and deep learning technologies in greenhouse conditions. J. Agric. Food Res. 2022, 9, 100325. [Google Scholar] [CrossRef]
- Vi Nguyen Thanh, L.; Ahderom, S.; Alameh, K. Performances of the LBP Based Algorithm over CNN Models for Detecting Crops and Weeds with Similar Morphologies. Sensors 2020, 20, 2193. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Chen, Y.; Zhao, B.; Kang, X.; Ding, Y. Review of weed detection methods based on computer vision. Sensors 2021, 21, 3647. [Google Scholar] [CrossRef]
- Alam, M.; Alam, M.S.; Roman, M.; Tufail, M.; Khan, M.U.; Khan, M.T. Real-time machine-learning based crop/weed detection and classification for variable-rate spraying in precision agriculture. In Proceedings of the 7th International Conference on Electrical and Electronics Engineering (ICEEE), Antalya, Turkey, 14–16 April 2020; pp. 273–280. [Google Scholar]
- Das, M.; Bais, A. DeepVeg: Deep learning model for segmentation of weed, canola, and canola flea beetle damage. IEEE Access 2021, 9, 119367–119380. [Google Scholar] [CrossRef]
- Ullah, H.S.; Asad, M.H.; Bais, A. End to end segmentation of canola field images using dilated U-Net. IEEE Access 2021, 9, 59741–59753. [Google Scholar] [CrossRef]
- Ganaie, M.A.; Hu, M.; Malik, A.K.; Tanveer, M.; Suganthan, P.N. Ensemble deep learning: A review. Eng. Appl. Artif. Intell. 2022, 115, 105151. [Google Scholar] [CrossRef]
- Rozendo, G.B.; Roberto, G.F.; do Nascimento, M.Z.; Alves Neves, L.; Lumini, A. Weeds Classification with Deep Learning: An Investigation Using CNN, Vision Transformers, Pyramid Vision Transformers, and Ensemble Strategy. In Proceedings of the Iberoamerican Congress on Pattern Recognition, Coimbra, Portugal, 27–30 November 2023; pp. 229–243. [Google Scholar]
- Gao, J.; French, A.P.; Pound, M.P.; He, Y.; Pridmore, T.P.; Pieters, J.G. Deep convolutional neural networks for image-based Convolvulus sepium detection in sugar beet fields. Plant Methods 2020, 16, 29. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Grains Research and Development Corporation. GRDC Grownotes—Section 4, Plant Growth and Physiology; GRDC: Barton, Australia, 2015. [Google Scholar]
- Harris, C.R.; Millman, K.J.; Van Der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- Kataoka, T.; Kaneko, T.; Okamoto, H.; Hata, S. Crop growth estimation system using machine vision. In Proceedings of the 2003 IEEE/ASME International Conference on Advanced Intelligent Mechatronics (AIM 2003), Chicago, IL, USA, 20–24 July 2003; Volume 1072, pp. b1079–b1083. [Google Scholar]
- Ostu, N. A threshold selection method from gray-level histograms. IEEE Trans SMC 1979, 9, 62. [Google Scholar]
- Bradski, G.; Kaehler, A. Learning OpenCV: Computer Vision with the OpenCV Library; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2008. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Howard, J.; Gugger, S. Fastai: A layered API for deep learning. Information 2020, 11, 108. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L. Pytorch: An imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 2019, 32, 8024–8035. [Google Scholar]
- Chechliński, Ł.; Siemiątkowska, B.; Majewski, M. A system for weeds and crops identification—Reaching over 10 fps on raspberry pi with the usage of mobilenets, densenet and custom modifications. Sensors 2019, 19, 3787. [Google Scholar] [CrossRef] [PubMed]
- Kaiming, H.; Xiangyu, Z.; Shaoqing, R.; Jian, S. Deep Residual Learning for Image Recognition. In Proceedings of the CVPR 2016, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Ikechukwu, A.V.; Murali, S.; Deepu, R.; Shivamurthy, R. ResNet-50 vs VGG-19 vs training from scratch: A comparative analysis of the segmentation and classification of Pneumonia from chest X-ray images. Glob. Transit. Proc. 2021, 2, 375–381. [Google Scholar] [CrossRef]
