Estimation of the Biogeochemical and Physical Properties of Lakes Based on Remote Sensing and Artificial Intelligence Applications
Abstract
:1. Introduction
2. Materials and Methods
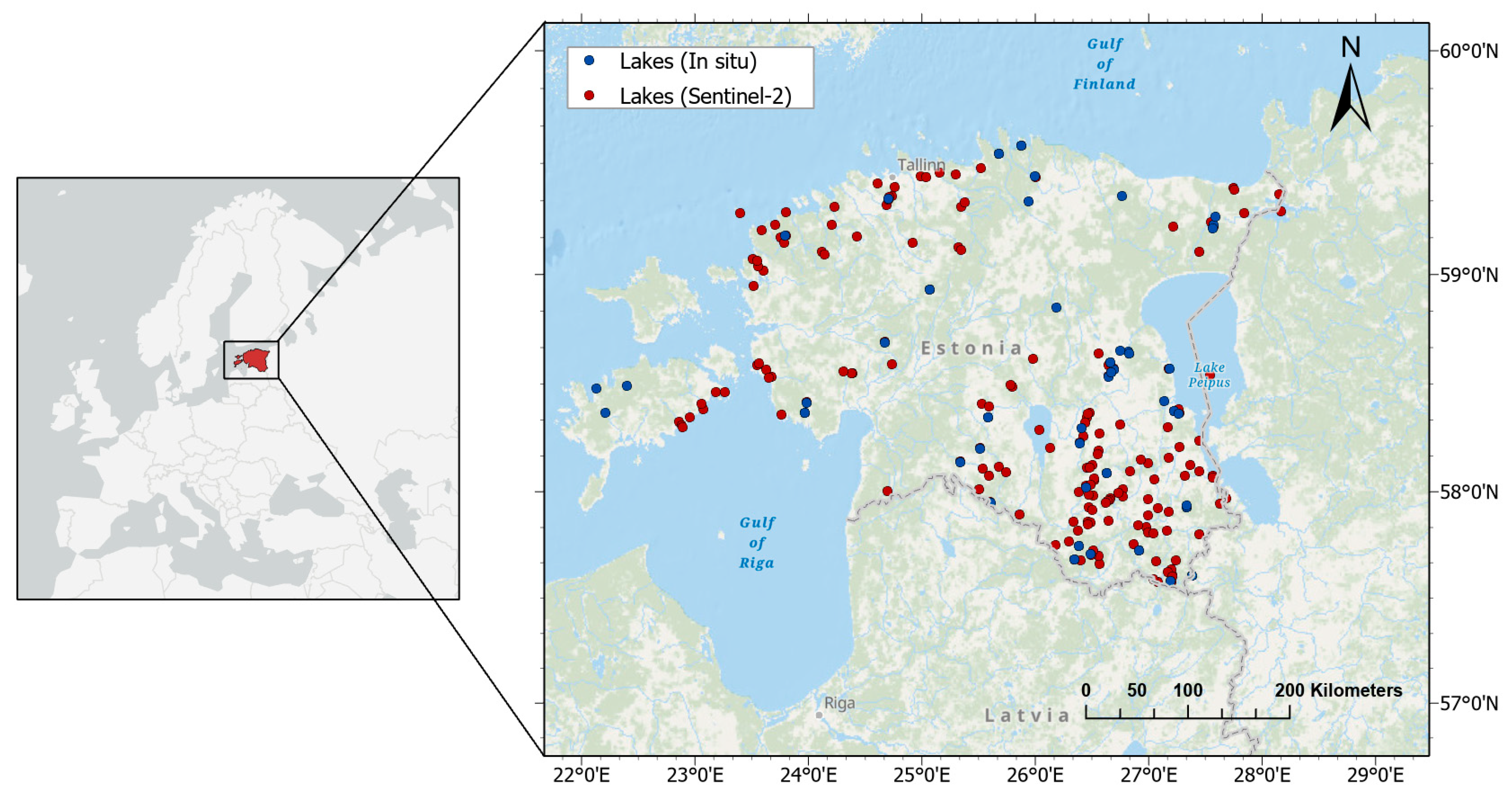
2.1. Study Sites
2.2. Biogeochemical and Physical Water Quality Parameters
2.3. Satellite Data
2.4. Retrieval of Biogeochemical and Physical Water Quality Parameters
2.5. Extreme Gradient Boosting Model and Genetic Algorithm
- (1)
- General parameters selection: Related to which booster to use for boosting. Gbtree booster that uses a tree-based model was selected;
- (2)
- Booster parameters:
- Step size shrinkage used in the update to avoid overfitting (learning_rate). Range 0–1.
- Maximum depth of a tree (max_depth). The higher the value the more complex the model and the probability of overfitting is higher. Range 0–∞.
- Minimum sum of instance weight (hessian) required in a child (min_child_weight). The larger min_child_weight is, the more conservative the algorithm. Range 0–∞.
- Subsample ratio of training instances (subsample). Setting it to 0.5 means that XGBoost will randomly sample half of the training data before trees grow, preventing overfitting. Subsampling occurs once in each boosting iteration. Range 0–1.
- The subsample ratio of columns when building each tree (colsample_bytree). Subsampling is performed once for each tree constructed. Range 0–1.
- (3)
- Learning task parameters: specify the learning task and the consistent learning objective. Objective reg:squarederror (regression with squared loss) was applied.
2.6. Accuracy Evaluation
3. Results
3.1. Correlations between Optically Active and Optically Non-Active Parameters
3.2. Reflectance Spectra of Sentinel-2 MSI
3.3. GA_XGBoost Model Performance and Evaluation
3.4. A Practical Demonstration of the Developed Inversion Models
4. Discussion
5. Conclusions
- GA_XGBoost exhibited strong predictive capabilities and it was able to accurately predict ten biogeochemical and two physical water quality parameters (TN, TP, PO4, BOD5, COD, CHL, CDOM, TSM, pH, O2, WT, and SD), showcasing its effectiveness in water quality and remote sensing applications.
- The observed increase in MAPE and RMSE, accompanied by a decrease in R2 during the transition from training to testing stages, highlighted the potential concern for overfitting, especially for specific parameters. This emphasizes the need for careful model selection, adjustments, and tuning in future studies.
- Despite the dynamic nature of lakes, our results demonstrated reliable estimates for multiple lakes simultaneously, considering the seasonal variations in water quality parameters.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Lake Name | Lat (N) | Lon (E) | Max Depth, m | Mean Depth, m | Catch. Area, km2 | Area, km2 | Trophic State | Sampling Date |
|---|---|---|---|---|---|---|---|---|
| Aheru järv/Kandsi järv | 57.68844 | 26.35283 | 7.4 | 3.4 | 52.4 | 2.34 | Eutrophic (hard water) | 12 September 2016 |
| Elistvere järv | 58.57139 | 26.70728 | 3.5 | 2 | 171 | 1.29 | Eutrophic (macrophyte) | 9 May 2016 |
| Elistvere järv | 58.57139 | 26.70728 | 3.5 | 2 | 171 | 1.29 | Eutrophic (macrophyte) | 15 September 2016 |
| Endla järv | 58.85357 | 26.19651 | 2.4 | 1.5 | 433 | 2.84 | Mixotrophic (hard water) | 16 May 2018 |
| Ermistu järv | 58.36923 | 23.98146 | 2.9 | 1.3 | 32.3 | 4.49 | Eutrophic (macrophyte) | 30 May 2017 |
| Hino järv | 57.58357 | 27.20177 | 10.4 | 3.1 | 2.12 | 1.99 | Oligotrophic | 6 May 2020 |
| Hino järv | 57.58357 | 27.20177 | 10.4 | 3.1 | 2.12 | 1.99 | Oligotrophic | 12 August 2020 |
| Hino järv | 57.58357 | 27.20177 | 10.4 | 3.1 | 2.12 | 1.99 | Oligotrophic | 7 September 2020 |
| Jõemõisa järv | 58.65372 | 26.82892 | 3.2 | 2.6 | 216 | 0.72 | Mixotrophic (hard water) | 5 August 2015 |
| Järise järv | 58.49416 | 22.41262 | 1.4 | 0.7 | 11.1 | 0.96 | Eutrophic (macrophyte) | 22 August 2018 |
| Kaiavere järv | 58.60383 | 26.67486 | 5 | 2.8 | 92.2 | 2.47 | Eutrophic (hard water) | 9 May 2016 |
| Kaiavere järv | 58.60383 | 26.67486 | 5 | 2.8 | 92.2 | 2.47 | Eutrophic (hard water) | 20 July 2016 |
| Kaiavere järv | 58.60383 | 26.67486 | 5 | 2.8 | 92.2 | 2.47 | Eutrophic (hard water) | 15 September 2016 |
| Kaisma järv | 58.69312 | 24.68132 | 2.1 | 1.25 | 16 | 1.4 | Mixotrophic (hard water) | 20 May 2019 |
| Kaisma järv | 58.69312 | 24.68132 | 2.1 | 1.25 | 16 | 1.4 | Mixotrophic (hard water) | 18 July 2019 |
| Kaiu järv | 58.64201 | 26.8389 | 3 | 2.6 | 216 | 1.34 | Mixotrophic (hard water) | 5 August 2015 |
| Kalli järv | 58.37695 | 27.23623 | 1.4 | 1.1 | 82.8 | 1.99 | Eutrophic (macrophyte) | 9 May 2020 |
| Karijärv | 58.29831 | 26.41993 | 14.5 | 5.7 | 11.1 | 0.82 | Eutrophic (hard water) | 14 September 2015 |
| Karijärv | 58.29831 | 26.41993 | 14.5 | 5.7 | 11.1 | 0.82 | Eutrophic (hard water) | 3 July 2019 |
| Karijärv | 58.29831 | 26.41993 | 14.5 | 5.7 | 11.1 | 0.82 | Eutrophic (hard water) | 4 September 2019 |
| Kariste järv | 58.14161 | 25.3484 | 7.2 | 3.3 | 128 | 0.61 | Eutrophic (hard water) | 30 May 2017 |
| Kariste järv | 58.14161 | 25.3484 | 7.2 | 3.3 | 128 | 0.61 | Eutrophic (hard water) | 25 September 2017 |
| Karujärv | 58.37102 | 22.2161 | 6 | 1.6 | 16.1 | 3.46 | Eutrophic (hard water) | 28 May 2018 |
| Karujärv | 58.37102 | 22.2161 | 6 | 1.6 | 16.1 | 3.46 | Eutrophic (hard water) | 22 August 2018 |
| Konsu järv | 59.22656 | 27.58052 | 10.2 | 5.8 | 27 | 1.39 | Mixotrophic (hard water) | 25 June 2019 |
| Konsu järv | 59.22656 | 27.58052 | 10.2 | 5.8 | 27 | 1.39 | Mixotrophic (hard water) | 22 April 2020 |
| Kooru järv | 58.48363 | 22.13946 | 1.2 | 0.3 | 38.7 | 0.85 | Eutrophic (halotrophic) | 16 August 2015 |
| Kooru järv | 58.48363 | 22.13946 | 1.2 | 0.3 | 38.7 | 0.85 | Eutrophic (halotrophic) | 27 September 2015 |
| Kooru järv | 58.48363 | 22.13946 | 1.2 | 0.3 | 38.7 | 0.85 | Eutrophic (halotrophic) | 28 September 2015 |
| Kooru järv | 58.48363 | 22.13946 | 1.2 | 0.3 | 38.7 | 0.85 | Eutrophic (halotrophic) | 29 May 2017 |
| Kooru järv | 58.48363 | 22.13946 | 1.2 | 0.3 | 38.7 | 0.85 | Eutrophic (halotrophic) | 28 May 2018 |
| Kooru järv | 58.48363 | 22.13946 | 1.2 | 0.3 | 38.7 | 0.85 | Eutrophic (halotrophic) | 28 July 2019 |
| Kooru järv | 58.48363 | 22.13946 | 1.2 | 0.3 | 38.7 | 0.85 | Eutrophic (halotrophic) | 29 August 2020 |
| Koosa järv | 58.4257 | 27.14411 | 1.9 | 1.2 | 75.9 | 2.83 | Mixotrophic (macrophyte) | 20 July 2020 |
| Käsmu järv | 59.58175 | 25.88399 | 3.3 | 2.2 | 16.5 | 0.49 | Mixotrophic (soft water) | 12 August 2015 |
| Käsmu järv | 59.58175 | 25.88399 | 3.3 | 2.2 | 16.5 | 0.49 | Mixotrophic (soft water) | 12 August 2020 |
| Köstrijärv | 57.75009 | 26.39461 | 4.4 | 3.3 | 1.8 | 0.12 | Eutrophic (macrophyte) | 7 May 2018 |
| Lahepera järv | 58.57375 | 27.19274 | 4.2 | 2.4 | 28.9 | 0.1 | Eutrophic (macrophyte) | 11 May 2020 |
| Lahepera järv | 58.57375 | 27.19274 | 4.2 | 2.4 | 28.9 | 0.1 | Eutrophic (macrophyte) | 20 July 2020 |
| Leegu järv | 58.36587 | 27.27614 | 1 | 0.6 | 5.6 | 0.86 | Eutrophic (macrophyte) | 20 July 2020 |
| Lohja järv | 59.54821 | 25.69092 | 3.7 | 2.2 | 12.3 | 0.56 | Mixotrophic (soft water) | 12 August 2015 |
| Lohja järv | 59.54821 | 25.69092 | 3.7 | 2.2 | 12.3 | 0.56 | Mixotrophic (soft water) | 8 July 2020 |
| Lohja järv | 59.54821 | 25.69092 | 3.7 | 2.2 | 12.3 | 0.56 | Mixotrophic (soft water) | 12 August 2020 |
| Loosalu järv | 58.93337 | 25.0777 | 5 | 3.7 | 1.6 | 0.35 | Dystrophic | 20 May 2018 |
| Mustjärv (Nohipalo Mustjärv) | 57.93201 | 27.34217 | 8.9 | 3.9 | 9.7 | 0.22 | Acidotrophic | 2 May 2016 |
| Mustjärv (Nohipalo Mustjärv) | 57.93201 | 27.34217 | 8.9 | 3.9 | 9.7 | 0.22 | Acidotrophic | 2 May 2017 |
| Mustjärv (Nohipalo Mustjärv) | 57.93201 | 27.34217 | 8.9 | 3.9 | 9.7 | 0.22 | Acidotrophic | 7 May 2020 |
| Männiku järv | 59.34583 | 24.71239 | 9 | 5 | 13 | 0.1 | Eutrophic (hard water) | 25 August 2015 |
| Ohepalu järv | 59.33395 | 25.95198 | 2.5 | 0.5 | 7.5 | 0.68 | Dystrophic | 23 July 2015 |
| Ohepalu järv | 59.33395 | 25.95198 | 2.5 | 0.5 | 7.5 | 0.68 | Dystrophic | 12 August 2020 |
| Pabra järv | 57.60901 | 27.39527 | 3.6 | 2.4 | 36.5 | 0.76 | Semidystrophic | 16 August 2017 |
