1. Introduction
Hyperspectral images (HSIs) consist of hundreds of contiguous narrow spectral bands extending across the electromagnetic spectrum, from visible to near-infrared wavelengths [
1], resulting in abundant SAS information. Effectively classifying SAS features is critical in HSI processing, which aims at categorizing the content of each pixel using a set of pre-defined classes. In recent years, HSIC has seen widespread adoption across various domains, including urban planning [
2], military reconnaissance [
3], agriculture monitoring [
4], and ocean monitoring [
5].
The advancement of deep learning (DL) in artificial intelligence has considerably improved the processing of remote sensing images. When compared with traditional machine learning techniques including support vector machines (SVMs) [
6], morphological profiles [
7],
k-nearest neighbor [
8], or random forests [
9], DL-based approaches exhibit a powerful feature extraction capability, thus being able to learn discriminative and high-level semantic information. Therefore, DL-based techniques are extensively employed for HSIC [
10]. For instance, a deep stacked autoencoder network has been suggested for the classification of HSIs by focusing on learning spectral features [
11]. Chen et al. [
12] employed a multi-layer deep neural network and a singular restricted Boltzmann machine for the purpose of capturing the spectral characteristics within HSI data. However, these approaches solely utilize spectral data information and overlook the importance of spatial-contextual information for enhancing classification performance. Hence, joint SAS feature extraction methods have been proposed to extract additional contextual semantic information from complex spatial structures, thus enhancing the model’s classification performance. Yang et al. [
13] presented a two-branch SAS characteristic extraction network that employed a 1D-CNN for spectral characteristic extraction and a 2D-CNN for spatial characteristic extraction. The learned SAS information is linked and channeled into a fully connected (FC) layer, which extracts spectral–spatial characteristics to facilitate further classification. Yet, the 2D-CNN architecture could potentially result in the loss of spectral information within HSI. To proficiently capture SAS features, a 3D-CNN coupled with a regularization model has been proposed [
14]. Roy et al. [
15] combined a 2D-CNN and 3D-CNN to acquire spectral–spatial characteristics jointly represented from spectral bands using 3D-CNN, and then further learned spatial feature representations using 2D-CNN. Guo et al. [
16] proposed a dual-view spectral and global spatial feature fusion network that utilized an encoder–decoder structure with channel and spatial attention to fully mine the global spatial characteristics, while utilizing a dual-view spectral feature aggregation model with a view attention for learning the diversity of the spectral characteristics and achieving a relatively good classification performance.
Despite the above CNN-based approaches achieving relatively good categorization results in the classification tasks, they did not exploit hierarchical SAS feature information across various layers. Furthermore, the excessive depth of convolutional layers may cause gradient vanishing and explosion problems. The dense connected convolutional network (DenseNet) offers an effective solution to mitigate these issues; it achieves this by promoting the maximal flow of information among different convolutional layers through connectivity operations, effectively fusing the hierarchical features between different layers [
17]. Based on this, a comprehensive deep multi-layer fusion DenseNet using 2D and 3D dense blocks was presented in [
18], which effectively improved the exploitation of HSI hierarchical signatures and handled the gradient vanishing problem. In [
19], a fast dense spectral–spatial convolution network (FDSSC) was introduced, which combines two separate dense blocks and increases the network’s depth, allowing for a more straightforward utilization of feature information across different layers. By combining the advantages of CNN and graph convolutional network (GCN), Zhou et al. [
20] proposed an attention fusion network based on multiscale convolution and multihop graph convolution to extract multi-level complex SAS features of HSI. Liang et al. [
21] presented a framework that integrates a multiscale DenseNet with bidirectional recurrent neural networks, which adopted the multiscale DenseNet (instead of traditional CNNs) to strengthen the utilization of spatial characteristics across different convolutional layers. Despite the powerful ability of the above DenseNet-based approaches to retrieve SAS characteristics in HSI classification tasks, they still suffer from the limitation that CNNs typically only consider local SAS information between features, while ignoring global SAS information (failing to establish global dependencies across long-range distances among HSI pixels).
