Woody Biomass Estimation in a Southwestern U.S. Juniper Savanna Using LiDAR-Derived Clumped Tree Segmentation and Existing Allometries
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Field Measurements
2.3. Airborne LiDAR Data
2.4. Canopy Segmentation and Statistics
2.5. Validation of Clump Level Allometries
2.6. Uncertainty Estimation
3. Results
3.1. Field Measurements
3.2. LiDAR Segmentation
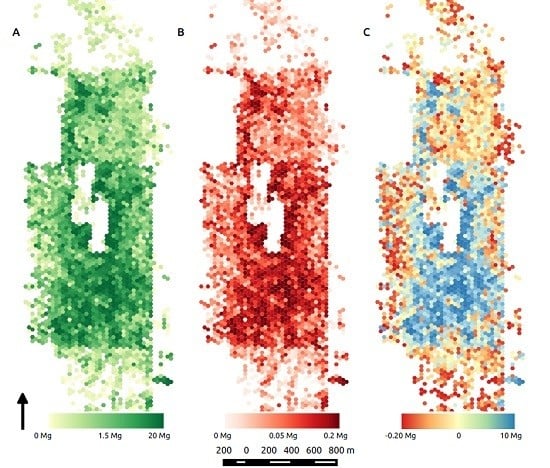
3.3. Segmentation-Derived Biomass and Uncertainty
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Savage, V.M.; Bentley, L.P.; Enquist, B.J.; Sperry, J.S.; Smith, D.D.; Reich, P.B.; von Allmen, E.I. Hydraulic trade-offs and space filling enable better predictions of vascular structure and function in plants. PNAS 2010, 107, 22722–22727. [Google Scholar] [CrossRef] [PubMed]
- West, G.B.; Enquist, B.J.; Brown, J.H. A general quantitative theory of forest structure and dynamics. PNAS 2009, 106, 7040–7045. [Google Scholar] [CrossRef] [PubMed]
- Swetnam, T.L.; Falk, D.A.; Lynch, A.M.; Yool, S.R. Estimating individual tree mid- and understory rank-size distributions from airborne laser scanning in semi-arid forests. Forest Ecol. Manag. 2014, 330, 271–282. [Google Scholar] [CrossRef]
- Poulter, B.; Frank, D.; Ciais, P.; Myneni, R.B.; Andela, N.; Bi, J.; Broquet, G.; Canadell, J.G.; Chevallier, F.; Liu, Y.Y.; et al. Contribution of semi-arid ecosystems to interannual variability of the global carbon cycle. Nature 2014, 509, 600–603. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, H.K.; Brown, S.; Niles, J.O.; Foley, J.A. Monitoring and estimating tropical forest carbon stocks: Making REDD a reality. Environ. Res. Lett. 2007, 2, 045023. [Google Scholar] [CrossRef]
- Jenkins, J.; Chojnacky, D.; Heath, L.; Birdsey, R. Comprehensive Database of Diameter-Based Biomass Regressions for North American Tree Species. Available online: http://svinet2.fs.fed.us/ne/durham/4104/papers/ne_gtr319_jenkins_and_others.pdf (assessed on 18 May 2016).
- Chojnacky, D.C.; Heath, L.S.; Jenkins, J.C. Updated generalized biomass equations for North American tree species. Forestry 2013, 87, 129–151. [Google Scholar] [CrossRef]
- Colgan, M.S.; Asner, G.P.; Swemmer, T. Harvesting tree biomass at the stand level to assess the accuracy of field and airborne biomass estimation in Savannas. Ecol. Appl. 2013, 23, 1170–1184. [Google Scholar] [CrossRef] [PubMed]
- Goetz, S.J.; Baccini, A.; Laporte, N.; Johns, T.; Walker, W.; Kellendorfer, J.; Houghton, R.; Sun, M. Mapping and monitoring carbon stocks with satellite observations: A comparison of methods. Carbon Balance Manag. 2009, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.; Potapov, P.; Moore, R.; Hancher, M.; Turubanova, S.; Tyukavina, A.; Thau, D.; Stehman, S.; Goetz, S.; Loveland, T.; et al. High-resolution global maps of 21st century forest cover change. Science 2013, 342, 6160. [Google Scholar] [CrossRef] [PubMed]
- Krofcheck, D.J.; Eitel, J.U.H.; Vierling, L.A.; Schulthess, U.; Hilton, T.M.; Dettweiler-Robinson, E.; Litvak, M.E. Detecting mortality induced structural and functional changes in a piñon-juniper woodland using Landsat and RapidEye time series. Remote Sens. Environ. 2014, 151, 102–113. [Google Scholar] [CrossRef]