| Peenjärv | 59.21379 | 27.57548 | - | - | - | 0.08 | Mixotrophic (hard water) | 25 June 2019 |
| Pikkjärv (Viitna Pikkjärv) | 59.4465 | 26.01005 | 6.2 | 3 | 1.1 | 0.16 | Oligotrophic | 14 August 2017 |
| Pikkjärv (Viitna Pikkjärv) | 59.4465 | 26.01005 | 6.2 | 3 | 1.1 | 0.16 | Oligotrophic | 15 May 2018 |
| Pikkjärv (Viitna Pikkjärv) | 59.4465 | 26.01005 | 6.2 | 3 | 1.1 | 0.16 | Oligotrophic | 19 May 2020 |
| Pikkjärv (Viitna Pikkjärv) | 59.4465 | 26.01005 | 6.2 | 3 | 1.1 | 0.16 | Oligotrophic | 17 August 2020 |
| Pühajärv | 58.02409 | 26.45667 | 8.5 | 4.3 | 44 | 2.98 | Eutrophic (hard water) | 2 May 2016 |
| Pühajärv | 58.02409 | 26.45667 | 8.5 | 4.3 | 44 | 2.98 | Eutrophic (hard water) | 2 May 2017 |
| Pühajärv | 58.02409 | 26.45667 | 8.5 | 4.3 | 44 | 2.98 | Eutrophic (hard water) | 2 May 2018 |
| Pühajärv | 58.02409 | 26.45667 | 8.5 | 4.3 | 44 | 2.98 | Eutrophic (hard water) | 7 August 2018 |
| Pühajärv | 58.02409 | 26.45667 | 8.5 | 4.3 | 44 | 2.98 | Eutrophic (hard water) | 1 July 2019 |
| Pühajärv | 58.02409 | 26.45667 | 8.5 | 4.3 | 44 | 2.98 | Eutrophic (hard water) | 2 September 2019 |
| Pühajärv | 58.02409 | 26.45667 | 8.5 | 4.3 | 44 | 2.98 | Eutrophic (hard water) | 4 May 2020 |
| Pühajärv | 58.02409 | 26.45667 | 8.5 | 4.3 | 44 | 2.98 | Eutrophic (hard water) | 7 May 2020 |
| Saadjärv | 58.53688 | 26.65778 | 25 | 8 | 31.9 | 7.23 | Eutrophic (hard water) | 9 May 2016 |
| Saadjärv | 58.53688 | 26.65778 | 25 | 8 | 31.9 | 7.23 | Eutrophic (hard water) | 14 July 2016 |
| Saare järv | 58.65489 | 26.7627 | 5.6 | 4.2 | 8.5 | 27.4 | Eutrophic (hard water) | 5 August 2015 |
| Soitsjärv | 58.55667 | 26.68168 | 8 | 1.2 | 15.2 | 1.58 | Mixotrophic (macrophyte) | 9 May 2016 |
| Suurjärv (Rouge Suurjärv) | 57.7275 | 26.92278 | 38 | 12 | 25.8 | 0.135 | Eutrophic (hard water) | 4 August 2015 |
| Suurjärv (Rouge Suurjärv) | 57.7275 | 26.92278 | 38 | 12 | 25.8 | 0.135 | Eutrophic (hard water) | 4 May 2017 |
| Suurjärv (Rouge Suurjärv) | 57.7275 | 26.92278 | 38 | 12 | 25.8 | 0.135 | Eutrophic (hard water) | 4 September 2019 |
| Suurjärv (Rouge Suurjärv) | 57.7275 | 26.92278 | 38 | 12 | 25.8 | 0.135 | Eutrophic (hard water) | 5 May 2020 |
| Suurjärv (Rouge Suurjärv) | 57.7275 | 26.92278 | 38 | 12 | 25.8 | 0.135 | Eutrophic (hard water) | 6 May 2020 |
| Suurjärv (Rouge Suurjärv) | 57.7275 | 26.92278 | 38 | 12 | 25.8 | 0.135 | Eutrophic (hard water) | 7 September 2020 |
| Tõhela järv | 58.41785 | 23.99619 | 1.5 | 1.3 | 21.7 | 4.07 | Eutrophic (macrophyte) | 30 May 2017 |
| Tõhela järv | 58.41785 | 23.99619 | 1.5 | 1.3 | 21.7 | 4.07 | Eutrophic (macrophyte) | 25 July 2017 |
| Tõhela järv | 58.41785 | 23.99619 | 1.5 | 1.3 | 21.7 | 4.07 | Eutrophic (macrophyte) | 21 July 2020 |
| Tänavjärv | 59.17897 | 23.80563 | 2.5 | 1.8 | 4.7 | 1.39 | Semidystrophic | 17 August 2015 |
| Tänavjärv | 59.17897 | 23.80563 | 2.5 | 1.8 | 4.7 | 1.39 | Semidystrophic | 29 May 2016 |
| Tänavjärv | 59.17897 | 23.80563 | 2.5 | 1.8 | 4.7 | 1.39 | Semidystrophic | 30 August 2016 |
| Tänavjärv | 59.17897 | 23.80563 | 2.5 | 1.8 | 4.7 | 1.39 | Semidystrophic | 26 September 2016 |
| Tänavjärv | 59.17897 | 23.80563 | 2.5 | 1.8 | 4.7 | 1.39 | Semidystrophic | 27 September 2016 |
| Tänavjärv | 59.17897 | 23.80563 | 2.5 | 1.8 | 4.7 | 1.39 | Semidystrophic | 20 May 2019 |
| Tänavjärv | 59.17897 | 23.80563 | 2.5 | 1.8 | 4.7 | 1.39 | Semidystrophic | 24 May 2020 |
| Tänavjärv | 59.17897 | 23.80563 | 2.5 | 1.8 | 4.7 | 1.39 | Semidystrophic | 18 July 2020 |
| Tänavjärv | 59.17897 | 23.80563 | 2.5 | 1.8 | 4.7 | 1.39 | Semidystrophic | 16 August 2020 |
| Tündre järv | 57.95075 | 25.61889 | 10.6 | 4.9 | 7.1 | 0.716 | Eutrophic (hard water) | 11 May 2016 |
| Uljaste järv | 59.3594 | 26.77396 | 6.4 | 2.2 | 1.1 | 0.63 | Semidystrophic | 14 August 2017 |
| Uljaste järv | 59.3594 | 26.77396 | 6.4 | 2.2 | 1.1 | 0.63 | Semidystrophic | 25 September 2017 |
| Uljaste järv | 59.3594 | 26.77396 | 6.4 | 2.2 | 1.1 | 0.63 | Semidystrophic | 15 May 2019 |
| Uljaste järv | 59.3594 | 26.77396 | 6.4 | 2.2 | 1.1 | 0.63 | Semidystrophic | 17 August 2020 |
| Valgejärv (Kurtna Valgejärv) | 59.26342 | 27.59712 | 10.5 | 4.2 | 1 | 0.08 | Semidystrophic | 15 May 2019 |
| Valgjärv | 58.08903 | 26.64033 | 5.5 | 3.2 | 4.9 | 0.65 | Eutrophic (hard water) | 4 May 2017 |
| Valgjärv | 58.08903 | 26.64033 | 5.5 | 3.2 | 4.9 | 0.65 | Eutrophic (hard water) | 5 July 2017 |
| Valgojärv (Nohipalo Valgojärv) | 57.9412 | 27.34662 | 12.5 | 6.2 | 2.2 | 0.07 | Oligotrophic | 2 May 2017 |
| Valgojärv (Nohipalo Valgojärv) | 57.9412 | 27.34662 | 12.5 | 6.2 | 2.2 | 0.07 | Oligotrophic | 1 August 2017 |
| Valgojärv (Nohipalo Valgojärv) | 57.9412 | 27.34662 | 12.5 | 6.2 | 2.2 | 0.07 | Oligotrophic | 2 September 2019 |
| Valgojärv (Nohipalo Valgojärv) | 57.9412 | 27.34662 | 12.5 | 6.2 | 2.2 | 0.07 | Oligotrophic | 7 May 2020 |
| Verevi järv | 58.23074 | 26.40464 | 11 | 3.6 | 1.1 | 0.12 | Hypertrophic | 8 August 2017 |
| Verevi järv | 58.23074 | 26.40464 | 11 | 3.6 | 1.1 | 0.12 | Hypertrophic | 6 May 2020 |
| Viljandi järv | 58.35027 | 25.59324 | 11 | 5.6 | 66.8 | 1.58 | Eutrophic (hard water) | 6 May 2020 |
| Õisu järv | 58.20532 | 25.52078 | 4.3 | 2.8 | 199 | 1.93 | Eutrophic (hard water) | 8 July 2019 |
| Ähijärv | 57.71297 | 26.49654 | 5.5 | 3.8 | 14.7 | 1.81 | Eutrophic (hard water) | 4 August 2015 |
| Ähijärv | 57.71297 | 26.49654 | 5.5 | 3.8 | 14.7 | 1.81 | Eutrophic (hard water) | 11 May 2016 |
| Ähijärv | 57.71297 | 26.49654 | 5.5 | 3.8 | 14.7 | 1.81 | Eutrophic (hard water) | 3 August 2016 |
| Ähijärv | 57.71297 | 26.49654 | 5.5 | 3.8 | 14.7 | 1.81 | Eutrophic (hard water) | 12 September 2016 |
| Ähijärv | 57.71297 | 26.49654 | 5.5 | 3.8 | 14.7 | 1.81 | Eutrophic (hard water) | 4 May 2017 |
| Ähijärv | 57.71297 | 26.49654 | 5.5 | 3.8 | 14.7 | 1.81 | Eutrophic (hard water) | 7 May 2018 |
| Ähijärv | 57.71297 | 26.49654 | 5.5 | 3.8 | 14.7 | 1.81 | Eutrophic (hard water) | 3 July 2019 |
| Ähijärv | 57.71297 | 26.49654 | 5.5 | 3.8 | 14.7 | 1.81 | Eutrophic (hard water) | 4 September 2019 |
| Ähijärv | 57.71297 | 26.49654 | 5.5 | 3.8 | 14.7 | 1.81 | Eutrophic (hard water) | 6 May 2020 |
| Ähijärv | 57.71297 | 26.49654 | 5.5 | 3.8 | 14.7 | 1.81 | Eutrophic (hard water) | 16 September 2020 |
Appendix B
| Water Quality Parameter | Model | R2 | MAE | RMSE | MAPE | Remote Sensing Platform/Sensor | Spatial Resolution | Waterbody | N | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| CHL | GA_XGBoost | 0.86 | 0.02 | 0.05 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| CHL | XGBoost | 0.82 | 0.03 | 0.05 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| CHL | XGBoost | - | 11.50 | 14.70 | 30.2 | Landsat 5 TM | 30 m | Lake Taihu | 163 | [119] |
| CHL | XGBoost | - | 7.20 | 12.90 | 34.8 | Landsat 7 ETM+ | 30 m | Lake Taihu | 163 | [119] |
| CHL | XGBoost | - | 11.60 | 15.70 | 35.2 | Landsat 8 OLI | 30 m | Lake Taihu | 163 | [119] |
| CHL | XGBoost | 0.42 | 1.52 | 2.07 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| CHL | XGBoost | 0.73 | - | 0.26 | 7.59 | Sentinel-2 MSI | 20 m | Q reservoir | 96 | [74] |
| CHL | XGBoost | 0.84 | - | 6.65 | - | Zhuhai-No.1, CMOS | 30 m | Dushan Lake, Weishan Lake | 99 | [123] |
| CHL | GA_RF | 0.80 | 0.03 | 0.05 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| CHL | RF | 0.74 | 0.04 | 0.06 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| CHL | RF | - | 8.90 | 14.40 | 18.3 | Landsat 5 TM | 30 m | Lake Taihu | 163 | [119] |
| CHL | RF | - | 7.70 | 13.80 | 44.1 | Landsat 7 ETM+ | 30 m | Lake Taihu | 163 | [119] |
| CHL | RF | - | 10.70 | 14.90 | 33.8 | Landsat 8 OLI | 30 m | Lake Taihu | 163 | [119] |
| CHL | RF | 0.32 | 1.51 | 1.94 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| CHL | RF | 0.67 | - | 0.30 | 13.13 | Sentinel-2 MSI | 20 m | Q reservoir | 96 | [74] |
| CHL | AdaBoost | 0.78 | 0.03 | 0.06 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| CHL | GA_AdaBoost | 0.83 | 0.03 | 0.05 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| CHL | SVR | - | 13.40 | 17.60 | 46.5 | Landsat 5 TM | 30 m | Lake Taihu | 163 | [119] |
| CHL | SVR | - | 8.40 | 18.70 | 37.7 | Landsat 7 ETM+ | 30 m | Lake Taihu | 163 | [119] |