- Nguyen, T.-H.; Nguyen, T.-N.; Ngo, B.-V. A VGG-19 model with transfer learning and image segmentation for classification of tomato leaf disease. AgriEngineering 2022, 4, 871–887. [Google Scholar] [CrossRef]
- Zhang, R.; Du, L.; Xiao, Q.; Liu, J. Comparison of backbones for semantic segmentation network. J. Phys. Conf. Ser. 2020, 1544, 012196. [Google Scholar] [CrossRef]
- Tao, T.; Wei, X. A hybrid CNN–SVM classifier for weed recognition in winter rape field. Plant Methods 2022, 18, 29. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Jadon, S. A survey of loss functions for semantic segmentation. In Proceedings of the 2020 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Vina del Mar, Chile, 27–29 October 2020; pp. 1–7. [Google Scholar]
- Zhang, Z. Improved adam optimizer for deep neural networks. In Proceedings of the 2018 IEEE/ACM 26th International Symposium on Quality of Service (IWQoS), Banff, AB, Canada, 4–6 June 2018; pp. 1–2. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 5998–6008. [Google Scholar]
- Sudre, C.H.; Li, W.; Vercauteren, T.; Ourselin, S.; Jorge Cardoso, M. Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 3rd International Workshop, DLMIA 2017, and 7th International Workshop, ML-CDS 2017, Held in Conjunction with MICCAI 2017, Québec City, QC, Canada, 14 September 2017; Proceedings 3. pp. 240–248. [Google Scholar]
- Zhang, H.; Goodfellow, I.; Metaxas, D.; Odena, A. Self-attention generative adversarial networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 7354–7363. [Google Scholar]
- Prakash, N.; Manconi, A.; Loew, S. Mapping Landslides on EO Data: Performance of Deep Learning Models vs. Traditional Machine Learning Models. Remote Sens. 2020, 12, 346. [Google Scholar] [CrossRef]
- Opitz, J.; Burst, S. Macro F1 and Macro F1. arXiv 2021, arXiv:1911.03347. [Google Scholar] [CrossRef]
- FastAi. Docs.fast.ai. Available online: https://docs.fast.ai/ (accessed on 20 April 2023).
- Waskom, M.L. Seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Ali, A.; Touvron, H.; Caron, M.; Bojanowski, P.; Douze, M.; Joulin, A.; Laptev, I.; Neverova, N.; Synnaeve, G.; Verbeek, J. Xcit: Cross-covariance image transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 20014–20027. [Google Scholar]
- El-Nouby, A.; Touvron, H.; Caron, M.; Bojanowski, P.; Douze, M.; Joulin, A.; Laptev, I.; Neverova, N.; Synnaeve, G.; Verbeek, J.; et al. XCiT: Cross-Covariance Image Transformers. arXiv 2021, arXiv:2106.09681. [Google Scholar] [CrossRef]
- Hasan, A.S.M.M.; Sohel, F.; Diepeveen, D.; Laga, H.; Jones, M.G.K. A survey of deep learning techniques for weed detection from images. Comput. Electron. Agric. 2021, 184, 106067. [Google Scholar] [CrossRef]
- Mascarenhas, S.; Agarwal, M. A comparison between VGG16, VGG19 and ResNet50 architecture frameworks for Image Classification. In Proceedings of the 2021 International Conference on Disruptive Technologies for Multi-Disciplinary Research and Applications (CENTCON), Bengaluru, India, 19–21 November 2021; pp. 96–99. [Google Scholar]
- Shah, S.R.; Qadri, S.; Bibi, H.; Shah, S.M.W.; Sharif, M.I.; Marinello, F. Comparing Inception V3, VGG 16, VGG 19, CNN, and ResNet 50: A Case Study on Early Detection of a Rice Disease. Agronomy 2023, 13, 1633. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Bah, M.D.; Hafiane, A.; Canals, R. Deep learning with unsupervised data labeling for weed detection in line crops in UAV images. Remote Sens. 2018, 10, 1690. [Google Scholar] [CrossRef]
- Suh, H.K.; Ijsselmuiden, J.; Hofstee, J.W.; van Henten, E.J. Transfer learning for the classification of sugar beet and volunteer potato under field conditions. Biosyst. Eng. 2018, 174, 50–65. [Google Scholar] [CrossRef]
- Grant, L. Diffuse and specular characteristics of leaf reflectance. Remote Sens. Environ. 1987, 22, 309–322. [Google Scholar] [CrossRef]
- Vrindts, E.; De Baerdemaeker, J.; Ramon, H. Weed detection using canopy reflection. Precis. Agric. 2002, 3, 63–80. [Google Scholar] [CrossRef]