- Mascaro, J.; Detto, M.; Asner, G.P.; Muller-Landau, H.C. Evaluating uncertainty in mapping forest carbon with airborne LiDAR. Remote Sens. Environ. 2011. [Google Scholar] [CrossRef]
- Eckert, S. Improved forest biomass and carbon estimations using texture measures from WorldView-2 satellite data. Remote Sens. 2011, 4, 810–829. [Google Scholar] [CrossRef]
- Asner, G.P.; Archer, S.; Hughes, R.F.; Ansley, R.J.; Wessman, C.A. Net changes in regional woody vegetation cover and carbon storage in Texas Drylands, 1937–1999. Glob. Chang. Biol. 2003, 9, 316–335. [Google Scholar] [CrossRef]
- Browning, D.M.; Archer, S.R.; Asner, G.P.; McClaran, M.P.; Wessman, C.A. Woody plants in grasslands: post-encroachment stand dynamics. Ecol. Appl. 2008, 18, 928–944. [Google Scholar] [CrossRef] [PubMed]
- Mirik, M.; Chaudhuri, S.; Surber, B.; Ale, S.; Ansley, R. Evaluating Biomass of Juniper Trees (Juniperus pinchotii) from Imagery-Derived Canopy Area Using the Support Vector Machine Classifier. Adv. Remote Sens. 2013, 2, 181–192. [Google Scholar] [CrossRef]
- Means, J.E.; Acker, S.A.; Fitt, B.J.; Renslow, M.; Emerson, L.; Hendrix, C.J. Predicting forest stand characteristics with airborne scanning LiDAR. Photogramm. Eng. Remote Sens. 2000, 66, 1367–1371. [Google Scholar]
- Næsset, E.; Gobakken, T. Estimation of above- and below-ground biomass across regions of the boreal forest zone using airborne laser. Remote Sens. Environ. 2008, 112, 3079–3090. [Google Scholar] [CrossRef]
- Packalen, P.; Vauhkonen, J.; Kallio, E.; Peuhkurinen, J.; Pitkanenm, J.; Pippuri, I.; Strunk, J.; Maltamo, M. Predicting the spatial pattern of trees by airborne laser scanning. Int. J. Remote Sens. 2013, 34, 5145–5165. [Google Scholar] [CrossRef]
- Swetnam, T.L.; Falk, D.A. Application of metabolic scaling theory to reduce error in local maxima tree segmentation from aerial LiDAR. Forest Ecol. Manag. 2014, 323, 158–167. [Google Scholar] [CrossRef]
- Sankey, T.T.; Glenn, N.; Ehinger, S.; Boehm, A.; Hardegree, S. Characterizing western juniper expansion via a fusion of Landsat 5 Thematic Mapper and LiDAR data. Rangel. Ecol. Manag. 2010, 63, 514–523. [Google Scholar] [CrossRef]
- Sankey, T.; and Glenn, N. Landsat-5 TM and LiDAR fusion for sub-pixel juniper tree cover estimates in a western rangeland. Photogr. Eng. Rempte Sens. 2011, 77, 1241–1248. [Google Scholar] [CrossRef]
- Wessels, K.J.; Erasmus, B.F.N.; Colgan, M.; Asner, G.P.; Mathieu, R.; Twine, W.; Van Aardt, J.A.N.; Smit, I. Impacts of communal fuelwood extraction on LiDAR-estimated biomass patterns of savanna woodlands. Int. Geosci. Remote Sens. 2012, 1676, 1679. [Google Scholar]
- Sankey, T.; Shrestha, R.; Sankey, J.B.; Hardegree, S.; Strand, E. LiDAR-derived estimate and uncertainty of carbon sink in successional phases of woody encroachment. J. Geophys. Res. Biogeosci. 2013, 118. [Google Scholar] [CrossRef]
- Swetnam, T.W.; Allen, C.D.; Betancourt, J.L. Applied historical ecology: Using the past to manage for the future. Ecol. Appl. 1999, 9, 1189–1206. [Google Scholar] [CrossRef]
- Miller, R.F.; Tausch, R.J. The role of fire in juniper and pinyon woodlands: A descriptive analysis. In Proceedings of the Invasive Species Workshop: Fire Conference 2000: The First National Congress on Fire Ecology, Prevention, and Management, San Diego, CA, USA, 27 November–1 December 2000.