Recently, vision transformers have witnessed a surge in popularity within numerous facets of computer vision, including target recognition, image classification, and instance segmentation [
22,
23]. Transformers are primarily composed of numerous self-attention and feed-forward layers that inherit the global receptive field, which allows them to efficiently establish long-range dependencies among HSI pixels, compensating for the lack of CNNs in global feature extraction. Hence, vision transformers have attracted widespread attention in HSIC, in which the MHSA serves as the primary characteristic extractor of the transformer for learning the remote locations of HSI pixels and global dependencies between spectral bands [
24,
25]. Furthermore, the transformer emphasizes prominent features while concealing less significant information. He et al. [
26] were pioneers in developing a bi-directional encoder representation of the transformer-based model for establishing global dependencies in HSIC. This approach primarily relies on the MHSA mechanism of the MHSA layer, where each head encodes a global contextual semantic-aware representation of the HSI for discriminative SAS characteristics. Hong et al. [
27] proposed a framework for learning the long-range dependence information between spectral signatures using group spectral embedding and transform encoders by treating HSI data as sequential information, while fusing “soft” residuals across layers to mitigate the loss of critical signature information in the process of hierarchical propagation. Xue et al. [
28] introduced a local transformer model in combination with the spatial partition restore network, which can effectively acquire the HSI global contextual dependencies and dynamically acquire the spatial attention weights through the local transformer to adapt to the intrinsic changes in HSI spatial pixels, thus augmenting the model’s ability to retrieve spatial–contextual pixel characteristics. Mei et al. [
29] introduced a group-aware hierarchical transformer (GAHT) for HSIC, which incorporates a new group pixel embedding module that highlights local relationships in each HSI spectral channel, thus modeling global–local dependencies from a spectral–spatial point of view.
Although the above transformer-based models exhibit excellent abilities to model long-range dependencies among HSI pixels, they still suffer from some limitations in terms of extracting HSI characteristic information: (1) MHSA falls short in effectively considering both the positional and spectral information of the input HSI blocks when establishing the global dependencies of the HSI, which renders that the network lacks the utilization of the positional information among HSI pixels, and (2) some discriminative local SAS characteristic information that is helpful for HSIC purposes is not sufficiently exploited. Given that CNNs exhibit strong local characteristic learning abilities, a convolutional transformer (CT) network was proposed in [
30], first employing central position coding to merge the spectral signatures and pixel positions to obtain the spatial positional signatures of the HSI patches, and then utilizing the CT block (containing two 2D-CNNs with 3 × 3 convolutional kernel sizes) to acquire the local–global characteristic information of HSIs, which significantly improved this model’s local–global feature acquisition ability. The spectral–spatial feature tokenization transformer (SSFTT) was introduced in [
31], which converts the SAS characteristics learned by a simple 3D-CNN and 2D-CNN layer into semantic tokens, and inputs them into a transformer encoder to perform spectral–spatial characteristic representation. Although the above methodologies try to employ CNNs to strengthen the local characteristic extraction capabilities of the network, the simple CNN structure fails to adequately extract hierarchical features in various network layers. In this regard, Yan et al. [
32] proposed a hybrid convolutional and ViT network classification approach, where one branch uses hybrid convolution and ViT to boost the capability of acquiring local–global spatial characteristics, and the other branch utilizes 3D-CNNs to retrieve spectral characteristics. However, separate extraction of SAS characteristics with a branch based on 2D-CNNs and a hybrid convolutional transformer network based on 3D-CNNs may ignore the intrinsic correlation between SAS signatures. A local semantic feature aggregation-based transformer approach was proposed in [
33], which utilizes 3D-CNNs to simultaneously extract shallow spectral–spatial characteristics, and then merges pixel-labeled features using a local pixel aggregation operation to provide multi-scale characteristic neighborhood representations for HSIC. A two-branch bottleneck spectral–spatial transformer (BS2T) method was introduced in [
34], which utilizes two 3D-CNNs DenseNet structures to separately abstract SAS properties to boost the extraction of the localized characteristics, as well as two transformers for establishing the long-range dependencies between HSI pixels. However, it may result in the model failing to adequately leverage the correlation between SAS information (this architecture contains two 3D-CNNs hierarchical structures and two transformers, and is relatively complex). Zu et al. [
35] proposed exploiting a cascaded convolutional feature token to obtain joint spectral–spatial information and incorporate certain inductive bias properties of CNNs into the transformer. The densely connected transformer is then utilized to improve the characteristic propagation, significantly boosting the model’s performance.