- Li, N.; Zhang, X.; Zhang, C.; Ge, L.; He, Y.; Wu, X. Review of machine-vision-based plant detection technologies for robotic weeding. In Proceedings of the 2019 IEEE International Conference on Robotics and Biomimetics (ROBIO), Dali, China, 6–8 December 2019; pp. 2370–2377. [Google Scholar]
- Sanders, J.T.; Jones, E.A.; Minter, A.; Austin, R.; Roberson, G.T.; Richardson, R.J.; Everman, W.J. Remote sensing for Italian ryegrass [Lolium perenne L. ssp. multiflorum (Lam.) Husnot] detection in winter wheat (Triticum aestivum L.). Front. Agron. 2021, 3, 687112. [Google Scholar] [CrossRef]
- Milioto, A.; Lottes, P.; Stachniss, C. Real-time blob-wise sugar beets vs weeds classification for monitoring fields using convolutional neural networks. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2017, 4, 41–48. [Google Scholar] [CrossRef]
- Bai, X.; Cao, Z.; Wang, Y.; Yu, Z.; Hu, Z.; Zhang, X.; Li, C. Vegetation segmentation robust to illumination variations based on clustering and morphology modelling. Biosyst. Eng. 2014, 125, 80–97. [Google Scholar] [CrossRef]
- Sharma, A.; Jain, A.; Gupta, P.; Chowdary, V. Machine learning applications for precision agriculture: A comprehensive review. IEEE Access 2020, 9, 4843–4873. [Google Scholar] [CrossRef]
- Faccini, D.; Puricelli, E. Efficacy of herbicide dose and plant growth stage on weeds present in fallow ground. Agriscientia 2007, 24, 29–35. [Google Scholar]
- Steckel, G.J.; Wax, L.M.; Simmons, F.W.; Phillips, W.H. Glufosinate efficacy on annual weeds is influenced by rate and growth stage. Weed Technol. 1997, 11, 484–488. [Google Scholar] [CrossRef]
- Su, D.; Qiao, Y.; Kong, H.; Sukkarieh, S. Real time detection of inter-row ryegrass in wheat farms using deep learning. Biosyst. Eng. 2021, 204, 198–211. [Google Scholar] [CrossRef]
- Buhrmester, V.; Münch, D.; Arens, M. Analysis of explainers of black box deep neural networks for computer vision: A survey. Mach. Learn. Knowl. Extr. 2021, 3, 966–989. [Google Scholar] [CrossRef]
- Zhao, H.; Jia, J.; Koltun, V. Exploring self-attention for image recognition. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 10076–10085. [Google Scholar]
- Khan, S.; Naseer, M.; Hayat, M.; Zamir, S.W.; Khan, F.S.; Shah, M. Transformers in vision: A survey. ACM Comput. Surv. 2022, 54, 1–41. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- De Camargo, T.; Schirrmann, M.; Landwehr, N.; Dammer, K.-H.; Pflanz, M. Optimized Deep Learning Model as a Basis for Fast UAV Mapping of Weed Species in Winter Wheat Crops. Remote Sens. 2021, 13, 1704. [Google Scholar] [CrossRef]
- Partel, V.; Kim, J.; Costa, L.; Pardalos, P.M.; Ampatzidis, Y. Smart Sprayer for Precision Weed Control Using Artificial Intelligence: Comparison of Deep Learning Frameworks. In Proceedings of the ISAIM, Fort Lauderdale, FL, USA, 6–8 January 2020. [Google Scholar]
- Quan, L.; Jiang, W.; Li, H.; Li, H.; Wang, Q.; Chen, L. Intelligent intra-row robotic weeding system combining deep learning technology with a targeted weeding mode. Biosyst. Eng. 2022, 216, 13–31. [Google Scholar] [CrossRef]
- Milioto, A.; Lottes, P.; Stachniss, C. Real-Time Semantic Segmentation of Crop and Weed for Precision Agriculture Robots Leveraging Background Knowledge in CNNs. In Proceedings of the 2018 IEEE International Conference on Robotics and Automation (ICRA), Brisbane, Australia, 21–25 May 2018; pp. 2229–2235. [Google Scholar]
- Beckie, H.J.; Flower, K.C.; Ashworth, M.B. Farming without glyphosate? Plants 2020, 9, 96. [Google Scholar] [CrossRef]
- Beckie, H.J.; Ashworth, M.B.; Flower, K.C. Herbicide resistance management: Recent developments and trends. Plants 2019, 8, 161. [Google Scholar] [CrossRef]
- Dayan, F.E. Current status and future prospects in herbicide discovery. Plants 2019, 8, 341. [Google Scholar] [CrossRef] [PubMed]
