Chrysanthemum Leaf Ethanol Extract Prevents Obesity and Metabolic Disease in Diet-Induced Obese Mice via Lipid Mobilization in White Adipose Tissue
Abstract
:1. Introduction
2. Material and Methods
2.1. Preparation of Chrysanthemum morifolium Ramat. Ethanol Extract (CLE)
2.2. Experimental Design
2.3. Measurement of Calorimetry and Oxygen Consumption
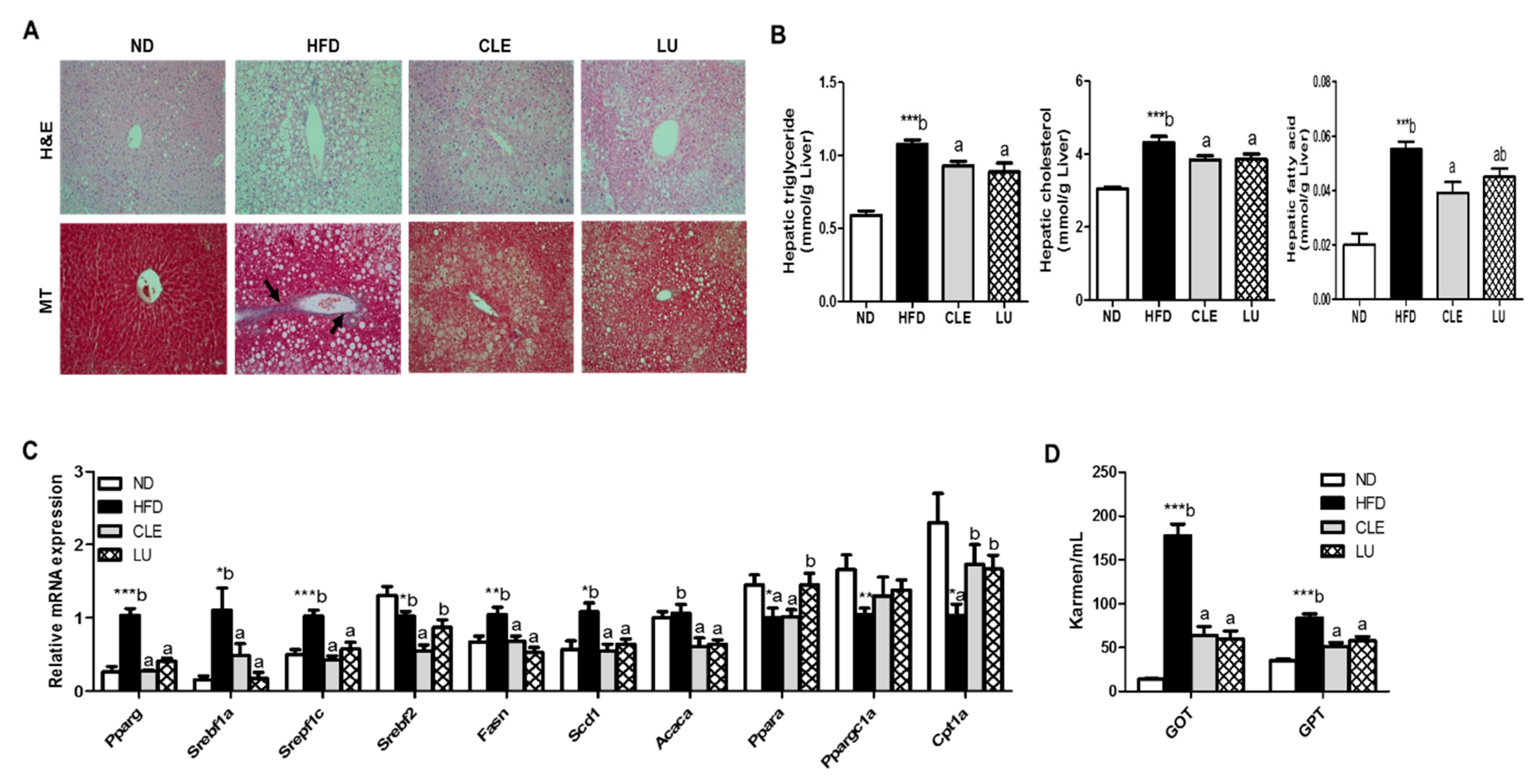
2.4. Plasma, Hepatic, and WAT Lipid Contents
2.5. Plasma Biochemical Profiles, Fasting Blood Glucose, Homeostatic Model Assessment of Insulin Resistance (HOMA-IR), and Hepatic Glycogen
2.6. Plasma Glutamic Oxalacetic Transaminase (GOT) and Glutamic Pyruvic Transaminase (GPT) Activities Measurement
2.7. Histological and Immunohistochemistry Analyses
2.8. mRNA-Seq and Molecular Network Analysis
2.9. Gene Expression Analysis
2.10. Statistical Analysis
3. Results
3.1. CLE Supplement Suppresses Increase in Body Weight Gain and Fat Deposition
3.2. CLE Supplement Regulates Lipid Metabolism and EE
3.3. CLE Supplement Prevents Dyslipidemia
3.4. CLE Supplement Suppresses Hepatic Steatosis and Hepatic Toxicity
3.5. CLE Supplement Improves Insulin Resistance
3.6. CLE Supplement Reduces Inflammatory Markers
3.7. CLE Supplement Alters the Transcriptional Response in eWAT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ADPOQ | adiponectin |
| ADIPOR | adiponectin receptor |
| ACADL | acyl-Coenzyme A dehydrogenase, long-chain |
| ACADM | acyl-Coenzyme A dehydrogenase, medium chain |
| ACC | acetyl-CoA carboxylase |
| ACOX1 | acyl-Coenzyme A oxidase 1 |
| ADRB3 | adrenergic receptor, beta 3 |
| BAT | brown adipose tissue |
| BMP | bone morphogenetic protein |
| C | cholesterol |
| CIDEA | cell death-inducing DNA fragmentation factor, alpha subunit-like effector A |
| CLE | chrysanthemum leaf ethanol extract |
| COX8B | cytochrome c oxidase subunit VIIIb |
| CPT2 | carnitine palmitoyltransferase 2 |
| FAO | fatty acid oxidation |
| FASN | fatty acid synthase |
| FFA | free fatty acid |
| FGFf21 | fibroblast growth factor 21 |
| GOT | glutamic oxalacetic transaminase |
| GPT | glutamic pyruvic transaminase |
| HDL | high-density lipoprotein |
| HFD | high-fat diet |
| HOMA-IR | homeostatic index of insulin resistance |
| HTR | ratio of HDL-C to Total-C |
| IL | Interleukin |
| IPA | ingenuity pathway analysis |
| LD | lipid droplet |
| LU | luteolin |
| MCP-1 | monocyte chemotactic protein 1 |
| MIP-1β | macrophage inflammatory protein-1β |
| ND | normal diet |
| PGC1 | peroxisome proliferative activated receptor, gamma, coactivator 1 |
| PPAR | peroxisome proliferator-activated receptor |