| CHL | SVR | - | 13.10 | 15.60 | 32.2 | Landsat 8 OLI | 30 m | Lake Taihu | 163 | [119] |
| CHL | SVR | 0.46 | - | 0.36 | 14.3 | Sentinel-2 MSI | 20 m | Q reservoir | 96 | [74] |
| CHL | ANN | 0.15 | - | 0.45 | 17.94 | Sentinel-2 MSI | 20 m | Q reservoir | 96 | [74] |
| CHL | DNN | 0.81 | 0.03 | 0.05 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| CHL | BP | 0.12 | 1.57 | 2.21 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| CHL | Lasso | 0.20 | 1.54 | 2.08 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| CHL | MLR | 0.10 | 1.60 | 2.24 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| CODMn | XGBoost | 0.11 | 0.79 | 0.86 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| CODMn | RF | 0.20 | 0.71 | 0.80 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| CODMn | BP | 0.22 | 0.69 | 0.80 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| CODMn | Lasso | 0.07 | 0.70 | 0.83 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| CODMn | MLR | 0.06 | 0.71 | 0.83 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| CODMn | ML-MLR | 0.19 | 0.72 | 0.82 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| EC | XGBoost | 0.27 | - | 1.23 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 0 | 159 | [133] |
| EC | XGBoost | 0.33 | - | 2.57 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 1 | 159 | [133] |
| EC | XGBoost | 0.21 | - | 2.85 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 2 | 159 | [133] |
| EC | XGBoost | 0.32 | - | 2.58 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 3 | 159 | [133] |
| NH3-N | GA_XGBoost | 0.69 | 0.14 | 0.16 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| NH3-N | XGBoost | 0.65 | 0.15 | 0.17 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| NH3-N | XGBoost | 0.82 | - | 0.14 | 28.6 | Sentinel-2 MSI | 20 m | Q reservoir | 96 | [74] |
| NH3-N | GA_RF | 0.62 | 0.15 | 0.17 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| NH3-N | RF | 0.60 | 0.15 | 0.19 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| NH3-N | RF | 0.12 | - | 0.22 | 73.53 | Sentinel-2 MSI | 20 m | Q reservoir | 96 | [74] |
| NH3-N | AdaBoost | 0.55 | 0.15 | 0.20 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| NH3-N | GA_AdaBoost | 0.67 | 0.15 | 0.17 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| NH3-N | SVR | 0.49 | - | 0.15 | 118.45 | Sentinel-2 MSI | 20 m | Q reservoir | 96 | [74] |
| NH3-N | ANN | 0.25 | - | 0.17 | 107.43 | Sentinel-2 MSI | 20 m | Q reservoir | 96 | [74] |
| NH3-N | DNN | 0.63 | 0.15 | 0.18 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| O2 | XGBoost | 0.97 | - | 0.01 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 0 | 159 | [133] |
| O2 | XGBoost | 0.93 | - | 0.01 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 1 | 159 | [133] |
| O2 | XGBoost | 0.90 | - | 0.01 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 2 | 159 | [133] |
| O2 | XGBoost | 0.96 | - | 0.01 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 3 | 159 | [133] |
| O2 | XGBoost | 0.90 | - | 0.14 | 0.07 | Sentinel-2 MSI | 20 m | Q reservoir | 96 | [74] |
| O2 | RF | 0.77 | - | 0.34 | 3.43 | Sentinel-2 MSI | 20 m | Q reservoir | 96 | [74] |
| O2 | SVR | 0.85 | - | 0.17 | 1.38 | Sentinel-2 MSI | 20 m | Q reservoir | 96 | [74] |
| O2 | ANN | 0.79 | - | 0.20 | 2.04 | Sentinel-2 MSI | 20 m | Q reservoir | 96 | [74] |
| pH | XGBoost | 0.78 | - | 0.08 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 0 | 159 | [133] |
| pH | XGBoost | 0.74 | - | 0.19 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 1 | 159 | [133] |
| pH | XGBoost | 0.74 | - | 0.26 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 2 | 159 | [133] |
| pH | XGBoost | 0.76 | - | 0.09 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 3 | 159 | [133] |
| SD | XGBoost | 0.84 | 0.64 | 1.14 | - | Landsat 5 TM | 30 | Different lake datasets from Europe, China, and America | 4099 | [123] |
| SD | XGBoost | 0.76 | 0.89 | 1.87 | - | Landsat 7 ETM+ | 30 | Different lake datasets from Europe, China, and America | 2420 | [123] |
| SD | XGBoost | 0.88 | 0.50 | 0.80 | - | Landsat 8 OLI | 30 | Different lake datasets from Europe, China, and America | 1249 | [123] |
| SD | XGBoost | 0.98 | 2.01 | 2.52 | - | UAV | 0.185 m | The Shahu Port channel, The Xunsi River | 72 | [120] |
| SD | RF | 0.97 | 1.98 | 2.81 | - | UAV | 0.185 m | The Shahu Port channel, The Xunsi River | 72 | [120] |
| SD | RF | 0.82 | 0.62 | 1.13 | - | Landsat 5 TM | 30 | Different lake datasets from Europe, China, and America | 4099 | [123] |
| SD | RF | 0.78 | 0.84 | 1.84 | - | Landsat 7 ETM+ | 30 m | Different lake datasets from Europe, China, and America | 2420 | [123] |
| SD | RF | 0.85 | 0.47 | 0.74 | - | Landsat 8 OLI | 30 m | Different lake datasets from Europe, China, and America | 1249 | [123] |
| SD | AdaBoost | 0.98 | 2.00 | 2.55 | - | UAV | 0.185 m | The Shahu Port channel, The Xunsi River | 72 | [120] |
| SD | GBDT | 0.91 | 3.62 | 4.75 | - | UAV | 0.185 m | The Shahu Port channel, The Xunsi River | 72 | [120] |
| SD | Exponential function | 0.45 | - | 12.48 | - | UAV | 0.185 m | The Shahu Port channel, The Xunsi River | 72 | [120] |
| SD | Linear function | 0.80 | - | 7.59 | - | UAV | 0.185 m | The Shahu Port channel, The Xunsi River | 72 | [120] |
| SD | Logarithmic function | 0.80 | - | 7.58 | - | UAV | 0.185 m | The Shahu Port channel, The Xunsi River | 72 | [120] |
| SD | Power function | 0.68 | - | 9.44 | - | UAV | 0.185 m | The Shahu Port channel, The Xunsi River | 72 | [120] |
| SD | Quadratic polynomial | 0.80 | - | 7.65 | - | UAV | 0.185 m | The Shahu Port channel, The Xunsi River | 72 | [120] |
| SiO2 | XGBoost | 0.98 | - | 0.01 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 0 | 159 | [133] |
| SiO2 | XGBoost | 0.96 | - | 0.01 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 1 | 159 | [133] |
| SiO2 | XGBoost | 0.97 | - | 0.00 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 2 | 159 | [133] |
| SiO2 | XGBoost | 0.97 | - | 0.00 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 3 | 159 | [133] |
| TN | GA_XGBoost | 0.79 | 0.74 | 1.09 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TN | XGBoost | 0.70 | 0.81 | 1.28 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TN | XGBoost | 0.71 | 1.03 | 1.33 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| TN | GA_RF | 0.67 | 0.91 | 1.35 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TN | RF | 0.67 | 0.90 | 1.36 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TN | RF | 0.70 | 1.13 | 1.50 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| TN | AdaBoost | 0.61 | 1.22 | 1.55 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TN | GA_AdaBoost | 0.67 | 0.89 | 1.36 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TN | DNN | 0.77 | 0.84 | 1.14 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TN | BP | 0.82 | 0.84 | 1.27 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| TN | Lasso | 0.64 | 1.28 | 1.45 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| TN | MLR | 0.64 | 1.27 | 1.46 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| TN | ML-MLR | 0.82 | 0.87 | 1.28 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| TP | GA_XGBoost | 0.70 | 0.03 | 0.03 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TP | XGBoost | 0.61 | 0.03 | 0.04 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TP | XGBoost | 0.28 | 0.05 | 0.07 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| TP | GA_RF | 0.55 | 0.03 | 0.04 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TP | RF | 0.46 | 0.03 | 0.05 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TP | RF | 0.35 | 0.04 | 0.06 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| TP | AdaBoost | 0.61 | 0.03 | 0.04 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TP | GA_AdaBoost | 0.64 | 0.03 | 0.04 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TP | DNN | 0.56 | 0.03 | 0.04 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TP | BP | 0.43 | 0.05 | 0.05 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| TP | Lasso | 0.38 | 0.05 | 0.06 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| TP | MLR | 0.38 | 0.05 | 0.06 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| TP | ML-MLR | 0.27 | 0.04 | 0.07 | - | UAV | 1600 × 1300 pixels | The Zhanghe River | 45 | [119] |