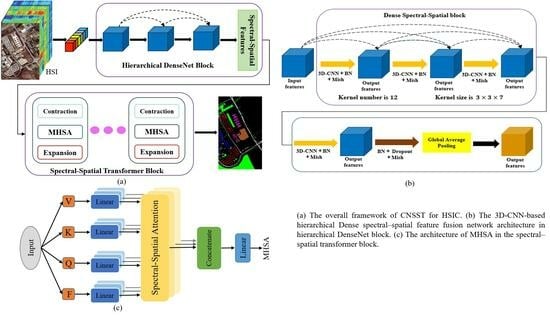
Inspired by the above, we propose a simply structured, end-to-end convolutional network and spectral–spatial transformer (CNSST) architecture for HSIC. It comprises two primary modules, a 3D-CNN-based hierarchical feature fusion network and a spectral–spatial transformer that introduces inductive bias properties information (i.e., localization, contextual position, and translation equivariance), which are used to boost the extraction of local feature information and establish global dependencies, respectively. Regarding the local spectral–spatial feature extraction, to acquire SAS hierarchical characteristic representations with more rich inductive bias information, a 3D-CNN-based hierarchical network strategy is utilized to capture SAS information simultaneously, so as to establish the correlation between the SAS information of HSI pixels and to obtain a more rich inductive bias (yet more discriminative spectral–spatial joint feature information). Meanwhile, the hierarchical feature fusion structure is utilized to boost the utilization of the HSI semantic feature information across different convolutional layers. In the spectral–spatial transformer network, the SAS hierarchical characteristics containing rich inductive bias information are introduced into the MHSA to make up for the shortcoming of insufficient inductive bias in the image features acquired by the transformer. This allows the transformer not only to effectively establish long-range dependencies among HSI pixels, but also to enhance the model’s location-aware and spectral-aware capabilities. Moreover, a Lion optimizer is exploited to enhance the performance of the model. A summary of the primary contributions of this research is as follows:
We propose a simply structured, end-to-end convolutional network and spectral–spatial transformer (CNSST) architecture based on a 3D-CNN hierarchical feature fusion network and a spectral–spatial transformer that introduces rich inductive bias information in the HSI classification process.
To obtain feature representations with richer inductive bias information, a 3D-CNN-based hierarchical network is utilized to capture SAS information simultaneously in order to establish the correlation between these two sources of information in HSI pixels, while the hierarchical structure is exploited to improve the utilization of the HSI semantic feature information in various convolutional layers.
Spectral–spatial hierarchical features containing rich inductive bias information are introduced into MHSA, which enables the transformer to effectively establish long-range dependencies among HSI pixels, and to be more location-aware and spectral-aware. Moreover, a Lion optimizer is exploited to boost the categorization performance of the network.
The rest of this article is structured as follows. The related work is briefly described in
Section 2.
Section 3 provides an in-depth description of the general framework of the CNSST.
Section 4 shows the experimental analysis and discussion. Finally,
Section 5 wraps up the paper with concluding remarks and hints at future research directions.
4. Experiments and Analysis
To assess the efficacy of the proposed CNSST approach, intensive experiments are performed using three familiar HSIC datasets. Next, we describe the datasets utilized, experimental settings, and then compare and experimentally analyze them in conjunction with several state-of-the-art models to exemplify the validity of the CNSST.
