Where Do We Stand in the Behavioral Pathogenesis of Inflammatory Bowel Disease? The Western Dietary Pattern and Microbiota—A Narrative Review
Abstract
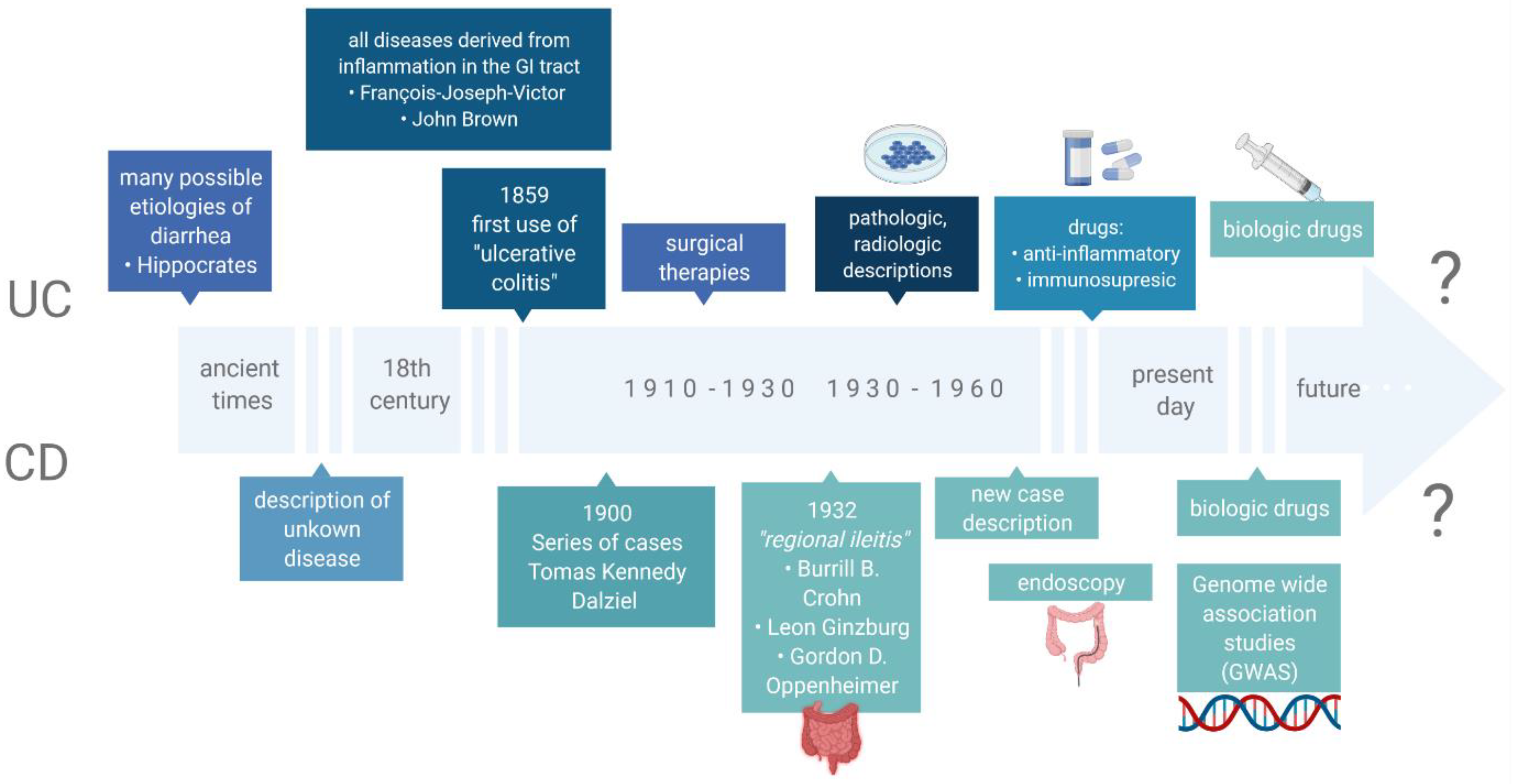
:1. Introduction
2. Inflammatory Bowel Disease
3. Risk Factors of IBD
3.1. Genetics
3.2. Immunology
3.3. Environment
3.4. Gut Microbiota
4. Characteristics of the Gut Microbial Community in IBD
4.1. Microbiota Composition
4.2. Genetic Identification of Microbiota
5. Nutrients of the Western Diet in the Course and Development of IBD
5.1. High Intake of Saturated Fatty Acid and Fast Food
5.2. Low Intake of Unsaturated Fatty Acid
5.3. High Intake of Carbohydrates, Sweets and Sweet Drinks
5.4. Low Intake of Fibre, Whole Grain, Vegetables and Fruits
6. Effects of the Western Diet on the Gut Microbiota Composition in IBD Patients
7. Is the Mediterranean Diet an Alternative to the Western Diet in IBD Patients?
8. Dietary Recommendations for IBD Patients That Are Beneficial for the Gut Microbiota
9. Summary and Conclusions
- The current theory regarding the pathogenesis of IBD states that enteritis results from an abnormal immune response in genetically predisposed individuals with coexisting disorders in the composition of the intestinal microbiota [193].
- The role of genetic predisposition in IBD is unquestionable, although it has not yet been fully defined. The current knowledge is incomplete with regard to the interdependencies and interactions of IBD factors, including genetics, their interactions and the ultimate impact on the human body.
- Environmental factors (including dietary habits, breastfeeding, air pollution and smoking) seem to be important in the development of IBD. However, further studies are necessary in order to fully understand these associations.
- Gut microbiota disbalance plays a role in IBD development. Additionally, the composition of microbiota may affect the course of the disease.
- There are a number of factors affecting both the risk and the course of inflammatory bowel disease. However, an increasing global prevalence of CD and UC is prompting further research aimed at gaining at better insight of these disease and improving patient care [118].
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mentella, M.C.; Scaldaferri, F.; Pizzoferrato, M.; Gasbarrini, A.; Miggiano, G.A.D. Nutrition, IBD and gut microbiota: A Review. Nutrients 2020, 12, 944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halme, L.; Paavola-Sakki, P.; Turunen, U.; Lappalainen, M.; Farkkila, M.; Kontula, K. Family and twin studies in inflammatory bowel disease. World J. Gastroenterol. 2006, 12, 3668–3672. [Google Scholar] [CrossRef]
- Khor, B.; Gardet, A.; Xavier, R.J. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011, 474, 307–317. [Google Scholar] [CrossRef] [Green Version]
- Dheer, R.; Santaolalla, R.; Davies, J.M.; Lang, J.K.; Phillips, M.C.; Pastorini, C.; Vazquez-Pertejo, M.T.; Abreu, M.T. Intestinal epithelial toll-like receptor 4 signaling affects epithelial function and colonic microbiota and promotes a risk for transmissible colitis. Infect. Immun. 2016, 84, 798–810. [Google Scholar] [CrossRef] [Green Version]
- Turpin, W.; Goethel, A.; Bedrani, L.; Croitoru Mdcm, K. Determinants of IBD heritability: Genes, bugs, and more. Inflamm. Bowel Dis. 2018, 24, 1133–1148. [Google Scholar] [CrossRef] [Green Version]
- Ko, Y.; Butcher, R.; Leong, R.W. Epidemiological studies of migration and environmental risk factors in the inflammatory bowel diseases. World J. Gastroenterol. 2014, 20, 1238–1247. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Adams, D.H.; Fava, F.; Hermes, G.D.A.; Hirschfield, G.M.; Hold, G.; Quraishi, M.N.; Kinross, J.; Smidt, H.; Tuohy, K.M.; et al. The gut microbiota and host health: A new clinical frontier. Gut 2016, 65, 330–339. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Butcher, J.; Mack, D.; Stintzi, A. Functional impacts of the intestinal microbiome in the pathogenesis of inflammatory bowel disease. Inflamm. Bowel Dis. 2015, 21, 139–153. [Google Scholar] [CrossRef]
- Kostic, A.D.; Xavier, R.J.; Gevers, D. The microbiome in inflammatory bowel disease: Current status and the future ahead. Gastroenterology 2014, 146, 1489–1499. [Google Scholar] [CrossRef] [Green Version]
- Sartor, R.B. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: Antibiotics, probiotics, and prebiotics. Gastroenterology 2004, 126, 1620–1633. [Google Scholar] [CrossRef] [Green Version]
- Mulder, D.J.; Noble, A.J.; Justinich, C.J.; Duffin, J.M. A tale of two diseases: The history of inflammatory bowel disease. J. Crohn’s Colitis 2014, 8, 341–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalili, H.; Håkansson, N.; Chan, S.S.; Chen, Y.; Lochhead, P.; Ludvigsson, J.F.; Chan, A.T.; Hart, A.R.; Olén, O.; Wolk, A. Adherence to a mediterranean diet is associated with a lower risk of later-onset Crohn’s disease: Results from two large prospective cohort studies. Gut 2020, 69, 1637–1644. [Google Scholar] [CrossRef]
- de Vries, J.H.M.; Dijkhuizen, M.; Tap, P.; Witteman, B.J.M. Patient’s dietary beliefs and behaviours in inflammatory bowel disease. Dig. Dis. 2019, 37, 131–139. [Google Scholar] [CrossRef]
- Freeman, H.J. Familial Crohn’s disease in single or multiple first-degree relatives. J. Clin. Gastroenterol. 2002, 35, 9–13. [Google Scholar] [CrossRef]
- Duerr, R.H. The genetics of inflammatory bowel disease. Gastroenterol. Clin. N. Am. 2002, 31, 63–76. [Google Scholar] [CrossRef]
- Leedham, S.J.; Jankowski, J.A.; Wright, N.A.; Tomlinson, I.P.M. Genetics of inflammatory bowel disease and associated cancers. Curr. Colorectal Cancer Rep. 2006, 2, 191–199. [Google Scholar] [CrossRef]
- Tysk, C.; Lindberg, E.; Järnerot, G.; Flodérus-Myrhed, B. Ulcerative colitis and crohn’s disease in an unselected population of monozygotic and dizygotic twins. A Study of heritability and the influence of smoking. Gut 1988, 29, 990–996. [Google Scholar] [CrossRef] [Green Version]
- Orholm, M.; Binder, V.; Sørensen, T.I.; Rasmussen, L.P.; Kyvik, K.O. Concordance of inflammatory bowel disease among danish twins. Results of a nationwide study. Scand. J. Gastroenterol. 2000, 35, 1075–1081. [Google Scholar] [CrossRef]
- Brant, S.R. Update on the heritability of inflammatory bowel disease: The importance of twin studies. Inflamm. Bowel Dis. 2011, 17, 1–5. [Google Scholar] [CrossRef]
- Hugot, J.P.; Laurent-Puig, P.; Gower-Rousseau, C.; Olson, J.M.; Lee, J.C.; Beaugerie, L.; Naom, I.; Dupas, J.L.; Van Gossum, A.; Orholm, M.; et al. Mapping of a susceptibility locus for Crohn’s disease on chromosome 16. Nature 1996, 379, 821–823. [Google Scholar] [CrossRef]
- Hugot, J.P.; Chamaillard, M.; Zouali, H.; Lesage, S.; Cézard, J.P.; Belaiche, J.; Almer, S.; Tysk, C.; O’Morain, C.A.; Gassull, M.; et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001, 411, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Ogura, Y.; Bonen, D.K.; Inohara, N.; Nicolae, D.L.; Chen, F.F.; Ramos, R.; Britton, H.; Moran, T.; Karaliuskas, R.; Duerr, R.H.; et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 2001, 411, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Brant, S.R.; Wang, M.-H.; Rawsthorne, P.; Sargent, M.; Datta, L.W.; Nouvet, F.; Shugart, Y.Y.; Bernstein, C.N. A population-based case-control study of CARD15 and other risk factors in Crohn’s disease and ulcerative colitis. Am. J. Gastroenterol. 2007, 102, 313–323. [Google Scholar] [CrossRef]