| TSM | XGBoost | 0.18 | 641.20 | 751.90 | - | Landsat 8 OLI | 30 m | Ebinur Lake, China | 102 | [121] |
| TSM | XGBoost | 0.24 | 798.85 | 884.85 | - | Sentinel-2 MSI | 20 m | Ebinur Lake, China | 102 | [121] |
| TSM | RF | 0.68 | 215.88 | 256.92 | - | Landsat 8 OLI | 30 m | Ebinur Lake, China | 102 | [121] |
| TSM | RF | 0.73 | 220.27 | 222.69 | - | Sentinel-2 MSI | 20 m | Ebinur Lake, China | 102 | [121] |
| TUB | GA_XGBoost | 0.60 | 9.82 | 10.13 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TUB | XGBoost | 0.52 | 9.97 | 11.47 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TUB | GA_RF | 0.45 | 10.27 | 12.16 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TUB | RF | 0.37 | 10.56 | 13.20 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TUB | AdaBoost | 0.39 | 10.36 | 12.67 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TUB | GA_AdaBoost | 0.45 | 10.28 | 12.26 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| TUB | DNN | 0.54 | 9.92 | 11.03 | - | UAV | 0.1 | Nanfei River | 67 | [72] |
| WT | XGBoost | 0.73 | - | 0.15 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 0 | 159 | [133] |
| WT | XGBoost | 0.89 | - | 0.10 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 1 | 159 | [133] |
| WT | XGBoost | 0.89 | - | 0.08 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 2 | 159 | [133] |
| WT | XGBoost | 0.90 | - | 0.01 | - | Landsat 8 OLI | 30 | The Ganga River Basin, Cluster 3 | 159 | [133] |
References
- Verpoorter, C.; Kutser, T.; Seekell, D.A.; Tranvik, L.J. A Global Inventory of Lakes Based on High-Resolution Satellite Imagery. Geophys. Res. Lett. 2014, 41, 6396–6402. [Google Scholar] [CrossRef]
- Postel, S.L. Entering an Era of Water Scarity: The Challeenges Ahead. Ecol. Appl. 2000, 10, 941–948. [Google Scholar] [CrossRef]
- Brönmark, C.; Hansson, L.A. Environmental Issues in Lakes and Ponds: Current State and Perspectives. Environ. Conserv. 2002, 29, 290–307. [Google Scholar] [CrossRef]
- Bastviken, D.; Tranvik, L.J.; Downing, J.A.; Crill, P.M.; Enrich-Prast, A. Freshwater Methane Emissions Offset the Continental Carbon Sink. Science 2011, 331, 50. [Google Scholar] [CrossRef]
- Tranvik, L.J.; Downing, J.A.; Cotner, J.B.; Loiselle, S.A.; Striegl, R.G.; Ballatore, T.J.; Dillon, P.; Finlay, K.; Fortino, K.; Knoll, L.B.; et al. Lakes and Reservoirs as Regulators of Carbon Cycling and Climate. Limnol. Ocean. 2009, 54, 2298–2314. [Google Scholar] [CrossRef]
- Tranvik, L.J.; Cole, J.J.; Prairie, Y.T. The Study of Carbon in Inland Waters-from Isolated Ecosystems to Players in the Global Carbon Cycle. Limnol. Ocean. Lett. 2018, 3, 41–48. [Google Scholar] [CrossRef]
- Adrian, R.; O’Reilly, C.M.; Zagarese, H.; Baines, S.B.; Hessen, D.O.; Keller, W.; Livingstone, D.M.; Sommaruga, R.; Straile, D.; van Donk, E.; et al. Lakes as Sentinels of Climate Change. Limnol. Ocean. 2009, 54, 2283–2297. [Google Scholar] [CrossRef]
- Papenfus, M.; Schaeffer, B.; Pollard, A.I.; Loftin, K. Exploring the Potential Value of Satellite Remote Sensing to Monitor Chlorophyll-a for US Lakes and Reservoirs. Environ. Monit. Assess. 2020, 192, 808. [Google Scholar] [CrossRef] [PubMed]
- Mumby, P.J.; Green, E.P.; Edwards, A.J.; Clark, C.D. The Cost-Effectiveness of Remote Sensing for Tropical Coastal Resources Assessment and Management. J. Environ. Manag. 1999, 55, 157–166. [Google Scholar] [CrossRef]
- Marcé, R.; George, G.; Buscarinu, P.; Deidda, M.; Dunalska, J.; de Eyto, E.; Flaim, G.; Grossart, H.P.; Istvanovics, V.; Lenhardt, M.; et al. Automatic High Frequency Monitoring for Improved Lake and Reservoir Management. Environ. Sci. Technol. 2016, 50, 10780–10794. [Google Scholar] [CrossRef] [PubMed]
- Palmer, S.C.J.; Kutser, T.; Hunter, P.D. Remote Sensing of Inland Waters: Challenges, Progress and Future Directions. Remote Sens. Environ. 2015, 157, 1–8. [Google Scholar] [CrossRef]
- Palmer, S.C.J.; Odermatt, D.; Hunter, P.D.; Brockmann, C.; Présing, M.; Balzter, H.; Tóth, V.R. Satellite Remote Sensing of Phytoplankton Phenology in Lake Balaton Using 10 years of MERIS Observations. Remote Sens. Environ. 2015, 158, 441–452. [Google Scholar] [CrossRef]
- Bresciani, M.; Vascellari, M.; Giardino, C.; Matta, E. Remote Sensing Supports the Definition of the Water Quality Status of Lake Omodeo (Italy). Eur. J. Remote Sens. 2012, 45, 349–360. [Google Scholar] [CrossRef]
- Dekker, A.G.; Peters, S.W.M. The Use of the Thematic Mapper for the Analysis of Eutrophic Lakes: A Case Study in the Netherlands. Int. J. Remote Sens. 1993, 14, 799–821. [Google Scholar] [CrossRef]
- Lee, Z.; Shang, S.; Qi, L.; Yan, J.; Lin, G. A Semi-Analytical Scheme to Estimate Secchi-Disk Depth from Landsat-8 Measurements. Remote Sens. Environ. 2016, 177, 101–106. [Google Scholar] [CrossRef]
- Toming, K.; Kutser, T.; Laas, A.; Sepp, M.; Paavel, B.; Nõges, T. First Experiences in Mapping Lakewater Quality Parameters with Sentinel-2 MSI Imagery. Remote Sens. 2016, 8, 640. [Google Scholar] [CrossRef]
- Kutser, T.; Paavel, B.; Verpoorter, C.; Ligi, M.; Soomets, T.; Toming, K.; Casal, G. Remote Sensing of Black Lakes and Using 810 Nm Reflectance Peak for Retrieving Water Quality Parameters of Optically Complex Waters. Remote Sens. 2016, 8, 497. [Google Scholar] [CrossRef]
- Chen, J.; Zhu, W.; Tian, Y.Q.; Yu, Q.; Zheng, Y.; Huang, L. Remote Estimation of Colored Dissolved Organic Matter and Chlorophyll-a in Lake Huron Using Sentinel-2 Measurements. J. Appl. Remote Sens. 2017, 11, 036007. [Google Scholar] [CrossRef]
- Liu, H.; Li, Q.; Shi, T.; Hu, S.; Wu, G.; Zhou, Q. Application of Sentinel 2 MSI Images to Retrieve Suspended Particulate Matter Concentrations in Poyang Lake. Remote Sens. 2017, 9, 761. [Google Scholar] [CrossRef]
- Ogashawara, I.; Kiel, C.; Jechow, A.; Kohnert, K.; Ruhtz, T.; Grossart, H.-P.; Hölker, F.; Nejstgaard, J.C.; Berger, S.A.; Wollrab, S. The Use of Sentinel-2 for Chlorophyll-a Spatial Dynamics Assessment: A Comparative Study on Different Lakes in Northern Germany. Remote Sens. 2021, 13, 1542. [Google Scholar] [CrossRef]
- Soomets, T.; Uudeberg, K.; Jakovels, D.; Brauns, A.; Zagars, M.; Kutser, T. Validation and Comparison of Water Quality Products in Baltic Lakes Using Sentinel-2 MSI and Sentinel-3 OLCI Data. Sensors 2020, 20, 742. [Google Scholar] [CrossRef]
- Ekstrand, S. Landsat TM Based Quantification of Chlorophyll-a during Algae Blooms in Coastal Waters. Int. J. Remote Sens. 1992, 13, 1913–1926. [Google Scholar] [CrossRef]
- Bresciani, M.; Stroppiana, D.; Odermatt, D.; Morabito, G.; Giardino, C. Assessing Remotely Sensed Chlorophyll-a for the Implementation of the Water Framework Directive in European Perialpine Lakes. Sci. Total Environ. 2011, 409, 3083–3091. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Zhang, J.; Tong, C.; Liu, R.J.; Mu, B.; Ding, J. Retrieval Algorithm of Chlorophyll-a Concentration in Turbid Waters from Satellite HY-1C Coastal Zone Imager Data. J. Coast. Res. 2019, 90, 146–155. [Google Scholar] [CrossRef]
- Kutser, T. Monitoring Long Time Trends in Lake Cdom Using Landsat Image Archive. In Proceedings of the 2010 IEEE International Geoscience and Remote Sensing Symposium, Honolulu, HI, USA, 25–30 July 2010; pp. 389–392. [Google Scholar]
- Kutser, T.; Tranvik, L.; Pierson, D.C. Variations in Colored Dissolved Organic Matter between Boreal Lakes Studied by Satellite Remote Sensing. J. Appl. Remote Sens. 2009, 3, 033538. [Google Scholar] [CrossRef]
- Kutser, T.; Pierson, D.C.; Tranvik, L.; Reinart, A.; Sobek, S.; Kallio, K. Using Satellite Remote Sensing to Estimate the Colored Dissolved Organic Matter Absorption Coefficient in Lakes. Ecosystems 2005, 8, 709–720. [Google Scholar] [CrossRef]
- Knaeps, E.; Ruddick, K.G.; Doxaran, D.; Dogliotti, A.I.; Nechad, B.; Raymaekers, D.; Sterckx, S. A SWIR Based Algorithm to Retrieve Total Suspended Matter in Extremely Turbid Waters. Remote Sens. Environ. 2015, 168, 66–79. [Google Scholar] [CrossRef]