4.1. Datasets Description
In the experimental evaluations, four HSIC datasets are adopted to assess the CNSST approach we introduced. These datasets include the University of Pavia (UP), Salinas Scene (SV), Indian Pines (IP), and ZaoYan region (ZY). The corresponding pseudo-color and ground-truth images for these three datasets are depicted in
Figure 8. Details about the categories and samples of the counterpart datasets are provided in
Table 1,
Table 2,
Table 3 and
Table 4. The details are shown below:
UP: It was acquired utilizing the ROSIS-3 sensor through an aerial survey performed over the Pavia region, Italy. It includes pixels, containing a combined count of 42,776 labeled samples distributed among 9 distinct classes. Notably, this dataset encompasses 103 spectral bands, spanning a wavelength range from 4.3 to 8.6 .
SV: It was gathered utilizing the AVIRIS sensor—equipped with 224 spectrum bands—over Salinas Valley, USA. The dimensions of the images within this dataset are pixels. It contains 54,129 sample pixels labeled samples distributed among 16 distinct classes and encompasses 204 bands in the range of 0.4 to 2.5 wavelengths.
IP: It was gathered utilizing the AVIRIS sensor over the region of Indiana, USA. It includes 16 distinct classes in total, spanning a wavelength ranging from 0.4 to 2.5 . The scene’s dimensions encompass pixels, 220 spectral bands, and a combined count of 10,249 samples are available within this dataset.
ZY: It was collected by the OMIS sensor over the Zaoyuan region, China. The sense contained pixels and 80 spectral bands with the first 64 spectral bands in the range of 0.4 to 1.1 and the last 16 covering the region of 1.06 to 1.7 . The available ground-truth map contains only 23,821 labeled samples and 8 landcover classes.
Figure 8.
Pseudo-color and ground-truth images of three datasets. Pseudo-color images of the UP, SV, IP, and ZY datasets are depicted in (a,c,e,g), while the counterpart ground-truth maps are displayed in (b,d,f,h).
Figure 8.
Pseudo-color and ground-truth images of three datasets. Pseudo-color images of the UP, SV, IP, and ZY datasets are depicted in (a,c,e,g), while the counterpart ground-truth maps are displayed in (b,d,f,h).
Table 1.
Details of the categories and sample numbers for UP dataset.
Table 1.
Details of the categories and sample numbers for UP dataset.
| Category | Name | Total Number | Category | Name | Total Number |
|---|
| N1 | Asphalt | 6631 | N6 | Bare Soil | 5029 |
| N2 | Meadows | 18,649 | N7 | Bitumen | 1330 |
| N3 | Gravel | 2099 | N8 | Self-Blocking Bicks | 3682 |
| N4 | Trees | 3064 | N9 | Shadows Bare Soil | 947 |
| N5 | Painted metal sheets | 1345 | | | |
Table 2.
Details of the categories and sample numbers for SV dataset.
Table 2.
Details of the categories and sample numbers for SV dataset.
| Category | Name | Total Number | Category | Name | Total Number |
|---|
| N1 | Broccoli-green-weeds-1 | 2009 | N9 | Soil-vinyard-develop | 6203 |
| N2 | Broccoli-green-weeds-2 | 3726 | N10 | Corn-senesced-green-weeds | 3278 |
| N3 | Fallow | 1976 | N11 | Lettuce-romaine-4wk | 1068 |
| N4 | Fallow-rough-plow | 1394 | N12 | Lettuce-romaine-5wk | 1927 |
| N5 | Fallow-smooth | 2678 | N13 | Lettuce-romaine-6wk | 916 |
| N6 | Stubble | 3959 | N14 | Lettuce-romaine-7wk | 1070 |
| N7 | Celery | 3579 | N15 | Vinyard-untrained | 7268 |
| N8 | Grapes-untrained | 11,271 | N16 | Vinyard-vertical-trellis | 1807 |
Table 3.
Details of the categories and sample numbers for the IP dataset.
Table 3.