- Lesage, S.; Zouali, H.; Cézard, J.-P.; Colombel, J.-F.; Belaiche, J.; Almer, S.; Tysk, C.; O’Morain, C.; Gassull, M.; Binder, V.; et al. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am. J. Hum. Genet. 2002, 70, 845–857. [Google Scholar] [CrossRef] [Green Version]
- Horowitz, J.E.; Warner, N.; Staples, J.; Crowley, E.; Gosalia, N.; Murchie, R.; Van Hout, C.; Fiedler, K.; Welch, G.; King, A.K.; et al. Mutation spectrum of NOD2 reveals recessive inheritance as a main driver of early onset Crohn’s disease. Sci. Rep. 2021, 11, 5595. [Google Scholar] [CrossRef]
- Kugathasan, S.; Loizides, A.; Babusukumar, U.; McGuire, E.; Wang, T.; Hooper, P.; Nebel, J.; Kofman, G.; Noel, R.; Broeckel, U.; et al. Comparative phenotypic and CARD15 mutational analysis among African American, Hispanic, and white children with Crohn’s disease. Inflamm. Bowel Dis. 2005, 11, 631–638. [Google Scholar] [CrossRef]
- Brant, S.R.; Okou, D.T.; Simpson, C.L.; Cutler, D.J.; Haritunians, T.; Bradfield, J.P.; Chopra, P.; Prince, J.; Begum, F.; Kumar, A.; et al. Genome-wide association study identifies African-specific susceptibility loci in African Americans with inflammatory bowel disease. Gastroenterology 2017, 152, 206–217. [Google Scholar] [CrossRef] [Green Version]
- Jostins, L.; Ripke, S.; Weersma, R.K.; Duerr, R.H.; McGovern, D.P.; Hui, K.Y.; Lee, J.C.; Schumm, L.P.; Sharma, Y.; Anderson, C.A.; et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012, 491, 119–124. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Tian, L.; Sleiman, P.; Ghosh, S.; Hakonarson, H. International IBD genetics consortium Bayesian analysis of genome-wide inflammatory bowel disease data sets reveals new risk loci. Eur. J. Hum. Genet. EJHG 2018, 26, 265–274. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.Z.; van Sommeren, S.; Huang, H.; Ng, S.C.; Alberts, R.; Takahashi, A.; Ripke, S.; Lee, J.C.; Jostins, L.; Shah, T.; et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015, 47, 979–986. [Google Scholar] [CrossRef]
- Frenkel, S.; Bernstein, C.N.; Sargent, M.; Kuang, Q.; Jiang, W.; Wei, J.; Thiruvahindrapuram, B.; Spriggs, E.; Scherer, S.W.; Hu, P. Genome-wide analysis identifies rare copy number variations associated with inflammatory bowel disease. PLoS ONE 2019, 14, e0217846. [Google Scholar] [CrossRef] [PubMed]
- McDermott, E.; Ryan, E.J.; Tosetto, M.; Gibson, D.; Burrage, J.; Keegan, D.; Byrne, K.; Crowe, E.; Sexton, G.; Malone, K.; et al. DNA methylation profiling in inflammatory bowel disease provides new insights into disease pathogenesis. J. Crohn’s Colitis 2016, 10, 77–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.-O.; Park, D.-I.; Han, Y.K.; Kang, K.; Park, S.-G.; Park, H.R.; Yi, J.M. Genome-wide analysis of the DNA methylation profile identifies the fragile histidine triad (FHIT) gene as a new promising biomarker of Crohn’s disease. J. Clin. Med. 2020, 9, 1338. [Google Scholar] [CrossRef] [PubMed]
- Kalla, R.; Adams, A.T.; Ventham, N.T.; Kennedy, N.A.; White, R.; Clarke, C.; Ivens, A.; Bergemalm, D.; Vatn, S.; Lopez-Jimena, B.; et al. Whole blood profiling of T-cell-derived microRNA allows the development of prognostic models in inflammatory bowel disease. J. Crohn’s Colitis 2020, 14, 1724–1733. [Google Scholar] [CrossRef]
- Thomas, J.P.; Ölbei, M.; Brooks-Warburton, J.; Korcsmaros, T.; Modos, D. Analysing MiRNA-target gene networks in inflammatory bowel disease and other complex diseases using transcriptomic data. Genes 2022, 13, 370. [Google Scholar] [CrossRef]
- Stolfi, C.; Maresca, C.; Monteleone, G.; Laudisi, F. Implication of intestinal barrier dysfunction in gut dysbiosis and diseases. Biomedicines 2022, 10, 289. [Google Scholar] [CrossRef]
- Guo, X.-K.; Ou, J.; Liang, S.; Zhou, X.; Hu, X. Epithelial Hes1 maintains gut homeostasis by preventing microbial dysbiosis. Mucosal Immunol. 2018, 11, 716–726. [Google Scholar] [CrossRef]
- Liu, X.; Lu, J.; Liu, Z.; Zhao, J.; Sun, H.; Wu, N.; Liu, H.; Liu, W.; Hu, Z.; Meng, G.; et al. Intestinal epithelial Cell–Derived LKB1 suppresses colitogenic microbiota. J. Immunol. 2018, 200, 1889–1900. [Google Scholar] [CrossRef]
- Laudisi, F.; Stolfi, C.; Bevivino, G.; Maresca, C.; Franzè, E.; Troncone, E.; Lolli, E.; Marafini, I.; Pietrucci, D.; Teofani, A.; et al. GATA6 deficiency leads to epithelial barrier dysfunction and enhances susceptibility to gut inflammation. J. Crohn’s Colitis 2022, 16, 301–311. [Google Scholar] [CrossRef]
- Riba, A.; Olier, M.; Lacroix-Lamandé, S.; Lencina, C.; Bacquié, V.; Harkat, C.; Gillet, M.; Baron, M.; Sommer, C.; Mallet, V.; et al. Paneth cell defects induce microbiota dysbiosis in mice and promote visceral hypersensitivity. Gastroenterology 2017, 153, 1594–1606. [Google Scholar] [CrossRef] [Green Version]
- Zeissig, S.; Burgel, N.; Gunzel, D.; Richter, J.; Mankertz, J.; Wahnschaffe, U.; Kroesen, A.J.; Zeitz, M.; Fromm, M.; Schulzke, J.-D. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn’s disease. Gut 2007, 56, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Kucharzik, T.; Walsh, S.V.; Chen, J.; Parkos, C.A.; Nusrat, A. Neutrophil transmigration in inflammatory bowel disease is associated with differential expression of epithelial intercellular junction proteins. Am. J. Pathol. 2001, 159, 2001–2009. [Google Scholar] [CrossRef] [Green Version]
- Laukoetter, M.G.; Nava, P.; Lee, W.Y.; Severson, E.A.; Capaldo, C.T.; Babbin, B.A.; Williams, I.R.; Koval, M.; Peatman, E.; Campbell, J.A.; et al. JAM-A regulates permeability and inflammation in the intestine in vivo. J. Exp. Med. 2007, 204, 3067–3076. [Google Scholar] [CrossRef]
- Bennett, E.P.; Mandel, U.; Clausen, H.; Gerken, T.A.; Fritz, T.A.; Tabak, L.A. Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology 2012, 22, 736–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padra, M.; Adamczyk, B.; Flahou, B.; Erhardsson, M.; Chahal, G.; Smet, A.; Jin, C.; Thorell, A.; Ducatelle, R.; Haesebrouck, F.; et al. Helicobacter suis infection alters glycosylation and decreases the pathogen growth inhibiting effect and binding avidity of gastric mucins. Mucosal Immunol. 2019, 12, 784–794. [Google Scholar] [CrossRef] [Green Version]
- Haberman, Y.; Karns, R.; Dexheimer, P.J.; Schirmer, M.; Somekh, J.; Jurickova, I.; Braun, T.; Novak, E.; Bauman, L.; Collins, M.H.; et al. Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response. Nat. Commun. 2019, 10, 38. [Google Scholar] [CrossRef] [Green Version]
- Özsoy, M.; Stummer, N.; Zimmermann, F.A.; Feichtinger, R.G.; Sperl, W.; Weghuber, D.; Schneider, A.M. Role of energy metabolism and mitochondrial function in inflammatory bowel disease. Inflamm. Bowel Dis. 2022, izac024. [Google Scholar] [CrossRef]
- Cobrin, G.M.; Abreu, M.T. Defects in mucosal immunity leading to Crohn’s disease. Immunol. Rev. 2005, 206, 277–295. [Google Scholar] [CrossRef]