- Giardino, C.; Pepe, M.; Brivio, P.A.; Ghezzi, P.; Zilioli, E. Detecting Chlorophyll, Secchi Disk Depth and Surface Temperature in a Sub-Alpine Lake Using Landsat Imagery. Sci. Total Environ. 2001, 268, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, J.; Zhang, B.; Lee, Z.; Spyrakos, E.; Feng, L.; Liu, C.; Zhao, H.; Wu, Y.; Zhu, L.; et al. Changes of Water Clarity in Large Lakes and Reservoirs across China Observed from Long-Term MODIS. Remote Sens. Environ. 2020, 247, 111949. [Google Scholar] [CrossRef]
- Harrington, J.A.; Schiebe, F.R.; Nix, J.F. Remote Sensing of Lake Chicot, Arkansas: Monitoring Suspended Sediments, Turbidity, and Secchi Depth with Landsat MSS Data. Remote Sens. Environ. 1992, 39, 15–27. [Google Scholar] [CrossRef]
- Huang, C.; Yunmei, L.; Liu, G.; Guo, Y.; Yang, H.; Zhu, A.; Song, T.; Huang, T.; Zhang, M.; Shi, K. Tracing High Time-Resolution Fluctuations in Dissolved Organic Carbon Using Satellite and Buoy Observations: Case Study in Lake Taihu, China. Int. J. Appl. Earth Obs. Geoinf. 2017, 62, 174–182. [Google Scholar] [CrossRef]
- Li, S.; Toming, K.; Nõges, T.; Kutser, T. Integrating Remote Sensing of Hydrological Processes and Dissolved Organic Carbon Fluxes in Long-Term Lake Studies. J. Hydrol. 2022, 605, 127331. [Google Scholar] [CrossRef]
- Chen, J.; Zhu, W.; Tian, Y.Q.; Yu, Q. Monitoring Dissolved Organic Carbon by Combining Landsat-8 and Sentinel-2 Satellites: Case Study in Saginaw River Estuary, Lake Huron. Sci. Total Environ. 2020, 718, 137374. [Google Scholar] [CrossRef]
- Cao, F.; Tzortziou, M. Capturing Dissolved Organic Carbon Dynamics with Landsat-8 and Sentinel-2 in Tidally Influenced Wetland–Estuarine Systems. Sci. Total Environ. 2021, 777, 145910. [Google Scholar] [CrossRef]
- Arenz, R.F.; Lewis, W.M.; Saunders, J.F. Determination of Chlorophyll and Dissolved Organic Carbon from Reflectance Data for Colorado Reservoirs. Int. J. Remote Sens. 1996, 17, 1547–1565. [Google Scholar] [CrossRef]
- Shuchman, R.A.; Leshkevich, G.; Sayers, M.J.; Johengen, T.H.; Brooks, C.N.; Pozdnyakov, D. An Algorithm to Retrieve Chlorophyll, Dissolved Organic Carbon, and Suspended Minerals from Great Lakes Satellite Data. J. Great Lakes Res. 2013, 39, 14–33. [Google Scholar] [CrossRef]
- Winn, N.; Williamson, C.E.; Abbitt, R.; Rose, K.; Renwick, W.; Henry, M.; Saros, J. Modeling Dissolved Organic Carbon in Subalpine and Alpine Lakes with GIS and Remote Sensing. Landsc. Ecol. 2009, 24, 807–816. [Google Scholar] [CrossRef]
- Alcântara, E.; Bernardo, N.; Rodrigues, T.; Watanabe, F. Modeling the Spatio-Temporal Dissolved Organic Carbon Concentration in Barra Bonita Reservoir Using OLI/Landsat-8 Images. Model. Earth Syst. Environ. 2017, 3, 11. [Google Scholar] [CrossRef]
- Hirtle, H.; Rencz, A. The Relation between Spectral Reflectance and Dissolved Organic Carbon in Lake Water: Kejimkujik National Park, Nova Scotia, Canada. Int. J. Remote Sens. 2003, 24, 953–967. [Google Scholar] [CrossRef]
- Liu, D.; Yu, S.; Xiao, Q.; Qi, T.; Duan, H. Satellite Estimation of Dissolved Organic Carbon in Eutrophic Lake Taihu, China. Remote Sens. Environ. 2021, 264, 112572. [Google Scholar] [CrossRef]
- Jiang, G.; Ma, R.; Loiselle, S.A.; Duan, H. Optical Approaches to Examining the Dynamics of Dissolved Organic Carbon in Optically Complex Inland Waters. Environ. Res. Lett. 2012, 7, 034014. [Google Scholar] [CrossRef]
- Cai, X.; Li, Y.; Lei, S.; Zeng, S.; Zhao, Z.; Lyu, H.; Dong, X.; Li, J.; Wang, H.; Xu, J.; et al. A Hybrid Remote Sensing Approach for Estimating Chemical Oxygen Demand Concentration in Optically Complex Waters: A Case Study in Inland Lake Waters in Eastern China. Sci. Total Environ. 2023, 856, 158869. [Google Scholar] [CrossRef]
- Luo, J.; Pu, R.; Ma, R.; Wang, X.; Lai, X.; Mao, Z.; Zhang, L.; Peng, Z.; Sun, Z. Mapping Long-Term Spatiotemporal Dynamics of Pen Aquaculture in a Shallow Lake: Less Aquaculture Coming along Better Water Quality. Remote Sens. 2020, 12, 1866. [Google Scholar] [CrossRef]
- Cai, J.; Meng, L.; Liu, H.; Chen, J.; Xing, Q. Estimating Chemical Oxygen Demand in Estuarine Urban Rivers Using Unmanned Aerial Vehicle Hyperspectral Images. Ecol. Indic. 2022, 139, 108936. [Google Scholar] [CrossRef]
- Guo, H.; Huang, J.J.; Zhu, X.; Wang, B.; Tian, S.; Xu, W.; Mai, Y. A Generalized Machine Learning Approach for Dissolved Oxygen Estimation at Multiple Spatiotemporal Scales Using Remote Sensing. Environ. Pollut. 2021, 288, 117734. [Google Scholar] [CrossRef]
- Sharaf El Din, E.; Zhang, Y. Estimation of Both Optical and Nonoptical Surface Water Quality Parameters Using Landsat 8 OLI Imagery and Statistical Techniques. J. Appl. Remote Sens. 2017, 11, 1. [Google Scholar] [CrossRef]
- Elsayed, S.; Ibrahim, H.; Hussein, H.; Elsherbiny, O.; Elmetwalli, A.H.; Moghanm, F.S.; Ghoneim, A.M.; Danish, S.; Datta, R.; Gad, M. Assessment of Water Quality in Lake Qaroun Using Ground-Based Remote Sensing Data and Artificial Neural Networks. Water 2021, 13, 3094. [Google Scholar] [CrossRef]
- Ha, N.-T.; Nguyen, H.Q.; Truong, N.C.Q.; Le, T.L.; Thai, V.N.; Pham, T.L. Estimation of Nitrogen and Phosphorus Concentrations from Water Quality Surrogates Using Machine Learning in the Tri and Reservoir, Vietnam. Environ. Monit. Assess. 2020, 192, 789. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Hu, Z.; Liu, X.; Fu, Y.; Zhang, W. Spatio-Temporal Variation of Total Nitrogen and Ammonia Nitrogen in the Water Source of the Middle Route of the South-To-North Water Diversion Project. Water 2020, 12, 2615. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, H.; Ma, X.; Zhang, Y. Long-Term Spatiotemporal Variation and Environmental Driving Forces Analyses of Algal Blooms in Taihu Lake Based on Multi-Source Satellite and Land Observations. Water 2020, 12, 1035. [Google Scholar] [CrossRef]
- Yu, X.; Yi, H.; Liu, X.; Wang, Y.; Liu, X.; Zhang, H. Remote-Sensing Estimation of Dissolved Inorganic Nitrogen Concentration in the Bohai Sea Using Band Combinations Derived from MODIS Data. Int. J. Remote Sens. 2016, 37, 327–340. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, F.; Ge, X.; Zhang, X.; Chan, N.W.; Qi, Y. Measurement of Total Nitrogen Concentration in Surface Water Using Hyperspectral Band Observation Method. Water 2020, 12, 1842. [Google Scholar] [CrossRef]
- Arango, J.G.; Nairn, R.W. Prediction of Optical and Non-Optical Water Quality Parameters in Oligotrophic and Eutrophic Aquatic Systems Using a Small Unmanned Aerial System. Drones 2019, 4, 1. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, S.; Fan, F.; Dong, Y.; Li, Y.; Lin, W.; Zhou, C. Spatiotemporal Dynamics and Anthropologically Dominated Drivers of Chlorophyll-a, TN and TP Concentrations in the Pearl River Estuary Based on Retrieval Algorithm and Random Forest Regression. Environ. Res. 2022, 215, 114380. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gong, C.; Ji, T.; Hu, Y.; Li, L. Inland Water Quality Parameters Retrieval Based on the VIP-SPCA by Hyperspectral Remote Sensing. J. Appl. Remote Sens. 2021, 15, 042609. [Google Scholar] [CrossRef]
- Vakili, T.; Amanollahi, J. Determination of Optically Inactive Water Quality Variables Using Landsat 8 Data: A Case Study in Geshlagh Reservoir Affected by Agricultural Land Use. J. Clean. Prod. 2020, 247, 119134. [Google Scholar] [CrossRef]
- Soomets, T.; Toming, K.; Jefimova, J.; Jaanus, A.; Põllumäe, A.; Kutser, T. Deriving Nutrient Concentrations from Sentinel-3 OLCI Data in North-Eastern Baltic Sea. Remote Sens. 2022, 14, 1487. [Google Scholar] [CrossRef]
- Guo, H.; Tian, S.; Jeanne Huang, J.; Zhu, X.; Wang, B.; Zhang, Z. Performance of Deep Learning in Mapping Water Quality of Lake Simcoe with Long-Term Landsat Archive. ISPRS J. Photogramm. Remote Sens. 2022, 183, 451–469. [Google Scholar] [CrossRef]
- Isenstein, E.M.; Park, M.H. Assessment of Nutrient Distributions in Lake Champlain Using Satellite Remote Sensing. J. Environ. Sci. 2014, 26, 1831–1836. [Google Scholar] [CrossRef]
- Sun, D.; Qiu, Z.; Li, Y.; Shi, K.; Gong, S. Detection of Total Phosphorus Concentrations of Turbid Inland Waters Using a Remote Sensing Method. Water Air Soil. Pollut. 2014, 225, 1953. [Google Scholar] [CrossRef]