Details of the categories and sample numbers for the IP dataset.
| Category | Name | Total Number | Category | Name | Total Number |
|---|
| N1 | Alfalfa | 46 | N9 | Oats | 20 |
| N2 | Corn-notill | 142 | N10 | Soybean-notill | 972 |
| N3 | Corn-mintill | 830 | N11 | Soybean-mintill | 2455 |
| N4 | Corn | 237 | N12 | Soybean-clean | 593 |
| N5 | Grass-pasture | 483 | N13 | Wheat | 205 |
| N6 | Grass-trees | 730 | N14 | Woods | 1265 |
| N7 | Grass-pasture-mowed | 28 | N15 | Buildings-Grass-Trees-Drives | 386 |
| N8 | Hay-windrowed | 478 | N16 | Stone-Steel-Towers | 93 |
Table 4.
Details of the categories and sample numbers for ZY dataset.
Table 4.
Details of the categories and sample numbers for ZY dataset.
| Category | Name | Total Number | Category | Name | Total Number |
|---|
| N1 | Vegetable | 2625 | N5 | Corn | 1425 |
| N2 | Grape | 1302 | N6 | Terrace/Grass | 1484 |
| N3 | Dry vegetable | 3442 | N7 | Bush-Lespedeza | 1808 |
| N4 | Pear | 10,243 | N8 | Peach | 1492 |
4.2. Experimental Settings
To better compare the classification performance (experimental classification accuracy and classification visual maps) of different methods, during the selection of experimental training data, 1% of labeled samples are uniformly chosen for training from the UP and SV datasets, which contain a substantial number of labeled samples (42,776 and 54,129 labeled samples), while the remainder is for testing. However, for the IP and ZY datasets, which have relatively fewer labeled samples (10,249 and 23,821 labeled samples), 10% and 2.5% of the samples are respectively chosen for training, while the rest serve for testing. It’s worth noting that all experimental samples were chosen randomly. To evaluate the CNSST model’s performance, we assessed the outcomes using three well-established metrics: overall accuracy (OA), average accuracy (AA), and the Kappa coefficient (Ka). Every phase of model training and testing was performed on a computer system equipped with 64 GB RAM, RTX 3070Ti GPU, and Pytorch framework.
In addition, we performed a comparative analysis of the CNSST model, comparing it to several state-of-the-art classification approaches, including SVM [
6], SSRN [
45], CDCNN [
46], FDSSC [
19], DBMA [
47], SF [
27], SSFTT [
31], GAHT [
29], and BS2T [
34]. The CNSST framework takes the original 3D HSI as input, without any pre-processing for dimensionality reduction. For optimizing the performance of CNSST, the optimal experimental parameters are empirically adopted. The batch size, epoch and learning rate are correspondingly set as 64, 200, and 0.0001. The convolution kernel size is set at
, and there are a total of 5 convolution layers in the architecture (the hierarchical Dense spectral-spatial feature fusion block consists of 4 layers, with each layer having 12 convolutional kernel channels). After repeating the test twenty times for each experimental method, the final classification outcome is determined by taking the average of the results from each test.
The spatial patch size has a significant influence on HSIC. As the size of the spatial patch in the CNN increases, the model can cover more pixel information. It helps to enhance the HSIC accuracy because a larger patch can collect more HSI characteristics and contextual information. However, too large spatial patches may also suffer from the problem of introducing too much irrelevant pixel information, which may cause confusion and misclassification [
21]. Hence, the sizes of spatial were set to
,
,
,
,
, and
to explore the influence on the categorization performance. The OA outcomes of the CNSST approach on UP, SV, IP and ZY datasets at various spatial sizes are reported in
Figure 9. According to the classification accuracies under different spatial patch sizes in three datasets, the patch size of the proposed CNSST set as
.