- Targan, S.R.; Karp, L.C. Defects in mucosal immunity leading to ulcerative colitis. Immunol. Rev. 2005, 206, 296–305. [Google Scholar] [CrossRef]
- Rovedatti, L.; Kudo, T.; Biancheri, P.; Sarra, M.; Knowles, C.H.; Rampton, D.S.; Corazza, G.R.; Monteleone, G.; Di Sabatino, A.; Macdonald, T.T. Differential regulation of interleukin 17 and interferon gamma production in inflammatory bowel disease. Gut 2009, 58, 1629–1636. [Google Scholar] [CrossRef]
- Kadivar, K.; Ruchelli, E.D.; Markowitz, J.E.; Defelice, M.L.; Strogatz, M.L.; Kanzaria, M.M.; Reddy, K.P.; Baldassano, R.N.; von Allmen, D.; Brown, K.A. Intestinal interleukin-13 in pediatric inflammatory bowel disease patients. Inflamm. Bowel Dis. 2004, 10, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.S.; Ramalingam, T.R.; Rivollier, A.; Shenderov, K.; Mentink-Kane, M.M.; Madala, S.K.; Cheever, A.W.; Artis, D.; Kelsall, B.L.; Wynn, T.A. Colitis and intestinal inflammation in IL10−/− mice results from IL-13Rα2-mediated attenuation of IL-13 activity. Gastroenterology 2011, 140, 254–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.-B.; Luo, M.-M.; Chen, Z.-Y.; He, B.-H. The function and role of the Th17/Treg cell balance in inflammatory bowel disease. J. Immunol. Res. 2020, 2020, 8813558. [Google Scholar] [CrossRef] [PubMed]
- Wehkamp, J.; Harder, J.; Weichenthal, M.; Mueller, O.; Herrlinger, K.R.; Fellermann, K.; Schroeder, J.M.; Stange, E.F. Inducible and constitutive beta-defensins are differentially expressed in Crohn’s disease and ulcerative colitis. Inflamm. Bowel Dis. 2003, 9, 215–223. [Google Scholar] [CrossRef]
- Abreu, M.T.; Fukata, M.; Arditi, M. TLR Signaling in the gut in health and disease. J. Immunol. Baltim. 2005, 174, 4453–4460. [Google Scholar] [CrossRef] [Green Version]
- Takatori, H.; Kanno, Y.; Watford, W.T.; Tato, C.M.; Weiss, G.; Ivanov, I.I.; Littman, D.R.; O’Shea, J.J. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J. Exp. Med. 2009, 206, 35–41. [Google Scholar] [CrossRef] [Green Version]
- Wehkamp, J.; Harder, J.; Weichenthal, M.; Schwab, M.; Schäffeler, E.; Schlee, M.; Herrlinger, K.R.; Stallmach, A.; Noack, F.; Fritz, P.; et al. NOD2 (CARD15) mutations in Crohn’s disease are associated with diminished mucosal alpha-defensin expression. Gut 2004, 53, 1658–1664. [Google Scholar] [CrossRef] [Green Version]
- Larabi, A.; Barnich, N.; Nguyen, H.T.T. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy 2020, 16, 38–51. [Google Scholar] [CrossRef] [Green Version]
- Burisch, J.; Pedersen, N.; Cukovic-Cavka, S.; Turk, N.; Kaimakliotis, I.; Duricova, D.; Bortlik, M.; Shonová, O.; Vind, I.; Avnstrøm, S.; et al. Environmental factors in a population-based inception cohort of inflammatory bowel disease patients in Europe—An ECCO-EpiCom study. J. Crohn’s Colitis 2014, 8, 607–616. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Feng, R.; Ben-Horin, S.; Zhuang, X.; Tian, Z.; Li, X.; Ma, R.; Mao, R.; Qiu, Y.; Chen, M. Systematic review with meta-analysis: Environmental and dietary differences of inflammatory bowel disease in eastern and western populations. Aliment. Pharmacol. Ther. 2022, 55, 266–276. [Google Scholar] [CrossRef]
- Maaser, C.; Langholz, E.; Gordon, H.; Burisch, J.; Ellul, P.; Ramirez, V.H.; Karakan, T.; Katsanos, K.H.; Krustins, E.; Levine, A.; et al. European Crohn’s and colitis organisation topical review on environmental factors in IBD. J. Crohn’s Colitis 2017, 11, 905–920. [Google Scholar] [CrossRef]
- Calkins, B.M. A meta-analysis of the role of smoking in inflammatory bowel disease. Dig. Dis. Sci. 1989, 34, 1841–1854. [Google Scholar] [CrossRef] [PubMed]
- To, N.; Gracie, D.J.; Ford, A.C. Systematic review with meta-analysis: The adverse effects of tobacco smoking on the natural history of Crohn’s disease. Aliment. Pharmacol. Ther. 2016, 43, 549–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuenzig, M.E.; Lee, S.M.; Eksteen, B.; Seow, C.H.; Barnabe, C.; Panaccione, R.; Kaplan, G.G. Smoking influences the need for surgery in patients with the inflammatory bowel diseases: A systematic review and meta-analysis incorporating disease duration. BMC Gastroenterol. 2016, 16, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cosnes, J.; Nion-Larmurier, I.; Afchain, P.; Beaugerie, L.; Gendre, J.-P. Gender differences in the response of colitis to smoking. Clin. Gastroenterol. Hepatol. 2004, 2, 41–48. [Google Scholar] [CrossRef]
- Sher, M.E.; Bank, S.; Greenberg, R.; Sardinha, T.C.; Weissman, S.; Bailey, B.; Gilliland, R.; Wexner, S.D. The influence of cigarette smoking on cytokine levels in patients with inflammatory bowel disease. Inflamm. Bowel Dis. 1999, 5, 73–78. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.N. Environmental risk factors for inflammatory bowel disease. Gastroenterol. Hepatol. 2013, 9, 367–374. [Google Scholar] [CrossRef]
- Andersson, R.E.; Olaison, G.; Tysk, C.; Ekbom, A. Appendectomy and protection against ulcerative colitis. N. Engl. J. Med. 2001, 344, 808–814. [Google Scholar] [CrossRef]
- Frisch, M.; Gridley, G. Appendectomy in adulthood and the risk of inflammatory bowel diseases. Scand. J. Gastroenterol. 2002, 37, 1175–1177. [Google Scholar] [CrossRef]
- Koutroubakis, I.E.; Vlachonikolis, I.G.; Kapsoritakis, A.; Spanoudakis, S.; Roussomoustakaki, M.; Mouzas, I.A.; Kouroumalis, E.A.; Manousos, O.N. Appendectomy, tonsillectomy, and risk of inflammatory bowel disease: Case-controlled study in Crete. Dis. Colon Rectum 1999, 42, 225–230. [Google Scholar] [CrossRef]
- Radford-Smith, G.L.; Edwards, J.E.; Purdie, D.M.; Pandeya, N.; Watson, M.; Martin, N.G.; Green, A.; Newman, B.; Florin, T.H.J. Protective role of appendicectomy on onset and severity of ulcerative colitis and Crohn’s disease. Gut 2002, 51, 808–813. [Google Scholar] [CrossRef] [PubMed]
- Khalili, H.; Higuchi, L.M.; Ananthakrishnan, A.N.; Richter, J.M.; Feskanich, D.; Fuchs, C.S.; Chan, A.T. Oral contraceptives, reproductive factors and risk of inflammatory bowel disease. Gut 2013, 62, 1153–1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tezel, A.; Dökmeci, G.; Eskiocak, M.; Umit, H.; Soylu, A.R. Epidemiological features of ulcerative colitis in Trakya, Turkey. J. Int. Med. Res. 2003, 31, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Card, T.; Logan, R.F.A.; Rodrigues, L.C.; Wheeler, J.G. Antibiotic use and the development of Crohn’s disease. Gut 2004, 53, 246–250. [Google Scholar] [CrossRef] [Green Version]
- Aniwan, S.; Tremaine, W.J.; Raffals, L.E.; Kane, S.V.; Loftus, E.V. Antibiotic use and new-onset inflammatory bowel disease in Olmsted county, Minnesota: A population-based case-control study. J. Crohns Colitis 2018, 12, 137–144. [Google Scholar] [CrossRef]
- Hildebrand, H.; Malmborg, P.; Askling, J.; Ekbom, A.; Montgomery, S.M. Early-life exposures associated with antibiotic use and risk of subsequent Crohn’s disease. Scand. J. Gastroenterol. 2008, 43, 961–966. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.N.; Higuchi, L.M.; Huang, E.S.; Khalili, H.; Richter, J.M.; Fuchs, C.S.; Chan, A.T. Aspirin, nonsteroidal anti-inflammatory drug use, and risk for Crohn disease and ulcerative colitis: A cohort study. Ann. Intern. Med. 2012, 156, 350–359. [Google Scholar] [CrossRef]