- Baban, S.M.J. Detecting Water Quality Parameters in the Norfolk Broads, U.K. Using Landsat Imagery. Int. J. Remote Sens. 1993, 14, 1247–1267. [Google Scholar] [CrossRef]
- Li, L.; Chen, X.; Zhang, M.; Zhang, W.; Wang, D.; Wang, H. The Spatial Variations of Water Quality and Effects of Water Landscape in Baiyangdian Lake, North China. Environ. Sci. Pollut. Res. 2022, 29, 16716–16726. [Google Scholar] [CrossRef]
- Gao, Y.; Gao, J.; Yin, H.; Liu, C.; Xia, T.; Wang, J.; Huang, Q. Remote Sensing Estimation of the Total Phosphorus Concentration in a Large Lake Using Band Combinations and Regional Multivariate Statistical Modeling Techniques. J. Environ. Manag. 2015, 151, 33–43. [Google Scholar] [CrossRef]
- Gholizadeh, M.H.; Melesse, A.M.; Reddi, L. A Comprehensive Review on Water Quality Parameters Estimation Using Remote Sensing Techniques. Sensors 2016, 16, 1298. [Google Scholar] [CrossRef]
- Mohseni, F.; Saba, F.; Mirmazloumi, S.M.; Amani, M.; Mokhtarzade, M.; Jamali, S.; Mahdavi, S. Ocean Water Quality Monitoring Using Remote Sensing Techniques: A Review. Mar. Environ. Res. 2022, 180, 105701. [Google Scholar] [CrossRef]
- Morel, A.Y.; Gordon, H.R. Report of the Working Group on Water Color. Bound. Layer. Meteorol. 1980, 18, 343–355. [Google Scholar] [CrossRef]
- Yang, H.; Kong, J.; Hu, H.; Du, Y.; Gao, M.; Chen, F. A Review of Remote Sensing for Water Quality Retrieval: Progress and Challenges. Remote Sens. 2022, 14, 1770. [Google Scholar] [CrossRef]
- Gordon, H.R.; Morel, A.Y. Remote Assessment of Ocean Color for Interpretation of Satellite Visible Imagery: A Review; Springer: New York, NY, USA, 1983; Volume 4. [Google Scholar] [CrossRef]
- Dekker, A.G.; Brando, V.E.; Anstee, J.M.; Pinnel, N.; Kutser, T.; Hoogenboom, E.J.; Peters, S.; Pasterkamp, R.; Vos, R.; Olbert, C.; et al. Imaging Spectrometry: Basic Principles and Prospective Applications; Springer: Berlin/Heidelberg, Germany, 2002; pp. 307–359. [Google Scholar] [CrossRef]
- Zhang, B.; Li, J.; Shen, Q.; Chen, D. A Bio-Optical Model Based Method of Estimating Total Suspended Matter of Lake Taihu from near-Infrared Remote Sensing Reflectance. Environ. Monit. Assess. 2008, 145, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Mu, X.; Chen, P.; Wang, B.; Choi, J.; Park, H.; Xu, S.; Wu, Y.; Yang, H. Machine Learning-Based Inversion of Water Quality Parameters in Typical Reach of the Urban River by UAV Multispectral Data. Ecol. Indic. 2021, 133, 108434. [Google Scholar] [CrossRef]
- Ruescas, A.; Hieronymi, M.; Mateo-Garcia, G.; Koponen, S.; Kallio, K.; Camps-Valls, G. Machine Learning Regression Approaches for Colored Dissolved Organic Matter (CDOM) Retrieval with S2-MSI and S3-OLCI Simulated Data. Remote Sens. 2018, 10, 786. [Google Scholar] [CrossRef]
- Tian, S.; Guo, H.; Xu, W.; Zhu, X.; Wang, B.; Zeng, Q.; Mai, Y.; Huang, J.J. Remote Sensing Retrieval of Inland Water Quality Parameters Using Sentinel-2 and Multiple Machine Learning Algorithms. Environ. Sci. Pollut. Res. 2022, 30, 18617–18630. [Google Scholar] [CrossRef]
- Xiao, Y.; Guo, Y.; Yin, G.; Zhang, X.; Shi, Y.; Hao, F.; Fu, Y. UAV Multispectral Image-Based Urban River Water Quality Monitoring Using Stacked Ensemble Machine Learning Algorithms—A Case Study of the Zhanghe River, China. Remote Sens. 2022, 14, 3272. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, J.; Wang, X. Recognizing the Relationship between Spatial Patterns in Water Quality and Land-Use/Cover Types: A Case Study of the Jinghe Oasis in Xinjiang, China. Water 2018, 10, 646. [Google Scholar] [CrossRef]
- Hafeez, S.; Wong, M.S.; Ho, H.C.; Nazeer, M.; Nichol, J.; Abbas, S.; Tang, D.; Lee, K.H.; Pun, L. Comparison of Machine Learning Algorithms for Retrieval of Water Quality Indicators in Case-Ii Waters: A Case Study of Hong Kong. Remote Sens. 2019, 11, 617. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: New York, NY, USA, 2016; pp. 785–794. [Google Scholar] [CrossRef]
- Ogunleye, A.; Wang, Q.-G. XGBoost Model for Chronic Kidney Disease Diagnosis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 17, 2131–2140. [Google Scholar] [CrossRef]
- Zamani Joharestani, M.; Cao, C.; Ni, X.; Bashir, B.; Talebiesfandarani, S. PM2.5 Prediction Based on Random Forest, XGBoost, and Deep Learning Using Multisource Remote Sensing Data. Atmosphere 2019, 10, 373. [Google Scholar] [CrossRef]
- Zhang, D.; Qian, L.; Mao, B.; Huang, C.; Huang, B.; Si, Y. A Data-Driven Design for Fault Detection of Wind Turbines Using Random Forests and XGboost. IEEE Access 2018, 6, 21020–21031. [Google Scholar] [CrossRef]
- Sheridan, R.P.; Wang, W.M.; Liaw, A.; Ma, J.; Gifford, E.M. Extreme Gradient Boosting as a Method for Quantitative Structure–Activity Relationships. J. Chem. Inf. Model. 2016, 56, 2353–2360. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Huang, L.; Xie, D.; Zhao, Q. EGBMMDA: Extreme Gradient Boosting Machine for MiRNA-Disease Association Prediction. Cell Death Dis. 2018, 9, 3. [Google Scholar] [CrossRef]
- Fan, J.; Wang, X.; Wu, L.; Zhou, H.; Zhang, F.; Yu, X.; Lu, X.; Xiang, Y. Comparison of Support Vector Machine and Extreme Gradient Boosting for Predicting Daily Global Solar Radiation Using Temperature and Precipitation in Humid Subtropical Climates: A Case Study in China. Energy Convers. Manag. 2018, 164, 102–111. [Google Scholar] [CrossRef]
- Bhagat, S.K.; Tiyasha, T.; Awadh, S.M.; Tung, T.M.; Jawad, A.H.; Yaseen, Z.M. Prediction of Sediment Heavy Metal at the Australian Bays Using Newly Developed Hybrid Artificial Intelligence Models. Environ. Pollut. 2021, 268, 115663. [Google Scholar] [CrossRef]
- Chen, L.; Tan, C.H.; Kao, S.J.; Wang, T.S. Improvement of Remote Monitoring on Water Quality in a Subtropical Reservoir by Incorporating Grammatical Evolution with Parallel Genetic Algorithms into Satellite Imagery. Water Res. 2008, 42, 296–306. [Google Scholar] [CrossRef]
- Ott, I.; Kõiv, T. Estonian Small Lakes: Special Features and Changes; Estonian Environment Information Centre: Tallinn, Estonia, 1999. [Google Scholar]
- EVS-EN ISO 11905-1:2003; Water Quality—Determination of Nitrogen—Part 1: Method Using Oxidative Digestion with Peroxodisulfate. International Organization for Standardization: Geneva, Switzerland, 2003.
- EVS-EN ISO 15681-2:2018; Water Quality—Determination of Orthophosphate and Total Phosphorus Contents by Flow Analysis—Part 2: Method by Continuous Flow Analysis. International Organization for Standardization: Geneva, Switzerland, 2018.
- EVS-EN ISO 6878:2004; Water Quality—Determination of Phosphorus—Ammonium Molybdate Spectrometric Method. International Organization for Standardization: Geneva, Switzerland, 2004.
- ISO 10304-1:2007; Water Quality—Determination of Dissolved Anions by Liquid Chromatography of Ions—Part 1: Determination of Bromide, Chloride, Fluoride, Nitrate, Nitrite, Phosphate and Sulfate. International Organization for Standardization: Geneva, Switzerland, 2007.
- ISO 7150-1:1984; Water Quality—Determination of Ammonium—Part 1: Manual Spectrometric Method. International Organization for Standardization: Geneva, Switzerland, 1984.
- EVS-EN ISO 5815-1:2019; Water Quality—Determination of Biochemical Oxygen Demand after n Days (BODn)—Part 1: Dilution and Seeding Method with Allylthiourea Addition. International Organization for Standardization: Geneva, Switzerland, 2019.
- EVS-ISO 15705:2004; Water Quality—Determination of the Chemical Qxygen Demand Index (ST-COD)—Small-Scale Sealed-Tube Method. International Organization for Standardization: Geneva, Switzerland, 2004.
- EVS-EN 15204:2006; Water Quality—Guidance Standard on the Enumeration of Phytoplankton Using Inverted Microscopy (Utermöhl Technique). International Organization for Standardization: Geneva, Switzerland, 1992.
- EVS-EN ISO 10523:2012; Water Quality—Determination of pH. International Organization for Standardization: Geneva, Switzerland, 2012.
- EVS-EN ISO 5814:2012; Water Quality—Determination of Dissolved Oxygen—Electrochemical Probe Method. International Organization for Standardization: Geneva, Switzerland, 2012.