4.3. Experiment Outcomes and Discussion Analysis
The results, categorized using various approaches for the UP dataset, are demonstrated in
Table 5, with the highest category-specific precision highlighted in bold. It is observed that CNSST has the highest categorization accuracy with 99.30%, 99.08%, and 99.07% for OA, AA and Ka, respectively. The OA categorization accuracy of SVM is 88.69%, which is 8.39%, 8.96%, 7.31%, 8.54%, 9.18%, 10.24%, and 10.61% lower than the DL-based SSRN, FDSSC, DBMA, SSFTT, GAHT, BS2T, and CNSST approaches, respectively. The reason is that the DL-based approaches (except for CDCNN and SF) can automatically extract the SAS characteristic information of HSI pixels and are superior in their characteristic extraction capability to the traditional SVM approach based on manual feature extraction. However, the classification accuracies of the DL-based methods, CDCNN and SF, are only 87.90% and 88.67% (similar to the classification accuracies of SVM and lower classification accuracies relative to other DL-based methods). The reason may be that there are limitations in the network structure design of CDCNN based on ResNet and multi-scale convolution, which results in CDCNN’s poor characteristic extraction capacity. The SF approach merely utilizes the group spectral embedding and transform encoder to acquire long-range dependency information, which fails to adequately use the local spectral–spatial feature information of HSI. In contrast, the classification accuracies of SSFTT, BS2T, and CNSST are 8.56%, 10.26%, and 10.63% higher, respectively, than that of SF, because they are not only able to utilize the transformer to efficiently establish long-range dependencies between HSI pixels, but also utilize CNN to efficiently augment the model’s ability to capture the local spectral–spatial characteristic information. Moreover, the accuracies of BS2T and CNSST are 98.93% and 99.30%, respectively, which are both higher than SSFTT. This is because SSFTT merely adopts one 3D-CNN and one 2D-CNN layer for extracting the local spectral–spatial signature information, which fails to extract the local signature information of HSI at a deeper level. However, BS2T and CNSST adopt the DenseNet-based structure, which can efficiently exploit the hierarchical local signature information from different convolutional layers, while also capturing the long-range dependency between HSI pixels with the transformer.
The classification maps for various approaches on UP are depicted in
Figure 10. FDSSC, BS2T, and CNSST have relatively fewer misclassified pixels and better intra-class homogeneity, generating relatively smoother classification visual maps. Meanwhile, the visual maps of the other methods have relatively more misclassified labels and poorer homogeneity. This may be because FDSSC using 3D-CNN dense SAS networks with various kernel sizes can adequately capture different hierarchical levels of detailed information on spectral–spatial characteristics. Meanwhile, BS2T and CNSST not only exploit the 3D-CNN DenseNet’s ability to efficiently extract local hierarchical features, but also the transformer’s ability to model the long-range global characteristics of HSI pixels, and thus their categorization performance is better than FDSSC. In addition, the categorization accuracy of the proposed CNSST is 0.37% higher than BS2T, and CNSST has significantly fewer misclassification labels than BS2T in the lower left corner of the classification map. This is because BS2T employs a two-branch DenseNet structure to acquire the SAS characteristics of HSI separately, which fails to efficiently build up the correlation between SAS characteristics, and may result in the loss of characteristic information. However, the proposed CNNST employs a single 3D-CNN-based hierarchical DenseNet structure to capture SAS information simultaneously, which not only establishes a correlation between the SAS information of the HSI pixels, and obtains richer inductive bias and more discriminative spectral–spatial joint feature information; this information (inductive bias and contextual positional information) is also input into the transformer, which enables the model to be more positional-aware and spectral-aware. In addition, CNNST also utilizes the new Lion optimizer to boost the categorization performance of the proposed CNSST.
From
Table 6, it can be seen that CNSST still achieves the optimal categorization accuracies of OA, AA and Ka, which are 99.35%, 99.52%, and 99.28%, respectively. Also, the classification accuracies of all the individual categories reached more than 99.04%, except for Vinyard-untrained (category N15) and Fallow-roughplow (category N4), which had classification accuracies of 97.78% and 98.13%, respectively. The classification accuracy of FDSSC based on 3D-CNN hierarchical DenseNet is 2.06% and 11.15% higher than SSRN and CDCNN based on the simple 3D-CNN structure, respectively. Similarly, the classification accuracies of CNSST and BS2T are significantly superior to SF, SSFTT, and GAHT in the transformer-based approaches. This further illustrates that the hierarchical DenseNet can effectively capture the characteristic information at different hierarchical levels, and has more powerful characteristic capture capabilities than methods based on simple CNN architectures. Moreover, the categorization accuracy of CNSST is 0.9% higher than BS2T on OA. This also illustrates that CNSST can effectively establish the correlation between the SAS feature information, reduce the loss of information, and obtain rich inductive bias information and spectral–spatial joint feature information. This information is then input into the spectral–spatial transformer with position encoding, which can effectively enhance the model’s spectral–spatial feature extraction capabilities.