- Molodecky, N.A.; Kaplan, G.G. Environmental risk factors for inflammatory bowel disease. Gastroenterol. Hepatol. 2010, 6, 339–346. [Google Scholar]
- Ho, S.-M.; Lewis, J.D.; Mayer, E.A.; Plevy, S.E.; Chuang, E.; Rappaport, S.M.; Croitoru, K.; Korzenik, J.R.; Krischer, J.; Hyams, J.S.; et al. Challenges in IBD research: Environmental triggers. Inflamm. Bowel Dis. 2019, 25, S13–S23. [Google Scholar] [CrossRef] [Green Version]
- Wintjens, D.S.J.; de Jong, M.J.; van der Meulen-de Jong, A.E.; Romberg-Camps, M.J.; Becx, M.C.; Maljaars, J.P.; van Bodegraven, A.A.; Mahmmod, N.; Markus, T.; Haans, J.; et al. Novel perceived stress and life events precede flares of inflammatory bowel disease: A prospective 12-month follow-up study. J. Crohn’s Colitis 2019, 13, 410–416. [Google Scholar] [CrossRef]
- Singh, S.; Graff, L.A.; Bernstein, C.N. Do NSAIDs, antibiotics, infections, or stress trigger flares in IBD? Am. J. Gastroenterol. 2009, 104, 1298–1313. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.H.; Pothoulakis, C.; Mayer, E.A. Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat. Rev. Gastroenterol. Hepatol. 2009, 6, 306–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzon, E.; Sturniolo, G.C.; Puzzolo, D.; Frisina, N.; Fries, W. Effect of stress on the paracellular barrier in the rat ileum. Gut 2002, 51, 507–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bitton, A.; Dobkin, P.L.; Edwardes, M.D.; Sewitch, M.J.; Meddings, J.B.; Rawal, S.; Cohen, A.; Vermeire, S.; Dufresne, L.; Franchimont, D.; et al. Predicting relapse in Crohn’s disease: A biopsychosocial model. Gut 2008, 57, 1386–1392. [Google Scholar] [CrossRef]
- Cámara, R.J.A.; Schoepfer, A.M.; Pittet, V.; Begré, S.; von Känel, R.; Swiss Inflammatory Bowel Disease Cohort Study (SIBDCS) Group. Mood and nonmood components of perceived stress and exacerbation of Crohn’s disease. Inflamm. Bowel Dis. 2011, 17, 2358–2365. [Google Scholar] [CrossRef]
- Mikhailov, T.A.; Furner, S.E. Breastfeeding and genetic factors in the etiology of inflammatory bowel disease in children. World J. Gastroenterol. 2009, 15, 270–279. [Google Scholar] [CrossRef]
- Nishida, A.; Inoue, R.; Inatomi, O.; Bamba, S.; Naito, Y.; Andoh, A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin. J. Gastroenterol. 2018, 11, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Jugder, B.-E.; Kamareddine, L.; Watnick, P.I. Microbiota-derived acetate activates intestinal innate immunity via the Tip60 histone acetyltransferase complex. Immunity 2021, 54, 1683–1697. [Google Scholar] [CrossRef]
- Bojanova, D.P.; Bordenstein, S.R. Fecal transplants: What is being transferred? PLoS Biol. 2016, 14, e1002503. [Google Scholar] [CrossRef]
- MetaHIT Consortium; Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Muñoz, M.; Heimesaat, M.M.; Danker, K.; Struck, D.; Lohmann, U.; Plickert, R.; Bereswill, S.; Fischer, A.; Dunay, I.R.; Wolk, K.; et al. Interleukin (IL)-23 mediates toxoplasma gondii-induced immunopathology in the gut via matrixmetalloproteinase-2 and IL-22 but independent of IL-17. J. Exp. Med. 2009, 206, 3047–3059. [Google Scholar] [CrossRef] [PubMed]
- Awane, M.; Andres, P.G.; Li, D.J.; Reinecker, H.-C. NF-ΚB-inducing kinase is a common mediator of IL-17-, TNF-α-, and IL-1β-induced chemokine promoter activation in intestinal epithelial cells. J. Immunol. 1999, 162, 5337. [Google Scholar] [PubMed]
- Bemark, M.; Boysen, P.; Lycke, N.Y. Induction of gut IgA production through T cell-dependent and T cell-independent pathways. Ann. N. Y. Acad. Sci. 2012, 1247, 97–116. [Google Scholar] [CrossRef] [PubMed]
- Bergqvist, P.; Stensson, A.; Lycke, N.Y.; Bemark, M. T cell-independent IgA class switch recombination is restricted to the galt and occurs prior to manifest germinal center formation. J. Immunol. 2010, 184, 3545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hooper, L.V.; Stappenbeck, T.S.; Hong, C.V.; Gordon, J.I. Angiogenins: A new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 2003, 4, 269–273. [Google Scholar] [CrossRef]
- Lamas, B.; Richard, M.L.; Leducq, V.; Pham, H.-P.; Michel, M.-L.; Da Costa, G.; Bridonneau, C.; Jegou, S.; Hoffmann, T.W.; Natividad, J.M.; et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016, 22, 598–605. [Google Scholar] [CrossRef]
- Longman, R.S.; Diehl, G.E.; Victorio, D.A.; Huh, J.R.; Galan, C.; Miraldi, E.R.; Swaminath, A.; Bonneau, R.; Scherl, E.J.; Littman, D.R. CX3CR1+ mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J. Exp. Med. 2014, 211, 1571–1583. [Google Scholar] [CrossRef]
- Huda-Faujan, N.; Abdulamir, A.S.; Fatimah, A.B.; Anas, O.M.; Shuhaimi, M.; Yazid, A.M.; Loong, Y.Y. The impact of the level of the intestinal short chain fatty acids in inflammatory bowel disease patients versus healthy subjects. Open Biochem. J. 2010, 4, 53–58. [Google Scholar] [CrossRef]
- Maslowski, K.M.; Vieira, A.T.; Ng, A.; Kranich, J.; Sierro, F.; Di, Y.; Schilter, H.C.; Rolph, M.S.; Mackay, F.; Artis, D.; et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009, 461, 1282–1286. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. the microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [Green Version]
- Khan, I.; Ullah, N.; Zha, L.; Bai, Y.; Khan, A.; Zhao, T.; Che, T.; Zhang, C. Alteration of gut microbiota in inflammatory bowel disease (IBD): Cause or consequence? IBD treatment targeting the gut microbiome. Pathogens 2019, 8, 126. [Google Scholar] [CrossRef] [Green Version]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.-L.; Wang, S.-N.; Miao, C.-Y. Influence of microbiota on intestinal immune system in ulcerative colitis and its intervention. Front. Immunol. 2017, 8, 1674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buttó, L.F.; Haller, D. Dysbiosis in Intestinal Inflammation: Cause or consequence. Int. J. Med. Microbiol. 2016, 306, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef]
- Littman, D.R.; Rudensky, A.Y. Th17 and regulatory T cells in mediating and restraining inflammation. Cell 2010, 140, 845–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayabe, T.; Satchell, D.P.; Pesendorfer, P.; Tanabe, H.; Wilson, C.L.; Hagen, S.J.; Ouellette, A.J. Activation of PANETH cell α-defensins in mouse small intestine. J. Biol. Chem. 2002, 277, 5219–5228. [Google Scholar] [CrossRef] [Green Version]
- Cash, H.L.; Whitham, C.V.; Behrendt, C.L.; Hooper, L.V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 2006, 313, 1126–1130. [Google Scholar] [CrossRef] [Green Version]
- Hapfelmeier, S.; Lawson, M.A.E.; Slack, E.; Kirundi, J.K.; Stoel, M.; Heikenwalder, M.; Cahenzli, J.; Velykoredko, Y.; Balmer, M.L.; Endt, K.; et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 2010, 328, 1705–1709. [Google Scholar] [CrossRef] [Green Version]
- Veltkamp, C.; Tonkonogy, S.L.; de Jong, Y.P.; Albright, C.; Grenther, W.B.; Balish, E.; Terhorst, C.; Sartor, R.B. Continuous stimulation by normal luminal bacteria is essential for the development and perpetuation of colitis in Tgϵ26 mice. Gastroenterology 2001, 120, 900–913. [Google Scholar] [CrossRef]