- Toming, K.; Kutser, T.; Tuvikene, L.; Viik, M.; Nõges, T. Dissolved Organic Carbon and Its Potential Predictors in Eutrophic Lakes. Water Res. 2016, 102, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, G.E. A Treatise on Limnology: Geography, Physics, and Chemistry. In A Treatise on Limnology; John Wiley and Sons: New York, NY, USA, 1957. [Google Scholar]
- Brockmann, C.; Doerffer, R.; Marco, P.; Stelzer, K.; Embacher, S.; Ruescas, A. Evolution of the C2RCC Neural Network for Sentinel 2 and 3 for the Retrieval of Ocean. In Proceedings of the ‘Living Planet Symposium 2016’, (ESA SP-740, August 2016), Prague, Czech Republic, 9–13 May 2016; pp. 9–13. [Google Scholar]
- Uudeberg, K.; Ansko, I.; Põru, G.; Ansper, A.; Reinart, A. Using Opticalwater Types to Monitor Changes in Optically Complex Inland and Coastalwaters. Remote Sens. 2019, 11, 2297. [Google Scholar] [CrossRef]
- XGBoost Tutorials—Xgboost 1.0.0-SNAPSHOT Documentation. Available online: https://xgboost.readthedocs.io/en/stable/tutorials/index.html (accessed on 30 December 2022).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Müller, A.; Nothman, J.; Louppe, G.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2012, 12, 2825–2830. [Google Scholar]
- Doxaran, D.; Ruddick, K.; McKee, D.; Gentili, B.; Tailliez, D.; Chami, M.; Babin, M. Spectral Variations of Light Scattering by Marine Particles in Coastal Waters, from Visible to near Infrared. Limnol. Ocean. 2009, 54, 1257–1271. [Google Scholar] [CrossRef]
- Doxaran, D.; Froidefond, J.M.; Lavender, S.; Castaing, P. Spectral Signature of Highly Turbid Waters: Application with SPOT Data to Quantify Suspended Particulate Matter Concentrations. Remote Sens. Environ. 2002, 81, 149–161. [Google Scholar] [CrossRef]
- Kutser, T. Quantitative Detection of Chlorophyll in Cyanobacterial Blooms by Satellite Remote Sensing. Limnol. Ocean. 2004, 49, 2179–2189. [Google Scholar] [CrossRef]
- Smith, V.H. Eutrophication of Freshwater and Coastal Marine Ecosystems: A Global Problem. Environ. Sci. Pollut. Res. 2003, 10, 126–139. [Google Scholar] [CrossRef]
- Xu, Z.; Xu, Y.J. Rapid Field Estimation of Biochemical Oxygen Demand in a Subtropical Eutrophic Urban Lake with Chlorophyll a Fluorescence. Environ. Monit. Assess. 2015, 187, 4171. [Google Scholar] [CrossRef]
- Hébert, M.P.; Soued, C.; Fussmann, G.F.; Beisner, B.E. Dissolved Organic Matter Mediates the Effects of Warming and Inorganic Nutrients on a Lake Planktonic Food Web. Limnol. Ocean. 2022, 68, S23–S38. [Google Scholar] [CrossRef]
- Erlandsson, M.; Cory, N.; Köhler, S.; Bishop, K. Direct and Indirect Effects of Increasing Dissolved Organic Carbon Levels on PH in Lakes Recovering from Acidification. J. Geophys. Res. Biogeosci. 2010, 115, 1–8. [Google Scholar] [CrossRef]
- Grayson, R.B.; Finlayson, B.L.; Gippel, C.J.; Hart, B.T. The Potential of Field Turbidity Measurements for the Computation of Total Phosphorus and Suspended Solids Loads. J. Environ. Manag. 1996, 47, 257–267. [Google Scholar] [CrossRef]
- Jones, A.S.; Stevens, D.K.; Horsburgh, J.S.; Mesner, N.O. Surrogate Measures for Providing High Frequency Estimates of Total Suspended Solids and Total Phosphorus Concentrations. J. Am. Water Resour. Assoc. 2011, 47, 239–253. [Google Scholar] [CrossRef]
- Kusari, L. Turbidity as a Surrogate for the Determination of Total Phosphorus, Using Relationship Based on Sub-Sampling Techniques. Ecol. Eng. Environ. Technol. 2022, 23, 88–93. [Google Scholar] [CrossRef]
- Lannergård, E.E.; Ledesma, J.L.J.; Fölster, J.; Futter, M.N. An Evaluation of High Frequency Turbidity as a Proxy for Riverine Total Phosphorus Concentrations. Sci. Total Environ. 2019, 651, 103–113. [Google Scholar] [CrossRef]
- Viso-Vázquez, M.; Acuña-Alonso, C.; Rodríguez, J.L.; Álvarez, X. Remote Detection of Cyanobacterial Blooms and Chlorophyll-a Analysis in a Eutrophic Reservoir Using Sentinel-2. Sustainability 2021, 13, 8570. [Google Scholar] [CrossRef]
- Buma, W.G.; Lee, S.-I. Evaluation of Sentinel-2 and Landsat 8 Images for Estimating Chlorophyll-a Concentrations in Lake Chad, Africa. Remote Sens. 2020, 12, 2437. [Google Scholar] [CrossRef]
- Shang, Y.; Liu, G.; Wen, Z.; Jacinthe, P.A.; Song, K.; Zhang, B.; Lyu, L.; Li, S.; Wang, X.; Yu, X. Remote Estimates of CDOM Using Sentinel-2 Remote Sensing Data in Reservoirs with Different Trophic States across China. J. Environ. Manag. 2021, 286, 112275. [Google Scholar] [CrossRef]
- Kutser, T. The Possibility of Using the Landsat Image Archive for Monitoring Long Time Trends in Coloured Dissolved Organic Matter Concentration in Lake Waters. Remote Sens. Environ. 2012, 123, 334–338. [Google Scholar] [CrossRef]
- Cao, Z.; Ma, R.; Melack, J.M.; Duan, H.; Liu, M.; Kutser, T.; Xue, K.; Shen, M.; Qi, T.; Yuan, H. Landsat Observations of Chlorophyll-a Variations in Lake Taihu from 1984 to 2019. Int. J. Appl. Earth Obs. Geoinf. 2022, 106, 102642. [Google Scholar] [CrossRef]
- Wei, L.; Wang, Z.; Huang, C.; Zhang, Y.; Wang, Z.; Xia, H.; Cao, L. Transparency Estimation of Narrow Rivers by UAV-Borne Hyperspectral Remote Sensing Imagery. IEEE Access 2020, 8, 168137–168153. [Google Scholar] [CrossRef]
- Liu, C.; Duan, P.; Zhang, F.; Jim, C.Y.; Tan, M.L.; Chan, N.W. Feasibility of the Spatiotemporal Fusion Model in Monitoring Ebinur Lake’s Suspended Particulate Matter under the Missing-Data Scenario. Remote Sens. 2021, 13, 3952. [Google Scholar] [CrossRef]
- Wang, C.L.; Shi, K.Y.; Ming, X.; Cong, M.Q.; Liu, X.Y.; Guo, W.J. A Comparative Study of the COD Hyperspectral Inversion Models in Water Based on the Maching Learning. Spectrosc. Spectr. Anal. 2022, 42, 2353–2358. [Google Scholar] [CrossRef]
- Zhang, Y.; Shi, K.; Sun, X.; Zhang, Y.; Li, N.; Wang, W.; Zhou, Y.; Zhi, W.; Liu, M.; Li, Y.; et al. Improving Remote Sensing Estimation of Secchi Disk Depth for Global Lakes and Reservoirs Using Machine Learning Methods. GIScience Remote Sens. 2022, 59, 1367–1383. [Google Scholar] [CrossRef]
- Sommer, U.; Gliwicz, Z.M.; Lampert, W.; Duncan, A. The PEG-Model of Seasonal Succession of Planktonic Events in Fresh Waters. Arch. Hydrobiol. 1986, 106, 433–471. [Google Scholar] [CrossRef]
- Welch, E.B. Should Nitrogen Be Reduced to Manage Eutrophication If It Is Growth Limiting? Evidence from Moses Lake. Lake Reserv. Manag. 2009, 25, 401–409. [Google Scholar] [CrossRef]
- Schindler, D.W.; Fee, E.J. Diurnal Variation of Dissolved Inorganic Carbon and Its Use in Estimating Primary Production and CO2 Invasion in Lake 227. J. Fish. Res. Board Can. 2011, 30, 1501–1510. [Google Scholar] [CrossRef]
- Toming, K.; Tuvikene, L.; Vilbaste, S.; Agasild, H.; Viik, M.; Kisand, A.; Feldmann, T.; Martma, T.; Jones, R.I.; Nõges, T. Contributions of Autochthonous and Allochthonous Sources to Dissolved Organic Matter in a Large, Shallow, Eutrophic Lake with a Highly Calcareous Catchment. Limnol. Ocean. 2013, 58, 1259–1270. [Google Scholar] [CrossRef]
- Brezonik, P.; Menken, K.D.; Bauer, M. Landsat-Based Remote Sensing of Lake Water Quality Characteristics, Including Chlorophyll and Colored Dissolved Organic Matter (CDOM). Lake Reserv. Manag. 2005, 21, 373–382. [Google Scholar] [CrossRef]
- Tilzer, M.M. Secchi Disk—Chlorophyll Relationships in a Lake with Highly Variable Phytoplankton Biomass. Hydrobiologia 1988, 162, 163–171. [Google Scholar] [CrossRef]
- Suursaar, Ü. Summer 2021 Marine Heat Wave in the Gulf of Finland from the Perspective of Climate Warming. Est. J. Earth Sci. 2022, 71, 1. [Google Scholar] [CrossRef]