The classification maps of various approaches on SV are depicted in
Figure 11. As FDSSC based on 3D-CNN hierarchical DenseNet can fully exploit the SAS characteristic information of various convolutional layers, it significantly outperforms SSRN, CDCNN, and DBMA in the classification maps. The classification maps of FDSSC based on 3D-CNN hierarchical DenseNet are significantly better than SSRN, CDCNN, and DBMA. Among the transformer-based approaches, the SF, SSFTT and GAHT approaches suffer from obvious misclassified pixels and relatively poor homogeneity. The reason for this is that SF merely exploits the transformer to capture long-range dependence information. GAHT merely utilizes the group-aware hierarchical transformer to constrain MHSA to the local spatial–spectral context. However, there are some limitations in these approaches based on the transformer structure alone in obtaining localized characteristic information. Although SSFTT adopts 3D-CNN and 2D-CNN to enhance the extraction of local feature information, its structure is relatively simple, resulting in a limited capacity for local feature extraction by the model. Comparatively, the BS2T and CNNST approaches, which combine the advantages of transformer and DenseNet, have a better performance in classification visual maps. Moreover, CNNST has fewer misclassified labels and better smoothing than BS2T. This further illustrates the effectiveness of CNNST in establishing the correlation between SAS feature information, reducing information loss and enhancing spectral–spatial transformer feature extraction.
From
Table 7, the proposed CNSST approach still achieves the highest accuracy of 98.84% on OA. SVM, CDCNN, and SF have the lowest accuracies on OA, which are 79.72%, 74.10%, and 87.46%, respectively. The classification maps of various approaches on IP are depicted in
Figure 12. It is also obvious that they contain a lot of noise and mislabels. This further indicates the limitations of the network structure design of ResNet and multiscale CNN-based CDCNN with a poor feature extraction capability, even lower than the traditional hand-crafted SVM. Furthermore, SF, which is based on group spectral embedding and a transform encoder, fails to efficiently utilize the local feature information of the HSI pixels, even though it can acquire the long-range dependency information among HSI pixels. The DenseNet-based FDSSC achieves a classification accuracy of 98.17% with relatively few misclassified pixels in the classification visual map. However, the 3D-CNN DenseNet-based FDSSC fails to exploit the long-distance dependency between HSI pixels. BS2T and CNSST combine the strengths of both hierarchical DenseNet and transformers, and effectively realize the extraction of local–global SAS features. Moreover, CNNST not only outperforms BS2T with 0.35% in classification accuracy, but also has fewer misclassified labels on the classified visual maps and is relatively smoother. It proves the effectiveness of CNSST in enhancing the correlation between SAS feature information and in introducing rich inductive bias information into the transformer with position coding to strengthen the local–global feature extraction of the model.
From
Table 8, it is obvious that the classification accuracy achieved by the proposed CNSST approach is still the highest, with OA, AA, and Ka of 98.27%, 97.70%, and 97.73%, respectively. In terms of OA, the classification accuracies of CNSST are higher than those of the GAHT, SSFTT, SF, and FDSSC approaches by 0.84%, 1.66%, 4.28%, and 0.86%, respectively. In addition to the classification results of the test labeled pixels in the reference map, we also considered background pixels (i.e., pixels that were not assigned any labels) for classification tests on the ZY dataset to show the consistency of the classification results from the classification visual map. From
Figure 13, the CNSST method has significantly fewer misclassified labels than them and has better edge detail information preservation. This further demonstrates that the CNSST approach combining the hierarchical DenseNet and Transformers can more adequately realize the local-global SAS feature extraction for HSI pixels. Moreover, the proposed CNSST method significantly outperforms BS2T both in terms of the classification accuracy and classification visual map, which further demonstrates the validity of CNNST in strengthening the correlation between SAS features as well as introducing location information and rich inductive bias information into the transformer to reinforce the feature extraction capability of the model.