- Schaubeck, M.; Clavel, T.; Calasan, J.; Lagkouvardos, I.; Haange, S.B.; Jehmlich, N.; Basic, M.; Dupont, A.; Hornef, M.; von Bergen, M.; et al. Dysbiotic gut microbiota causes transmissible Crohn’s disease-like ileitis independent of failure in antimicrobial defence. Gut 2016, 65, 225–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Britton, G.J.; Contijoch, E.J.; Mogno, I.; Vennaro, O.H.; Llewellyn, S.R.; Ng, R.; Li, Z.; Mortha, A.; Merad, M.; Das, A.; et al. Microbiotas from humans with inflammatory bowel disease alter the balance of gut Th17 and RORγt+ regulatory T cells and exacerbate colitis in mice. Immunity 2019, 50, 212–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinoso Webb, C.; den Bakker, H.; Koboziev, I.; Jones-Hall, Y.; Rao Kottapalli, K.; Ostanin, D.; Furr, K.L.; Mu, Q.; Luo, X.M.; Grisham, M.B. Differential susceptibility to T cell-induced colitis in mice: Role of the intestinal microbiota. Inflamm. Bowel Dis. 2018, 24, 361–379. [Google Scholar] [CrossRef]
- D’Haens, G.R.; Geboes, K.; Peeters, M.; Baert, F.; Penninckx, F.; Rutgeerts, P. Early lesions of recurrent Crohn’s disease caused by infusion of intestinal contents in excluded ileum. Gastroenterology 1998, 114, 262–267. [Google Scholar] [CrossRef]
- Kumar, S.; Kumar, A. Microbial pathogenesis in inflammatory bowel diseases. Microb. Pathog. 2022, 163, 105383. [Google Scholar] [CrossRef]
- Jandhyala, S.M. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787. [Google Scholar] [CrossRef] [PubMed]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.-I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef] [Green Version]
- IBDMDB Investigators; Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef]
- Vich Vila, A.; Imhann, F.; Collij, V.; Jankipersadsing, S.A.; Gurry, T.; Mujagic, Z.; Kurilshikov, A.; Bonder, M.J.; Jiang, X.; Tigchelaar, E.F.; et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci. Transl. Med. 2018, 10, eaap8914. [Google Scholar] [CrossRef] [Green Version]
- Walker, A.W.; Sanderson, J.D.; Churcher, C.; Parkes, G.C.; Hudspith, B.N.; Rayment, N.; Brostoff, J.; Parkhill, J.; Dougan, G.; Petrovska, L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011, 11, 7. [Google Scholar] [CrossRef] [Green Version]
- Frank, D.N.; St. Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokol, H.; Seksik, P.; Furet, J.P.; Firmesse, O.; Nion-Larmurier, I.; Beaugerie, L.; Cosnes, J.; Corthier, G.; Marteau, P.; Doré, J. Low counts of faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 2009, 15, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermudez-Humaran, L.G.; Gratadoux, J.-J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.; Huang, C.; Xu, J.; Xu, H.; Liu, L.; Zhao, H.; Wang, J.; Huang, W.; Peng, W.; Chen, Y.; et al. Gut microbiota is a potential biomarker in inflammatory bowel disease. Front. Nutr. 2022, 8, 818902. [Google Scholar] [CrossRef]
- Zhang, M.; Qiu, X.; Zhang, H.; Yang, X.; Hong, N.; Yang, Y.; Chen, H.; Yu, C. Faecalibacterium prausnitzii inhibits interleukin-17 to ameliorate colorectal colitis in rats. PLoS ONE 2014, 9, e109146. [Google Scholar] [CrossRef] [Green Version]
- Martín, R.; Chain, F.; Miquel, S.; Lu, J.; Gratadoux, J.-J.; Sokol, H.; Verdu, E.F.; Bercik, P.; Bermúdez-Humarán, L.G.; Langella, P. The commensal bacterium faecalibacterium prausnitzii is protective in DNBS-induced chronic moderate and severe colitis models. Inflamm. Bowel Dis. 2014, 20, 417–430. [Google Scholar] [CrossRef]
- Quévrain, E.; Maubert, M.A.; Michon, C.; Chain, F.; Marquant, R.; Tailhades, J.; Miquel, S.; Carlier, L.; Bermúdez-Humarán, L.G.; Pigneur, B.; et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 2016, 65, 415–425. [Google Scholar] [CrossRef] [Green Version]
- Pittayanon, R.; Lau, J.T.; Leontiadis, G.I.; Tse, F.; Yuan, Y.; Surette, M.; Moayyedi, P. Differences in gut microbiota in patients with vs without inflammatory bowel diseases: A systematic review. Gastroenterology 2020, 158, 930–946. [Google Scholar] [CrossRef]
- Zakerska-Banaszak, O.; Tomczak, H.; Gabryel, M.; Baturo, A.; Wolko, L.; Michalak, M.; Malinska, N.; Mankowska-Wierzbicka, D.; Eder, P.; Dobrowolska, A.; et al. Dysbiosis of gut microbiota in polish patients with ulcerative colitis: A pilot study. Sci. Rep. 2021, 11, 2166. [Google Scholar] [CrossRef]
- Vrakas, S.; Mountzouris, K.C.; Michalopoulos, G.; Karamanolis, G.; Papatheodoridis, G.; Tzathas, C.; Gazouli, M. Intestinal bacteria composition and translocation of bacteria in inflammatory bowel disease. PLoS ONE 2017, 12, e0170034. [Google Scholar] [CrossRef] [Green Version]
- Santoru, M.L.; Piras, C.; Murgia, A.; Palmas, V.; Camboni, T.; Liggi, S.; Ibba, I.; Lai, M.A.; Orrù, S.; Blois, S.; et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci. Rep. 2017, 7, 9523. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, S.; Rossen, N.G.; van der Spek, M.J.; Hartman, J.H.; Huuskonen, L.; Korpela, K.; Salojärvi, J.; Aalvink, S.; de Vos, W.M.; D’Haens, G.R.; et al. Microbial shifts and signatures of long-term remission in ulcerative colitis after faecal microbiota transplantation. ISME J. 2017, 11, 1877–1889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef] [Green Version]
- Albenberg, L.; Esipova, T.V.; Judge, C.P.; Bittinger, K.; Chen, J.; Laughlin, A.; Grunberg, S.; Baldassano, R.N.; Lewis, J.D.; Li, H.; et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 2014, 147, 1055–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirsepasi-Lauridsen, H.C.; Vallance, B.A.; Krogfelt, K.A.; Petersen, A.M. Escherichia coli pathobionts associated with inflammatory bowel disease. Clin. Microbiol. Rev. 2019, 32, e00060-18. [Google Scholar] [CrossRef] [Green Version]
- Kang, C.; Ban, M.; Choi, E.-J.; Moon, H.-G.; Jeon, J.-S.; Kim, D.-K.; Park, S.-K.; Jeon, S.G.; Roh, T.-Y.; Myung, S.-J.; et al. Extracellular vesicles derived from gut microbiota, especially akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS ONE 2013, 8, e76520. [Google Scholar] [CrossRef] [Green Version]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L.; et al. A purified membrane protein from akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef] [Green Version]
- Pascal, V.; Pozuelo, M.; Borruel, N.; Casellas, F.; Campos, D.; Santiago, A.; Martinez, X.; Varela, E.; Sarrabayrouse, G.; Machiels, K.; et al. A microbial signature for Crohn’s disease. Gut 2017, 66, 813–822. [Google Scholar] [CrossRef]
- Ryan, F.J.; Ahern, A.M.; Fitzgerald, R.S.; Laserna-Mendieta, E.J.; Power, E.M.; Clooney, A.G.; O’Donoghue, K.W.; McMurdie, P.J.; Iwai, S.; Crits-Christoph, A.; et al. Colonic microbiota is associated with inflammation and host epigenomic alterations in inflammatory bowel disease. Nat. Commun. 2020, 11, 1512. [Google Scholar] [CrossRef] [Green Version]
- Willing, B.P.; Dicksved, J.; Halfvarson, J.; Andersson, A.F.; Lucio, M.; Zheng, Z.; Järnerot, G.; Tysk, C.; Jansson, J.K.; Engstrand, L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010, 139, 1844–1854. [Google Scholar] [CrossRef]