| SCD1 | stearoyl-CoA desaturase 1 |
| SREBPs | sterol regulatory element-binding proteins |
| TG | triglyceride |
| TLR | toll-like receptor |
| TNF-α | tumor necrosis factor-α |
| UCP1 | uncoupling protein 1 |
| VCO2 | carbon dioxide production |
| VLDL | very low density lipoprotein |
| VO2 | whole-body oxygen consumption |
| WAT | white adipose tissue |
References
- Coelho, M.; Oliveira, T.; Fernandes, R. Biochemistry of adipose tissue: An endocrine organ. Archives of medical science. Arch. Med. Sci. 2013, 9, 19–200. [Google Scholar] [CrossRef]
- Rodríguez, A.; Ezquerro, S.; Méndez-Giménez, L.; Becerril, S.; Frühbeck, G. Revisiting the adipocyte: A model for integration of cytokine signaling in the regulation of energy metabolism. Am. J. Physiol. Endocrinol. Metab. 2015, 309, E691–E714. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Ji, Y.; Kersten, S.; Qi, L. Mechanisms of inflammatory responses in obese adipose tissue. Annu. Rev. Nutr. 2012, 32, 261–286. [Google Scholar] [CrossRef] [PubMed]
- Tataranni, P.A.; Ortega, E. A burning question: Does an adipokine-induced activation of the immune system mediate the effect of overnutrition on type 2 diabetes? Diabetes 2005, 54, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Trayhurn, P.; Wood, I.S. Adipokines: Inflammation and the pleiotropic role of white adipose tissue. Br. J. Nutr. 2004, 92, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Czech, M.P. Insulin action and resistance in obesity and type 2 diabetes. Nat. Med. 2017, 23, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Guilherme, A.; Virbasius, J.V.; Puri, V.; Czech, M.P. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat. Rev. Mol. Cell Biol. 2008, 9, 367–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, R.; Jeong, T.-S.; Kim, Y.J.; Choi, J.-Y.; Cho, S.-J.; Kwon, E.-Y.; Jung, U.J.; Ji, H.-S.; Shin, D.-H.; Choi, M.-S. Beneficial Effects of Pterocarpan-High Soybean Leaf Extract on Metabolic Syndrome in Overweight and Obese Korean Subjects: Randomized Controlled Trial. Nutrients 2016, 8, 734. [Google Scholar] [CrossRef]
- Bjørndal, B.; Burri, L.; Staalesen, V.; Skorve, J.; Berge, R.K. Different adipose depots: Their role in the development of metabolic syndrome and mitochondrial response to hypolipidemic agents. J. Obes. 2011, 2011. [Google Scholar] [CrossRef]
- Shimomura, I.; Bashmakov, Y.; Horton, J.D. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J. Biol. Chem. 1999, 274, 30028–30032. [Google Scholar] [CrossRef]