4.4. Performance with Various Percentages of Training Samples
To further verify the sample sensitivity of the CNSST method, a comparison experiment of the different methods at varying sample proportions was conducted. In the experiments, labeled samples amounting to 0.5%, 0.75%, 1%, 2%, and 3% were randomly chosen from the UP and SV datasets for training. Similarly, 6%, 7%, 8%, 9%, and 10% of the samples were randomly chosen from the IP dataset. For the ZY dataset, 0.5%, 1%, 1.5%, 2%, and 2.5% samples were randomly selected. The classification accuracies of the various approaches with various percentages of training samples on the UP, SV, IP and ZY datasets are presented in
Figure 14. Notably, the SVM and CDCNN methods are too low (even below 80.0%) to achieve their classification accuracy on the ZY dataset under small samples. Therefore, some curves in subfigure (d) are not shown for a better visual comparison. As depicted in
Figure 14, the categorization accuracy of all models rises with the increase in training samples. With a decrease in training samples, the classification accuracies of all models continue to decrease, and the curves of the other models (apart from CNSST) have relatively large variations on the UP and SV datasets. However, CNSST has a relatively smooth change and still maintains the optimal categorization accuracy on all four datasets. It also demonstrates that CNSST is relatively insensitive to the proportion of training samples and has a relatively good robustness.
4.5. Parameter Sizes and Runtimes
The parameter sizes and runtimes of the various methods on four datasets are shown in
Table 9, where Par denotes the size of the parameter. Obviously, it is shown that the good classification performance of our proposed CNSST method on the four datasets is obtained at the expense of the computational complexity of the model. Notably, despite the relatively large parameters of the CNSST model, its training time is not the longest. This is because, to reduce the training cost of the model and avoid model overfitting, the proposed CNSST method employs an early stopping strategy. Furthermore, the batch size of the proposed CNSST is 64, while that of the BoS2T and FDSSC methods is 16. Meanwhile, a larger batch size usually implies that more samples can be processed in parallel, allowing them to be simultaneously modeled by the forward propagation process, thus effectively reducing the inference time.
4.6. Ablation Experiments
To better validate the efficacy of the modules in the proposed CNSST approach, ablation experiments were conducted. The ablation experiment outcomes on various datasets are presented in
Figure 15. Among them, SCNSST indicates that the proposed CNSST does not utilize hierarchical dense blocks to obtain hierarchical spectral–spatial characteristics from different convolutional layers, but rather utilizes the simple 3D-CNN structure (as shown in
Figure 5, only the second stage is used, while the first stage is replaced with a conventional CNN network). The no-RPE means that the proposed CNSST does not utilize relative position encoding in the transformer. The no-RPT means that the proposed CNSST does not utilize the transformer with relative position encoding (considering only the first stage without the existence of the second stage, as seen in
Figure 5, solely employing the first stage without the presence of the second stage). Also, no-Lion indicates that the traditional Adam optimizer is employed in the proposed CNSST instead of the new Lion optimizer.
From
Figure 15, the CNSST approach achieves a significantly higher categorization accuracy than SCNSST, no-RPE, and no-RPT, which further demonstrates that CNSST with hierarchical DenseNet can adequately exploit the spectral–spatial joint characteristic information at various levels and acquire richer inductive bias information. Secondly, its introduction into the transformer with 2D-relative position encoding allows for a better characterization of the spatial position information of HSI pixels and strengthens the position-aware and spectral-aware capabilities of the model. Moreover, the spectral–spatial transformer with relative position encoding can effectively establish long-range dependencies between HSI pixels and enhance the feature extraction capabilities of the model. Moreover, the classification outcome of the proposed CNSST outperforms the no-Lion, which further demonstrates the effectiveness of the new Lion optimizer employed in this work in enhancing the model’s categorization performance.