- Khaliq, A.; Matloob, A.; Hafiz, M.S.; Cheema, Z.A.; Wahid, A. Evaluating sequential application of pre and post emergence herbicides in dry seeded fine rice. Pak. J. Weed Sci. Res. 2011, 17, 111–123. [Google Scholar]
- Yadav, D.B.; Yadav, A.; Punia, S.S.; Chauhan, B.S. Management of herbicide-resistant Phalaris minor in wheat by sequential or tank-mix applications of pre-and post-emergence herbicides in north-western Indo-Gangetic Plains. Crop Prot. 2016, 89, 239–247. [Google Scholar] [CrossRef]
- Jin, X.; Liu, T.; McCullough, P.E.; Chen, Y.; Yu, J. Evaluation of convolutional neural networks for herbicide susceptibility-based weed detection in turf. Front. Plant Sci. 2023, 14, 1096802. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.; Konovalov, D.A.; Philippa, B.; Ridd, P.; Wood, J.C.; Johns, J.; Banks, W.; Girgenti, B.; Kenny, O.; Whinney, J.; et al. DeepWeeds: A Multiclass Weed Species Image Dataset for Deep Learning. Nature 2019, 9, 2058. [Google Scholar] [CrossRef]
- Délye, C.; Jasieniuk, M.; Le Corre, V. Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 2013, 29, 649–658. [Google Scholar] [CrossRef]
- Heap, I.; Spafford, J.; Dodd, J.; Moore, J. Herbicide resistance—Australia vs. the rest of the world. World 2013, 200, 250. [Google Scholar]
- Hashem, A.; Wilkins, N. Competitiveness and persistence of wild radish (Raphanus raphanistrum L.) in a wheat-lupin rotation. In Proceedings of the 13th Australian Weeds Conference, Perth, Western Australia, 8–13 September 2002; pp. 712–715. [Google Scholar]
- Shaikh, T.A.; Rasool, T.; Lone, F.R. Towards leveraging the role of machine learning and artificial intelligence in precision agriculture and smart farming. Comput. Electron. Agric. 2022, 198, 107119. [Google Scholar] [CrossRef]
- Srinivasan, A. Handbook of Precision Agriculture: Principles and Applications; Food Products Press, Haworth Press Inc.: New York, NY, USA, 2006. [Google Scholar]

| Dataset | Resnet-18 | Resnet-34 | VGG-16 | Models’ Average | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prec | Rec | IoU | F1 | Prec | Rec | IoU | F1 | Prec | Rec | IoU | F1 | Prec | Rec | IoU | F1 | |
| York Canola | 0.83 | 0.89 | 0.77 | 0.85 | 0.84 | 0.89 | 0.78 | 0.85 | 0.81 | 0.83 | 0.73 | 0.82 | 0.83 | 0.87 | 0.76 | 0.84 |
| T1 Miling ARG | 0.84 | 0.85 | 0.75 | 0.84 | 0.84 | 0.86 | 0.76 | 0.85 | 0.80 | 0.81 | 0.70 | 0.79 | 0.83 | 0.84 | 0.74 | 0.83 |
| T2 Miling ARG | 0.84 | 0.86 | 0.75 | 0.84 | 0.84 | 0.87 | 0.76 | 0.85 | 0.80 | 0.81 | 0.70 | 0.80 | 0.83 | 0.85 | 0.74 | 0.83 |
| Metrics Average | 0.84 | 0.87 | 0.76 | 0.84 | 0.84 | 0.87 | 0.77 | 0.85 | 0.80 | 0.82 | 0.71 | 0.80 | 0.83 | 0.85 | 0.75 | 0.83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mckay, M.; Danilevicz, M.F.; Ashworth, M.B.; Rocha, R.L.; Upadhyaya, S.R.; Bennamoun, M.; Edwards, D. Focus on the Crop Not the Weed: Canola Identification for Precision Weed Management Using Deep Learning. Remote Sens. 2024, 16, 2041. https://doi.org/10.3390/rs16112041
Mckay M, Danilevicz MF, Ashworth MB, Rocha RL, Upadhyaya SR, Bennamoun M, Edwards D. Focus on the Crop Not the Weed: Canola Identification for Precision Weed Management Using Deep Learning. Remote Sensing. 2024; 16(11):2041. https://doi.org/10.3390/rs16112041
Chicago/Turabian StyleMckay, Michael, Monica F. Danilevicz, Michael B. Ashworth, Roberto Lujan Rocha, Shriprabha R. Upadhyaya, Mohammed Bennamoun, and David Edwards. 2024. "Focus on the Crop Not the Weed: Canola Identification for Precision Weed Management Using Deep Learning" Remote Sensing 16, no. 11: 2041. https://doi.org/10.3390/rs16112041
APA StyleMckay, M., Danilevicz, M. F., Ashworth, M. B., Rocha, R. L., Upadhyaya, S. R., Bennamoun, M., & Edwards, D. (2024). Focus on the Crop Not the Weed: Canola Identification for Precision Weed Management Using Deep Learning. Remote Sensing, 16(11), 2041. https://doi.org/10.3390/rs16112041