- Choi, S.H.; Ginsberg, H.N. Increased very low density lipoprotein (VLDL) secretion, hepatic steatosis, and insulin resistance. Trends Endocrinol. Metab. 2011, 22, 353–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tseng, Y.-H.; Cypess, A.M.; Kahn, C.R. Cellular bioenergetics as a target for obesity therapy. Nat. Rev. Drug Discov. 2019, 9, 465–482. [Google Scholar] [CrossRef] [PubMed]
- Talevi, A. Multi-target pharmacology: Possibilities and limitations of the “skeleton key approach” from a medicinal chemist perspective. Front. Pharmacol. 2015, 6, 205. [Google Scholar] [CrossRef] [PubMed]
- Lii, C.-K.; Lei, Y.-P.; Yao, H.-T.; Hsieh, Y.-S.; Tsai, C.-W.; Liu, K.-L.; Chen, H.-W. Chrysanthemum morifolium Ramat. reduces the oxidized LDL-induced expression of intercellular adhesion molecule-1 and E-selectin in human umbilical vein endothelial cells. J. Ethnopharmacol. 2010, 128, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, J.; Tadaishi, M.; Yamane, T.; Oishi, Y.; Shimizu, M.; Kobayashi-Hattori, K. Hot water extracts of edible Chrysanthemum morifolium Ramat. exert antidiabetic effects in obese diabetic KK-Ay mice. Biosci. Biotechnol. Biochem. 2015, 79, 1147–1154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, L.-Z.; Harnly, J.M. Identification of the phenolic components of chrysanthemum flower (Chrysanthemum morifolium Ramat). Food Chem. 2010, 120, 319–326. [Google Scholar] [CrossRef]
- Kwon, E.-Y.; Jung, U.J.; Park, T.; Yun, J.W.; Choi, M.-S. Luteolin attenuates hepatic steatosis and insulin resistance through the interplay between the liver and adipose tissue in diet-induced obese mice. Diabetes 2015, 64, 1658–1669. [Google Scholar] [CrossRef] [PubMed]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar]
- Leng, N.; Dawson, J.A.; Thomson, J.A.; Ruotti, V.; Rissman, A.I.; Smits, B.M.; Haag, J.D.; Gould, M.N.; Stewart, R.M.; Kendziorski, C. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 2013, 29, 1035–1043. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Hegarty, B.D.; Furler, S.M.; Oakes, N.D.; Kraegen, E.W.; Cooney, G.J. Peroxisome proliferator-activated receptor (PPAR) activation induces tissue-specific effects on fatty acid uptake and metabolism in vivo—A study using the novel PPARα/γ agonist tesaglitazar. Endocrinology 2004, 145, 3158–3164. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Curto, E.; Milligan, G. Metabolism meets immunity: The role of free fatty acid receptors in the immune system. Biochem. Pharmacol. 2016, 114, 3–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pærregaard, S.I.; Agerholm, M.; Serup, A.K.; Ma, T.; Kiens, B.; Madsen, L.; Kristiansen, K.; Jensen, B.A.H. FFAR4 (GPR120) signaling is not required for anti-inflammatory and insulin-sensitizing effects of Omega-3 fatty acids. Mediat. Inflamm. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Boden, G. Obesity and free fatty acids. Endocrinol. Metab. Clin. N. Am. 2008, 37, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Postic, C.; Girard, J. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: Lessons from genetically engineered mice. J. Clin. Investig. 2008, 118, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Ohtake, T.; Motomura, W.; Takahashi, N.; Hosoki, Y.; Miyoshi, S.; Suzuki, Y.; Saito, H.; Kohgo, Y.; Okumura, T. Increased expression of PPARγ in high fat diet-induced liver steatosis in mice. Biochem. Biophys. Res. Commun. 2005, 336, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Wahli, W.; Michalik, L. PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol. Metab. 2012, 23, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Gavrilova, O.; Haluzik, M.; Matsusue, K.; Cutson, J.J.; Johnson, L.; Dietz, K.R.; Nicol, C.J.; Vinson, C.; Gonzalez, F.J.; Reitman, M.L. Liver peroxisome proliferator-activated receptor γ contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J. Biol. Chem. 2003, 278, 34268–34276. [Google Scholar] [CrossRef]
- Xu, N.; Zhang, L.; Dong, J.; Zhang, X.; Chen, Y.G.; Bao, B.; Liu, J. Low-dose diet supplement of a natural flavonoid, luteolin, ameliorates diet-induced obesity and insulin resistance in mice. Mol. Nutr. Food Res. 2014, 58, 1258–1268. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Jin, D.; Chen, X. Luteolin enhances insulin sensitivity via activation of PPARγ transcriptional activity in adipocytes. J. Nutr. Biochem. 2010, 21, 941–947. [Google Scholar] [CrossRef] [PubMed]
- Choe, S.S.; Huh, J.Y.; Hwang, I.J.; Kim, J.I.; Kim, J.B. Adipose tissue remodeling: Its role in energy metabolism and metabolic disorders. Front. Endocrinol. 2016, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Kadowaki, T. Adiponectin receptor as a key player in healthy longevity and obesity-related diseases. Cell Metab. 2013, 17, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Coppack, S.W. Pro-inflammatory cytokines and adipose tissue. Proc. Nutr. Soc. 2001, 60, 349–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chinetti, G.; Fruchart, J.-C.; Staels, B. Peroxisome proliferator-activated receptors (PPARs): Nuclear receptors at the crossroads between lipid metabolism and inflammation. Inflamm. Res. 2000, 49, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Glass, C.K. Nuclear receptors and inflammation control: Molecular mechanisms and pathophysiological relevance. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1542–1549. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.H.; Wang, Z.; Quispe, G.; Yanymee, N.; Lim, S.S.; Yu, J.M. Evaluation of Aldose Reductase, Protein Glycation, and Antioxidant Inhibitory Activities of Bioactive Flavonoids in Matricaria recutita L. and Their Structure-Activity Relationship. J. Diabetes Res. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Cao, J.; Feng, Q.; Peng, J.; Hu, Y. Roles of chlorogenic acid on regulating glucose and lipids metabolism: A review. Evid.-Based Complement. Altern. Med. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]






| Constituent | ND | HFD | CLE | LU |
|---|---|---|---|---|
| FFAs (mmol/L) | 0.69 ± 0.03 | 0.82 ± 0.06 * | 0.75 ± 0.04 # | 0.76 ± 0.05 |
| TGs (mmol/L) | 0.82 ± 0.06 | 0.98 ± 0.02 * | 1.05 ± 0.05 | 1.05 ± 0.06 |
| PLs (mg/dL) | 276.74 ± 9.61 | 376.23 ± 16.90 ***b | 329.75 ± 11.72 a | 327.28 ± 12.71 a |
| Total-C (mmol/L) | 3.95 ± 0.19 | 7.01 ± 0.38 ***b | 5.59 ± 0.25 a | 5.40 ± 0.25 a |
| HDL-C (mmol/L) | 1.56 ± 0.04 | 1.88 ± 0.04 ***a | 2.08 ± 0.05 b | 1.82 ± 0.05 a |
| Non-HDL-C (mmol/L) | 2.54 ± 0.15 | 5.48 ± 0.50 ***b | 3.75 ± 0.25 a | 4.23 ± 0.23 a |
| HTR | 39.90 ± 1.15 | 26.87 ± 1.60 ***a | 36.94 ± 2.11 b | 34.87 ± 1.78 b |
| Apo A1 (mg/dL) | 20.24 ± 0.27 | 19.18 ± 0.18 **a | 20.85 ± 0.20 b | 20.43 ± 0.27 b |
| Apo B (mg/dL) | 3.61 ± 0.76 | 5.88 ± 0.51 ***b | 3.71 ± 0.42 a | 4.08 ± 0.40 a |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ryu, R.; Kwon, E.-Y.; Choi, J.-Y.; Shon, J.C.; Liu, K.-H.; Choi, M.-S. Chrysanthemum Leaf Ethanol Extract Prevents Obesity and Metabolic Disease in Diet-Induced Obese Mice via Lipid Mobilization in White Adipose Tissue. Nutrients 2019, 11, 1347. https://doi.org/10.3390/nu11061347
Ryu R, Kwon E-Y, Choi J-Y, Shon JC, Liu K-H, Choi M-S. Chrysanthemum Leaf Ethanol Extract Prevents Obesity and Metabolic Disease in Diet-Induced Obese Mice via Lipid Mobilization in White Adipose Tissue. Nutrients. 2019; 11(6):1347. https://doi.org/10.3390/nu11061347
Chicago/Turabian StyleRyu, Ri, Eun-Young Kwon, Ji-Young Choi, Jong Cheol Shon, Kwang-Hyeon Liu, and Myung-Sook Choi. 2019. "Chrysanthemum Leaf Ethanol Extract Prevents Obesity and Metabolic Disease in Diet-Induced Obese Mice via Lipid Mobilization in White Adipose Tissue" Nutrients 11, no. 6: 1347. https://doi.org/10.3390/nu11061347
APA StyleRyu, R., Kwon, E.-Y., Choi, J.-Y., Shon, J. C., Liu, K.-H., & Choi, M.-S. (2019). Chrysanthemum Leaf Ethanol Extract Prevents Obesity and Metabolic Disease in Diet-Induced Obese Mice via Lipid Mobilization in White Adipose Tissue. Nutrients, 11(6), 1347. https://doi.org/10.3390/nu11061347