- Metwaly, A.; Dunkel, A.; Waldschmitt, N.; Raj, A.C.D.; Lagkouvardos, I.; Corraliza, A.M.; Mayorgas, A.; Martinez-Medina, M.; Reiter, S.; Schloter, M.; et al. Integrated microbiota and metabolite profiles link Crohn’s disease to sulfur metabolism. Nat. Commun. 2020, 11, 4322. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, S.; Nagao-Kitamoto, H.; Jiao, Y.; Gillilland, M.G.; Hayashi, A.; Imai, J.; Sugihara, K.; Miyoshi, M.; Brazil, J.C.; Kuffa, P.; et al. The intermucosal connection between the mouth and gut in commensal pathobiont-driven colitis. Cell 2020, 182, 447–462. [Google Scholar] [CrossRef] [PubMed]
- Clooney, A.G.; Sutton, T.D.S.; Shkoporov, A.N.; Holohan, R.K.; Daly, K.M.; O’Regan, O.; Ryan, F.J.; Draper, L.A.; Plevy, S.E.; Ross, R.P.; et al. Whole-virome analysis sheds light on viral dark matter in inflammatory bowel disease. Cell Host Microbe 2019, 26, 764–778. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Lu, X.-J.; Zhang, Y.; Cheung, C.P.; Lam, S.; Zhang, F.; Tang, W.; Ching, J.Y.L.; Zhao, R.; Chan, P.K.S.; et al. Gut mucosal virome alterations in ulcerative colitis. Gut 2019, 68, 1169–1179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.-P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal microbiota dysbiosis in IBD. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knox, N.C.; Forbes, J.D.; Peterson, C.-L.; Van Domselaar, G.; Bernstein, C.N. The gut microbiome in inflammatory bowel disease: Lessons learned from other immune-mediated inflammatory diseases. Am. J. Gastroenterol. 2019, 114, 1051–1070. [Google Scholar] [CrossRef] [PubMed]
- Schierbeek, A. The collected letters of antoni van leeuwenhoek: An appeal to the scientific world. Antonie Van Leeuwenhoek 1953, 19, 181–188. [Google Scholar] [CrossRef] [Green Version]
- Blevins, S.M.; Bronze, M.S. Robert koch and the ‘golden age’ of bacteriology. Int. J. Infect. Dis. 2010, 14, e744–e751. [Google Scholar] [CrossRef] [Green Version]
- Ackert, L. Sergei Vinogradskii and the Cycle of Life: From the Thermodynamics of Life to Ecological Microbiology, 1850–1950; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Woese, C.R.; Fox, G.E. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc. Natl. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef] [Green Version]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA Sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [Green Version]
- Hugenholtz, P.; Goebel, B.M.; Pace, N.R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 1998, 180, 4765–4774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campmans-Kuijpers, M.J.E.; Dijkstra, G. Food and food groups in inflammatory bowel disease (IBD): The design of the groningen anti-inflammatory diet (GrAID). Nutrients 2021, 13, 1067. [Google Scholar] [CrossRef] [PubMed]
- Basson, A.R.; Chen, C.; Sagl, F.; Trotter, A.; Bederman, I.; Gomez-Nguyen, A.; Sundrud, M.S.; Ilic, S.; Cominelli, F.; Rodriguez-Palacios, A. Regulation of intestinal inflammation by dietary fats. Front. Immunol. 2021, 11, 604989. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-C.; Lee, H.-Y.; Kim, T.K.; Kim, M.-S.; Park, Y.M.; Kim, J.; Park, K.; Kweon, M.-N.; Kim, S.-H.; Bae, J.-W.; et al. Obesogenic diet-induced gut barrier dysfunction and pathobiont expansion aggravate experimental colitis. PLoS ONE 2017, 12, e0187515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Wei, X.; Sun, Y.; Du, J.; Li, X.; Xun, Z.; Li, Y.C. High-fat diet promotes experimental colitis by inducing oxidative stress in the colon. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 317, G453–G462. [Google Scholar] [CrossRef]
- Kim, I.-W.; Myung, S.-J.; Do, M.Y.; Ryu, Y.-M.; Kim, M.J.; Do, E.-J.; Park, S.; Yoon, S.M.; Ye, B.D.; Byeon, J.-S.; et al. Western-style diets induce macrophage infiltration and contribute to colitis-associated carcinogenesis. J. Gastroenterol. Hepatol. 2010, 25, 1785–1794. [Google Scholar] [CrossRef]
- Okada, Y.; Tsuzuki, Y.; Sato, H.; Narimatsu, K.; Hokari, R.; Kurihara, C.; Watanabe, C.; Tomita, K.; Komoto, S.; Kawaguchi, A.; et al. Trans fatty acids exacerbate dextran sodium sulphate-induced colitis by promoting the up-regulation of macrophage-derived proinflammatory cytokines involved in T helper 17 cell polarization. Clin. Exp. Immunol. 2013, 174, 459–471. [Google Scholar] [CrossRef]
- Cheng, L.; Jin, H.; Qiang, Y.; Wu, S.; Yan, C.; Han, M.; Xiao, T.; Yan, N.; An, H.; Zhou, X.; et al. High fat diet exacerbates dextran sulfate sodium induced colitis through disturbing mucosal dendritic cell homeostasis. Int. Immunopharmacol. 2016, 40, 1–10. [Google Scholar] [CrossRef]
- Enos, R.T.; Davis, J.M.; Velázquez, K.T.; McClellan, J.L.; Day, S.D.; Carnevale, K.A.; Murphy, E.A. Influence of dietary saturated fat content on adiposity, macrophage behavior, inflammation, and metabolism: Composition matters. J. Lipid Res. 2013, 54, 152–163. [Google Scholar] [CrossRef] [Green Version]
- Devkota, S.; Wang, Y.; Musch, M.W.; Leone, V.; Fehlner-Peach, H.; Nadimpalli, A.; Antonopoulos, D.A.; Jabri, B.; Chang, E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012, 487, 104–108. [Google Scholar] [CrossRef] [Green Version]
- Muhomah, T.A.; Nishino, N.; Katsumata, E.; Haoming, W.; Tsuruta, T. High-fat diet reduces the level of secretory immunoglobulin a coating of commensal gut microbiota. Biosci. Microbiota Food Health 2019, 38, 55–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, E.L.; Nestor, M.; Onyewadume, L.; de Silva, P.S.; Korzenik, J.R.; Aguilar, H.; Bailen, L.; Berman, A.; Bhaskar, S.K.; Brown, M.; et al. High dietary intake of specific fatty acids increases risk of flares in patients with ulcerative colitis in remission during treatment with aminosalicylates. Clin. Gastroenterol. Hepatol. 2017, 15, 1390–1396.e1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niewiadomski, O.; Studd, C.; Wilson, J.; Williams, J.; Hair, C.; Knight, R.; Prewett, E.; Dabkowski, P.; Alexander, S.; Allen, B.; et al. Influence of food and lifestyle on the risk of developing inflammatory bowel disease. Intern. Med. J. 2016, 46, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Persson, P.G.; Ahlbom, A.; Hellers, G. Diet and inflammatory bowel disease: A case-control study. Epidemiol. Camb. Mass 1992, 3, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Piotrowska, M.; Binienda, A.; Fichna, J. The role of fatty acids in Crohn’s disease pathophysiology—An overview. Mol. Cell. Endocrinol. 2021, 538, 111448. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Choi, J.; Hwang, S.H.; Kim, J.K.; Kim, E.K.; Lee, S.Y.; Lee, B.I.; Park, S.H.; Cho, M.L. Cottonseed oil protects against intestinal inflammation in dextran sodium sulfate-induced inflammatory bowel disease. J. Med. Food 2019, 22, 672–679. Available online: https://www.liebertpub.com/doi/10.1089/jmf.2018.4323# (accessed on 11 March 2022). [CrossRef]