- Allen, C.D.; Macalady, A.K.; Chenchouni, H.; Bachelet, D.; McDowell, N.; Vennetier, M.; Kitzberger, T.; Rigling, A.; Breshears, D.D.; Gonzalez, P.; et al. A global overview of drought and heat-induced tree mortality reveals emerging climate change risks for forests. Forest Ecol. Manag. 2010, 259, 660–684. [Google Scholar] [CrossRef]
- Breshears, D.; Rich, P. Overstory-imposed heterogeneity in solar radiation and soil moisture in a semiarid woodland. Ecol. Appl. 1997, 4, 1201–1215. [Google Scholar] [CrossRef]
- Clifford, M.; Rocca, M.; Delph, R. Drought induced tree mortality and ensuing Bark beetle outbreaks in Southwestern pinyon-juniper woodlands. In USDA Forest Service Proceedings; US Forest Service: Washington, DC, USA, 2008. [Google Scholar]
- Clifford, M.; Royer, P.; Cobb, N. Precipitation thresholds and drought-induced tree die-off: Insights from patterns of Pinus edulis mortality along an environmental stress gradient. New Phytol. 2013, 200, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Anderson-Teixeira, K.J.; Delong, J.P.; Fox, A.M.; Brese, D.A.; Litvak, M.E. Differential responses of production and respiration to temperature and moisture drive the carbon balance across a climatic gradient in New Mexico. Glob. Chang. Biol. 2011, 17, 410–424. [Google Scholar] [CrossRef]
- Grier, C.C.; Elliott, K.J.; Mccullough, D.G. Biomass distribution and productivity of Pinus edulis-Juniperus monosperma woodlands of north-central Arizona. Forest Ecol. Manag. 1992, 50, 331–350. [Google Scholar] [CrossRef]
- Stevenson, T.H.; Magruder, L.A.; Neuenschwander, A.L.; Bradford, B. Automated bare earth extraction technique for complex topography in light detection and ranging surveys. J. Appl. Remote Sens. 2013, 7, 073560. [Google Scholar] [CrossRef]
- Gonzalez, P.; Asner, G.P.; Battles, J.J.; Lefsky, M.A.; Waring, K.M.; Palace, M. Forest carbon densities and uncertainties from LiDAR, Quickbird, and field measurements in California. Remote Sens. Environ. 2010, 114, 1561–1575. [Google Scholar] [CrossRef]
- Weltz, M.A.; Ritchie, C.; Fox, H.D. Comparison of laser and field measurements of vegetation height and canopy cover watershed precision vegetation properties height and canopy. Water Resour. Res. 1994, 30, 1311–1319. [Google Scholar] [CrossRef]
- Colgan, M.S.; Swemmer, T.; Asner, G.P. Structural relationships between form factor, wood density, and biomass in African savanna woodlands. Trees 2013, 28, 91–102. [Google Scholar] [CrossRef]
- Fernandez, D.P.; Neff, J.C.; Huang, C.-Y.; Asner, G.P.; Barger, N.N. Twentieth century carbon stock changes related to Piñon-Juniper expansion into a black sagebrush community. Carbon Balance Manag. 2013, 8, 8. [Google Scholar] [CrossRef] [PubMed]










| Segment Statistic | Min | Max | Mean | Std |
|---|---|---|---|---|
| Max Elevation (m) | 1.55 | 8.31 | 4.14 | 0.76 |
| Min Elevation (m) | 0 | 3.44 | 0.3 | 0.09 |
| Avg Elevation (m) | 0.57 | 5.67 | 2.12 | 0.37 |
| Med Elevation (m) | 0.22 | 7.36 | 2.28 | 1.06 |
| Std Elevation (m) | 0.28 | 2.38 | 1.02 | 0.23 |
| RMS Elevation (m) | 0.64 | 5.77 | 2.36 | 0.4 |
| Perimeter (m) | 10.97 | 290.45 | 42.08 | 24.43 |
| Projected Area (m2) | 12.67 | 2158.13 | 94.35 | 117.73 |
| Volume (m3) | 41.33 | 5302.16 | 399.52 | 611.66 |
| Density (unitless) | 0.08 | 0.99 | 0.44 | 0.21 |
| Closure (unitless) | 0.17 | 1 | 0.75 | 0.15 |
| ESD = b + α (x1) + β (x2) + γ (x1 × x2) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Model | x1 | x2 | b | α | β | γ | R2ADJ | p |
| HCHM | CHM Height | - | −3.05 | 14.04 | - | - | 0.22 | 0.046 |
| HTEX | TEX Height | - | −10.07 | 15.73 | - | - | 0.24 | 0.038 |
| Vol | Volume | - | 31.43 | 0.08 | - | - | 0.89 | <0.0001 |
| VolD | Volume | Density | 6.41 | 0.16 | 42.17 | −0.15 | 0.91 | <0.0001 |
| VolC | Volume | Closure | 11.21 | 11.21 | 26.65 | −0.18 | 0.89 | <0.0001 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krofcheck, D.J.; Litvak, M.E.; Lippitt, C.D.; Neuenschwander, A. Woody Biomass Estimation in a Southwestern U.S. Juniper Savanna Using LiDAR-Derived Clumped Tree Segmentation and Existing Allometries. Remote Sens. 2016, 8, 453. https://doi.org/10.3390/rs8060453
Krofcheck DJ, Litvak ME, Lippitt CD, Neuenschwander A. Woody Biomass Estimation in a Southwestern U.S. Juniper Savanna Using LiDAR-Derived Clumped Tree Segmentation and Existing Allometries. Remote Sensing. 2016; 8(6):453. https://doi.org/10.3390/rs8060453
Chicago/Turabian StyleKrofcheck, Dan J., Marcy E. Litvak, Christopher D. Lippitt, and Amy Neuenschwander. 2016. "Woody Biomass Estimation in a Southwestern U.S. Juniper Savanna Using LiDAR-Derived Clumped Tree Segmentation and Existing Allometries" Remote Sensing 8, no. 6: 453. https://doi.org/10.3390/rs8060453
APA StyleKrofcheck, D. J., Litvak, M. E., Lippitt, C. D., & Neuenschwander, A. (2016). Woody Biomass Estimation in a Southwestern U.S. Juniper Savanna Using LiDAR-Derived Clumped Tree Segmentation and Existing Allometries. Remote Sensing, 8(6), 453. https://doi.org/10.3390/rs8060453