- Stefan, H.G.; Fang, X. Dissolved Oxygen Model for Regional Lake Analysis. Ecol. Model. 1994, 71, 37–68. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Z.; Liu, M.; He, J.; Shi, K.; Zhou, Y.; Wang, M.; Liu, X. Dissolved Oxygen Stratification and Response to Thermal Structure and Long-Term Climate Change in a Large and Deep Subtropical Reservoir (Lake Qiandaohu, China). Water Res. 2015, 75, 249–258. [Google Scholar] [CrossRef]
- Krishnaraj, A.; Honnasiddaiah, R. Remote Sensing and Machine Learning Based Framework for the Assessment of Spatio-Temporal Water Quality in the Middle Ganga Basin. Environ. Sci. Pollut. Res. 2022, 29, 64939–64958. [Google Scholar] [CrossRef]





| Parameter | Abbreviation | Unit | Reference/Standard |
|---|---|---|---|
| Total nitrogen | TN | mgN/L | ISO, 2003 [88] |
| Total phosphorus | TP | mgP/L | ISO, 2018 [89] |
| Phosphate | PO4 | mg/L | ISO, 2004 [90] |
| Sulfate | SO4 | mg/L | ISO, 2007 [91] |
| Ammonium nitrogen | NH4N | mg/L | ISO, 1984 [92] |
| 5-day biochemical oxygen demand | BOD5 | mgO₂/L | ISO, 2019 [93] |
| Dichromatic chemical oxygen demand | COD | mgO₂/L | ISO, 2004 [94] |
| Biomass of phytoplankton | FPBM | mg/L | ISO, 1992 [95] |
| Biomass of cyanobacteria | CYBM | mg/L | ISO, 1992 [95] |
| pH | pH | ISO, 2012a [96] | |
| Dissolved oxygen | O2 | mg/L | ISO, 2012b [97] |
| Water temperature | WT | °C | [98] |
| Secchi disk depth | SD | m | [99] |
| Chlorophyll a | CHL | µg/L | ISO, 1992 [95] |
| Colored dissolved organic matter | CDOM | mg/L | [98] |
| Total suspended matter | TSM | mg/L | [98] |
| Count | Mean | Std | Min | 25% | 50% | 75% | Max | Skewness | Kurtosis | |
|---|---|---|---|---|---|---|---|---|---|---|
| TN | 102 | 0.90 | 0.61 | 0.15 | 0.51 | 0.73 | 1.10 | 3.90 | 2.23 | 6.77 |
| TP | 102 | 0.06 | 0.19 | 0.01 | 0.02 | 0.03 | 0.05 | 1.60 | 7.02 | 50.4 |
| PO4 | 99 | 0.008 | 0.007 | 0.002 | 0.003 | 0.006 | 0.01 | 0.05 | 2.90 | 10.4 |
| SO4 | 100 | 7.70 | 7.28 | 0.10 | 1.70 | 4.65 | 12.0 | 31.0 | 1.13 | 0.58 |
| NH4N | 102 | 0.023 | 0.021 | 0.01 | 0.01 | 0.02 | 0.024 | 0.14 | 3.57 | 15.9 |
| BOD5 | 102 | 2.15 | 1.39 | 0.70 | 1.30 | 1.70 | 2.68 | 7.50 | 1.77 | 3.45 |
| COD | 87 | 42.1 | 29.2 | 15.0 | 23.0 | 36.0 | 48.0 | 160 | 1.99 | 4.20 |
| CHL | 102 | 13.6 | 14.9 | 1.00 | 3.45 | 8.30 | 17.5 | 100 | 2.60 | 10.4 |
| CDOM | 102 | 10.9 | 15.9 | 0.85 | 3.10 | 5.55 | 10.8 | 81.0 | 3.22 | 10.7 |
| TSM | 38 | 156 | 95.9 | 8.23 | 99.1 | 154 | 223 | 371 | 0.24 | −0.6 |
| FPBM | 80 | 4.73 | 5.40 | 0.16 | 0.76 | 2.60 | 6.78 | 21.7 | 1.36 | 0.82 |
| CYBM | 58 | 1.81 | 3.24 | 0.00 | 0.03 | 0.33 | 2.05 | 13.0 | 2.22 | 4.29 |
| PH | 83 | 7.98 | 1.07 | 3.65 | 7.85 | 8.21 | 8.53 | 9.40 | −2.16 | 5.05 |
| O2 | 84 | 8.62 | 2.42 | 2.63 | 7.21 | 8.80 | 10.1 | 15.6 | −0.06 | 0.41 |
| WT | 85 | 17.1 | 5.08 | 5.20 | 13.7 | 18.0 | 20.6 | 26.9 | −0.40 | −0.35 |
| SD | 98 | 1.88 | 1.24 | 0.25 | 0.70 | 1.75 | 2.60 | 5.00 | 0.67 | −0.37 |
| Formula |
|---|
| 1. Ba + Bb |
| 2. Ba − Bb |
| 3. Ba/Bb |
| 4. Ba * Bb |
| 5. Ba + Bb + Bc |
| 6. Ba + Bb * Bc |
| 7. (Ba + Bb) * Bc |
| 8. (Ba − Bb) * Bc |
| 9. (Ba + Bb)/Bc |
| 10. Ba * Bb/Bc |
| 11. (Ba − Bb)/(Ba + Bb) |
| 12. (Ba/Bb) * (Ba/Bb) |
| 13. Ba/Bb − Ba/Bc |
| 14. Ba − (Bb + Bc)/2 |
| 15. Ba/(Bb + Bc) |
| Water Quality Parameter (y) | x |
|---|---|
| TP | ‘B2 * B6’, ‘(B1 − B5) * B3’, ‘(B7/B3)*(B7/B3)’, ‘B4/B2-B4/B7’, ‘(B7 + B2)/B3’, ‘(B2 − B4)*B6’, ‘(B3 + B5) * B1’ |
| TN | ‘B5 − (B4 + B3)/2’, ‘B7/(B2 + B4)’, ‘B7 − (B4 + B8A)/2’, ‘(B4 + B1)/B3’, ‘B1 − (B7 + B5)/2’, ‘(B1 + B8A)/B3’ |
| PO4 | ‘B2 * B6/B1’, ‘B3 * B6/B2’, ‘B7 * B3/B2’, ‘B2/(B7 + B6)’, ‘B2 − (B6 + B4)/2’, ‘B5 − (B2 + B3)/2’, ‘(B2 − B7) * B4’, ‘(B2 − B6)/(B2 − B6)’, ‘(B2/B6) * (B2/B6)’ |
| NH4 | ‘B2 − (B3 + B4)/2’, ‘B3 − (B6 + B1)/2’, ‘(B6 − B8A) * B5’, ‘B2/B4 − B2/B6’, ‘B2/B6 − B2/B4’, ‘B4/B8A − B4/B1’ |
| SO4 | ‘B3 * B8A/B4’, ‘B4 * B1/B7’, ‘B4/B2 − B4/B1’,’B1/(B7 + B2)’, ‘(B3 + B5)/B1’, ‘(B7 + B2)/B1’, ‘B1 − (B7 + B6)/2’, ‘(B1 − B3) * B5’, ‘(B8A − B7) * B6’ |
| O2 | ‘B5 * B2/B3’, ‘(B4 + B8A) * B3’, ‘B5 − (B4 + B8A)/2’, ‘(B1 − B8A) * B6) ‘ |
| pH | ‘B2 − B1’, ‘(B6 + B8A)/B7’, ‘B2 − (B1 + B3)/2’, ‘B4 − (B3 + B5)/2’, ‘(B4 − B5) * B3’, ‘B4/B2 − B4/B1’ |
| WT | ‘(B1 − B3) * B6’, ‘(B1 − B4) * B6’, ‘(B2 − B3) * B4’ |
| COD | ‘B8A * B6/B1’, ‘B7 + B4 * B5’, ‘B7 + B5 * B4’, ‘B4 − (B5 + B8A)/2’, ‘(B5 − B6)/(B5 − B6)’, ‘B1/B3 − B1/B4’, ‘B1/B4 − B1/B3’ |
| BOD5 | ‘B4/B5’, ‘(B5 + B6)/B4’, ‘B6 − (B1 + B8A)/2’, ‘(B5/B4) * (B5/B4)’ |
| SD | ‘B6 * B5/B4’, ‘(B1 − B2) * B6’, ‘(B2 − B1) * B6’, ‘(B2 − B5) * B6’ |
| FPBM | ‘B6 * B2/B1’, ‘B7 + B3 * B2’, ‘(B7 + B6) * B8A’, ‘(B8A + B4) * B6’ |
| CYBM | ‘B4 + B3 * B7’, ‘(B5 + B4) * B8A’, ‘(B8A + B4) * B7’ |
| CHL | ‘(B2 + B4) * B8A’, ‘(B2/B6) * (B2/B6)’, ‘B4/B1 − B4/B5’ |
| CDOM | ‘B2/B5 − B2/B6’, ‘B2/B6 − B2/B4’, ‘B4/B6 − B4/B5’, ‘B6/B7 − B6/B5’, ‘B7/B6 − B7/B5’ |
| TSM | ‘B6-B2’, ‘B2-(B6 + B7)/2’, ‘(B3 − B4) * B1’, ‘(B4 − B3) * B1’, ‘(B5 − B4) * B8A’, ‘(B6 − B8A) * B7’ |
| Water Quality Parameter | Training | Testing | Validation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total n | n | R2 | MAPE(%) | RMSE | n | R2 | MAPE(%) | RMSE | n | R2 | MAPE(%) | RMSE | |
| TP | 102 | 60 | 0.99 | 0.16 | 0.00 | 21 | 0.90 | 36.5 | 0.02 | 21 | 0.60 | 34.4 | 0.02 |
| TN | 102 | 60 | 0.99 | 0.21 | 0.00 | 21 | 0.68 | 36.0 | 0.24 | 21 | 0.46 | 32.0 | 0.32 |
| PO4 | 99 | 59 | 0.99 | 7.24 | 0.0005 | 20 | 0.87 | 43.9 | 0.003 | 20 | 0.45 | 43.8 | 0.004 |
| NH4 | 102 | 60 | 0.99 | 3.39 | 0.0008 | 21 | 0.79 | 75.5 | 0.02 | 21 | 0.68 | 161 | 0.19 |
| SO4 | 100 | 60 | 0.99 | 0.89 | 0.03 | 20 | 0.69 | 168 | 3.26 | 20 | 0.58 | 123 | 5.20 |
| O2 | 84 | 50 | 0.99 | 1.98 | 0.21 | 17 | 0.62 | 15.2 | 1.31 | 17 | 0.62 | 46.1 | 4.54 |
| pH | 83 | 49 | 0.99 | 0.59 | 0.05 | 17 | 0.72 | 7.02 | 0.64 | 17 | 0.71 | 7.27 | 0.67 |
| WT | 85 | 51 | 0.99 | 1.37 | 0.78 | 17 | 0.63 | 14.1 | 3.08 | 17 | 0.58 | 17.3 | 3.96 |
| COD | 87 | 51 | 0.99 | 0.27 | 0.17 | 18 | 0.49 | 29.6 | 12.9 | 18 | 0.42 | 43.9 | 17.9 |
| BOD5 | 102 | 60 | 0.99 | 0.03 | 0.0005 | 21 | 0.90 | 17.8 | 0.56 | 21 | 0.85 | 30.1 | 0.66 |
| SD | 98 | 58 | 0.99 | 7.70 | 0.12 | 20 | 0.58 | 37.9 | 0.86 | 20 | 0.57 | 38.7 | 0.81 |
| FPBM | 80 | 48 | 0.99 | 1.32 | 0.01 | 16 | 0.79 | 169 | 2.01 | 16 | 0.79 | 109 | 2.19 |
| CYBM | 58 | 34 | 0.99 | 4.94 | 0.0008 | 12 | 0.85 | 684 | 1.64 | 12 | 0.88 | 532 | 1.81 |
| CHL | 102 | 60 | 0.96 | 17.3 | 3.41 | 21 | 0.80 | 71.5 | 9.87 | 21 | 0.82 | 48.8 | 4.78 |
| CDOM | 102 | 60 | 0.99 | 0.01 | 0.001 | 21 | 0.94 | 41.5 | 3.77 | 21 | 0.92 | 40.7 | 6.72 |
| TSM | 38 | 22 | 0.99 | 0.0007 | 0.001 | 8 | 0.94 | 20.3 | 22.3 | 8 | 0.83 | 43.8 | 32.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toming, K.; Liu, H.; Soomets, T.; Uuemaa, E.; Nõges, T.; Kutser, T. Estimation of the Biogeochemical and Physical Properties of Lakes Based on Remote Sensing and Artificial Intelligence Applications. Remote Sens. 2024, 16, 464. https://doi.org/10.3390/rs16030464
Toming K, Liu H, Soomets T, Uuemaa E, Nõges T, Kutser T. Estimation of the Biogeochemical and Physical Properties of Lakes Based on Remote Sensing and Artificial Intelligence Applications. Remote Sensing. 2024; 16(3):464. https://doi.org/10.3390/rs16030464
Chicago/Turabian StyleToming, Kaire, Hui Liu, Tuuli Soomets, Evelyn Uuemaa, Tiina Nõges, and Tiit Kutser. 2024. "Estimation of the Biogeochemical and Physical Properties of Lakes Based on Remote Sensing and Artificial Intelligence Applications" Remote Sensing 16, no. 3: 464. https://doi.org/10.3390/rs16030464
APA StyleToming, K., Liu, H., Soomets, T., Uuemaa, E., Nõges, T., & Kutser, T. (2024). Estimation of the Biogeochemical and Physical Properties of Lakes Based on Remote Sensing and Artificial Intelligence Applications. Remote Sensing, 16(3), 464. https://doi.org/10.3390/rs16030464