- Shoda, R.; Matsueda, K.; Yamato, S.; Umeda, N. Epidemiologic analysis of Crohn disease in Japan: Increased dietary intake of n-6 polyunsaturated fatty acids and animal protein relates to the increased incidence of Crohn disease in Japan. Am. J. Clin. Nutr. 1996, 63, 741–745. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.N.; Khalili, H.; Konijeti, G.G.; Higuchi, L.M.; de Silva, P.; Fuchs, C.S.; Willett, W.C.; Richter, J.M.; Chan, A.T. Long-term intake of dietary fat and risk of ulcerative colitis and Crohn’s disease. Gut 2014, 63, 776–784. [Google Scholar] [CrossRef] [Green Version]
- Amre, D.K.; D’Souza, S.; Morgan, K.; Seidman, G.; Lambrette, P.; Grimard, G.; Israel, D.; Mack, D.; Ghadirian, P.; Deslandres, C.; et al. Imbalances in dietary consumption of fatty acids, vegetables, and fruits are associated with risk for Crohn’s disease in children. Am. J. Gastroenterol. 2007, 102, 2016–2025. [Google Scholar] [CrossRef]
- IBD in EPIC Study Investigators; Tjonneland, A.; Overvad, K.; Bergmann, M.M.; Nagel, G.; Linseisen, J.; Hallmans, G.; Palmqvist, R.; Sjodin, H.; Hagglund, G.; et al. Linoleic acid, a dietary n-6 polyunsaturated fatty acid, and the aetiology of ulcerative colitis: A nested case-control study within a european prospective cohort study. Gut 2009, 58, 1606–1611. [Google Scholar] [CrossRef]
- John, S.; Luben, R.; Shrestha, S.S.; Welch, A.; Khaw, K.-T.; Hart, A.R. Dietary N-3 polyunsaturated fatty acids and the aetiology of ulcerative colitis: A UK prospective cohort study. Eur. J. Gastroenterol. Hepatol. 2010, 22, 602–606. [Google Scholar] [CrossRef] [PubMed]
- Marton, L.T.; Goulart, R.d.A.; de Carvalho, A.C.A.; Barbalho, S.M. Omega fatty acids and inflammatory bowel diseases: An overview. Int. J. Mol. Sci. 2019, 20, 4851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mozaffari, H.; Daneshzad, E.; Larijani, B.; Bellissimo, N.; Azadbakht, L. Dietary intake of fish, n-3 polyunsaturated fatty acids, and risk of inflammatory bowel disease: A systematic review and meta-analysis of observational studies. Eur. J. Nutr. 2020, 59, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Ajabnoor, S.M.; Thorpe, G.; Abdelhamid, A.; Hooper, L. Long-term effects of increasing omega-3, omega-6 and total polyunsaturated fats on inflammatory bowel disease and markers of inflammation: A systematic review and meta-analysis of randomized controlled trials. Eur. J. Nutr. 2021, 60, 2293–2316. [Google Scholar] [CrossRef]
- Wang, F.; Lin, X.; Zhao, Q.; Li, J. Fat intake and risk of ulcerative colitis: Systematic review and dose-response meta-analysis of epidemiological studies. J. Gastroenterol. Hepatol. 2017, 32, 19–27. [Google Scholar] [CrossRef]
- Sakamoto, N.; Kono, S.; Wakai, K.; Fukuda, Y.; Satomi, M.; Shimoyama, T.; Inaba, Y.; Miyake, Y.; Sasaki, S.; Okamoto, K.; et al. Dietary risk factors for inflammatory bowel disease: A multicenter case-control study in Japan. Inflamm. Bowel Dis. 2005, 11, 154–163. [Google Scholar] [CrossRef]
- Preda, C.M.; Manuc, T.; Chifulescu, A.; Istratescu, D.; Louis, E.; Baicus, C.; Sandra, I.; Diculescu, M.-M.; Reenaers, C.; van Kemseke, C.; et al. Diet as an environmental trigger in inflammatory bowel disease: A retrospective comparative study in two European cohorts. Revista Espanola de Enfermedades Digestivas 2020, 112, 440–447. [Google Scholar] [CrossRef]
- Zhou, Y.Q.; Xu, R.Y.; Wan, Y.P. The role of dietary factors in inflammatory bowel diseases: New perspectives. J. Dig. Dis. 2019, 20, 11–17. [Google Scholar] [CrossRef] [Green Version]
- Zeng, L.; Hu, S.; Chen, P.; Wei, W.; Tan, Y. Macronutrient intake and risk of Crohn’s disease: Systematic review and dose-response meta-analysis of epidemiological studies. Nutrients 2017, 9, 500. [Google Scholar] [CrossRef]
- Racine, A.; Carbonnel, F.; Chan, S.S.M.; Hart, A.R.; Bueno-de-Mesquita, H.B.; Oldenburg, B.; van Schaik, F.D.M.; Tjønneland, A.; Olsen, A.; Dahm, C.C.; et al. Dietary patterns and risk of inflammatory bowel disease in Europe: Results from the EPIC study. Inflamm. Bowel Dis. 2016, 22, 345–354. [Google Scholar] [CrossRef] [Green Version]
- Khademi, Z.; Milajerdi, A.; Larijani, B.; Esmaillzadeh, A. Dietary intake of total carbohydrates, sugar and sugar-sweetened beverages, and risk of inflammatory bowel disease: A systematic review and meta-analysis of prospective cohort studies. Front. Nutr. 2021, 8, 707795. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.R.; Lee, J.H.; Lee, J.H.; Na, G.Y.; Lee, K.-H.; Lee, Y.-B.; Jung, G.-H.; Kim, O.Y. Low-FODMAP formula improves diarrhea and nutritional status in hospitalized patients receiving enteral nutrition: A randomized, multicenter, double-blind clinical trial. Nutr. J. 2015, 14, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, S.R.; Prince, A.C.; Myers, C.E.; Irving, P.M.; Lindsay, J.O.; Lomer, M.C.; Whelan, K. Fermentable carbohydrates [FODMAPs] exacerbate functional gastrointestinal symptoms in patients with inflammatory bowel disease: A randomised, double-blind, placebo-controlled, cross-over, re-challenge trial. J. Crohn’s Colitis 2017, 11, 1420–1429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, F.; Park, S.; Liu, Y.; Greenlund, K.J. Dietary intake patterns among adults with inflammatory bowel disease in the United States, 2015. PLoS ONE 2021, 16, e0250441. [Google Scholar] [CrossRef] [PubMed]
- Brotherton, C.S.; Martin, C.A.; Long, M.D.; Kappelman, M.D.; Sandler, R.S. Avoidance of fiber is associated with greater risk of Crohn’s disease flare in a 6-month period. Clin. Gastroenterol. Hepatol 2016, 14, 1130–1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, P.S.; Rhodes, J. Maintenance of remission in ulcerative colitis with sulphasalazine or a high-fibre diet: A clinical trial. Br. Med. J. 1978, 1, 1524–1525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, J.K.; Abraham, B.; El-Serag, H. Dietary intake and risk of developing inflammatory bowel disease: A systematic review of the literature. Am. J. Gastroenterol. 2011, 106, 563–573. [Google Scholar] [CrossRef]
- Jowett, S.L.; Seal, C.J.; Pearce, M.S.; Phillips, E.; Gregory, W.; Barton, J.R.; Welfare, M.R. Influence of dietary factors on the clinical course of ulcerative colitis: A prospective cohort study. Gut 2004, 53, 1479–1484. [Google Scholar] [CrossRef]
- Morton, H.; Pedley, K.C.; Stewart, R.J.C.; Coad, J. Inflammatory bowel disease: Are symptoms and diet linked? Nutrients 2020, 12, 2975. [Google Scholar] [CrossRef]
- Li, F.; Liu, X.; Wang, W.; Zhang, D. Consumption of vegetables and fruit and the risk of inflammatory bowel disease: A meta-analysis. Eur. J. Gastroenterol. Hepatol. 2015, 27, 623–630. [Google Scholar] [CrossRef]
- Sandefur, K.; Kahleova, H.; Desmond, A.N.; Elfrink, E.; Barnard, N.D. Crohn’s disease remission with a plant-based diet: A case report. Nutrients 2019, 11, 1385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lane, E.R.; Zisman, T.; Suskind, D. The microbiota in inflammatory bowel disease: Current and therapeutic insights. J. Inflamm. Res. 2017, 10, 63–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, E.M.; Sadarangani, M.; Finlay, B.B. The role of the immune system in governing host-microbe interactions in the intestine. Nat. Immunol. 2013, 14, 660–667. [Google Scholar] [CrossRef] [PubMed]
- Chassaing, B.; Koren, O.; Goodrich, J.K.; Poole, A.C.; Srinivasan, S.; Ley, R.E.; Gewirtz, A.T. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 2015, 519, 92–96. [Google Scholar] [CrossRef] [Green Version]
- Russell, W.R.; Gratz, S.W.; Duncan, S.H.; Holtrop, G.; Ince, J.; Scobbie, L.; Duncan, G.; Johnstone, A.M.; Lobley, G.E.; Wallace, R.J.; et al. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am. J. Clin. Nutr. 2011, 93, 1062–1072. [Google Scholar] [CrossRef] [PubMed]
- Blachier, F.; Beaumont, M.; Andriamihaja, M.; Davila, A.-M.; Lan, A.; Grauso, M.; Armand, L.; Benamouzig, R.; Tomé, D. Changes in the luminal environment of the colonic epithelial cells and physiopathological consequences. Am. J. Pathol. 2017, 187, 476–486. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-Y.; Cevallos, S.A.; Byndloss, M.X.; Tiffany, C.R.; Olsan, E.E.; Butler, B.P.; Young, B.M.; Rogers, A.W.L.; Nguyen, H.; Kim, K.; et al. High-fat diet and antibiotics cooperatively impair mitochondrial bioenergetics to trigger dysbiosis that exacerbates pre-inflammatory bowel disease. Cell Host Microbe 2020, 28, 273–284. [Google Scholar] [CrossRef]
- Byndloss, M.X.; Olsan, E.E.; Rivera-Chávez, F.; Tiffany, C.R.; Cevallos, S.A.; Lokken, K.L.; Torres, T.P.; Byndloss, A.J.; Faber, F.; Gao, Y.; et al. Microbiota-activated ppar-γ signaling inhibits dysbiotic enterobacteriaceae expansion. Science 2017, 357, 570–575. [Google Scholar] [CrossRef]
- Wills, E.S.; Jonkers, D.M.A.E.; Savelkoul, P.H.; Masclee, A.A.; Pierik, M.J.; Penders, J. Fecal microbial composition of ulcerative colitis and Crohn’s disease patients in remission and subsequent exacerbation. PLoS ONE 2014, 9, e90981. [Google Scholar] [CrossRef] [Green Version]
- Molodecky, N.A.; Soon, I.S.; Rabi, D.M.; Ghali, W.A.; Ferris, M.; Chernoff, G.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Barkema, H.W.; et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 2012, 142, 46–54. [Google Scholar] [CrossRef] [Green Version]
- Falcinelli, S.; Rodiles, A.; Unniappan, S.; Picchietti, S.; Gioacchini, G.; Merrifield, D.L.; Carnevali, O. Probiotic treatment reduces appetite and glucose level in the zebrafish model. Sci. Rep. 2016, 6, 18061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rychter, A.M.; Ratajczak, A.E.; Zawada, A.; Dobrowolska, A.; Krela-Kaźmierczak, I. Non-systematic review of diet and nutritional risk factors of cardiovascular disease in obesity. Nutrients 2020, 12, 814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bendall, C.L.; Mayr, H.L.; Opie, R.S.; Bes-Rastrollo, M.; Itsiopoulos, C.; Thomas, C.J. Central obesity and the mediterranean diet: A systematic review of intervention trials. Crit. Rev. Food Sci. Nutr. 2018, 58, 3070–3084. [Google Scholar] [CrossRef] [PubMed]
- Esposito, K.; Giugliano, D. Mediterranean diet and Type 2 diabetes. Diabetes Metab. Res. Rev. 2014, 30, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Esposito, K.; Marfella, R.; Ciotola, M.; Di Palo, C.; Giugliano, F.; Giugliano, G.; D’Armiento, M.; D’Andrea, F.; Giugliano, D. Effect of a Mediterranean-style diet on endothelial dysfunction and markers of vascular inflammation in the metabolic syndrome: A randomized trial. JAMA 2004, 292, 1440–1446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Álvarez-Álvarez, I.; Martínez-González, M.Á.; Sánchez-Tainta, A.; Corella, D.; Díaz-López, A.; Fitó, M.; Vioque, J.; Romaguera, D.; Martínez, J.A.; Wärnberg, J.; et al. Adherence to an energy-restricted mediterranean diet score and prevalence of cardiovascular risk factors in the predimed-plus: A cross-sectional study. Rev. Espanola Cardiol. Engl. Ed. 2019, 72, 925–934. [Google Scholar] [CrossRef]
- Guasch-Ferré, M.; Salas-Salvadó, J.; Ros, E.; Estruch, R.; Corella, D.; Fitó, M.; Martínez-González, M.A.; Arós, F.; Gómez-Gracia, E.; Fiol, M.; et al. The PREDIMED trial, mediterranean diet and health outcomes: How strong is the evidence? Nutr. Metab. Cardiovasc. Dis. 2017, 27, 624–632. [Google Scholar] [CrossRef] [Green Version]
- Nuts and CVD—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/26148914/ (accessed on 6 December 2021).
- Castiglione, D.; Platania, A.; Conti, A.; Falla, M.; D’Urso, M.; Marranzano, M. Dietary micronutrient and mineral intake in the mediterranean healthy eating, ageing, and lifestyle (MEAL) study. Antioxidants 2018, 7, 79. [Google Scholar] [CrossRef] [Green Version]
- Alcalay, R.; Gu, Y.; Mejia-Santana, H.; Cote, L.; Marder, K.; Scarmeas, N. The association between mediterranean diet adherence and Parkinson’s disease. Mov. Disord. 2012, 27, 771–774. [Google Scholar] [CrossRef] [Green Version]
- Donovan, M.G.; Selmin, O.I.; Doetschman, T.C.; Romagnolo, D.F. Mediterranean diet: Prevention of colorectal cancer. Front. Nutr. 2017, 4, 59. [Google Scholar] [CrossRef]
- Larussa, T.; Imeneo, M.; Luzza, F. Olive tree biophenols in inflammatory bowel disease: When bitter is better. Int. J. Mol. Sci. 2019, 20, 1390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cândido, F.G.; Valente, F.X.; Grześkowiak, Ł.M.; Moreira, A.P.B.; Rocha, D.M.U.P.; Alfenas, R.d.C.G. Impact of dietary fat on gut microbiota and low-grade systemic inflammation: Mechanisms and clinical implications on obesity. Int. J. Food Sci. Nutr. 2018, 69, 125–143. [Google Scholar] [CrossRef] [PubMed]
- García-Montero, C.; Fraile-Martínez, O.; Gómez-Lahoz, A.M.; Pekarek, L.; Castellanos, A.J.; Noguerales-Fraguas, F.; Coca, S.; Guijarro, L.G.; García-Honduvilla, N.; Asúnsolo, A.; et al. Nutritional components in western diet versus mediterranean diet at the gut microbiota–immune system interplay. implications for health and disease. Nutrients 2021, 13, 699. [Google Scholar] [CrossRef] [PubMed]
- Bibiloni, M.D.M.; Julibert, A.; Argelich, E.; Aparicio-Ugarriza, R.; Palacios, G.; Pons, A.; Gonzalez-Gross, M.; Tur, J.A. Western and Mediterranean dietary patterns and physical activity and fitness among spanish older adults. Nutrients 2017, 9, 704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz, M.-A.; Fíto, M.; Marrugat, J.; Covas, M.-I.; Schröder, H. REGICOR and HERMES investigators adherence to the Mediterranean diet is associated with better mental and physical health. Br. J. Nutr. 2009, 101, 1821–1827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClure, R.; Villani, A. Greater adherence to a Mediterranean diet is associated with better gait speed in older adults with type 2 diabetes mellitus. Clin. Nutr. ESPEN 2019, 32, 33–39. [Google Scholar] [CrossRef]
- Galan-Lopez, P.; Sánchez-Oliver, A.J.; Ries, F.; González-Jurado, J.A. Mediterranean diet, physical fitness and body composition in Sevillian adolescents: A Healthy lifestyle. Nutrients 2019, 11, 2009. [Google Scholar] [CrossRef] [Green Version]
- Filippi, J.; Al-Jaouni, R.; Wiroth, J.-B.; Hébuterne, X.; Schneider, S.M. Nutritional deficiencies in patients with Crohn’s disease in remission. Inflamm. Bowel Dis. 2006, 12, 185–191. [Google Scholar] [CrossRef]
- Eder, P.; Niezgódka, A.; Krela-Kaźmierczak, I.; Stawczyk-Eder, K.; Banasik, E.; Dobrowolska, A. Dietary support in elderly patients with inflammatory bowel disease. Nutrients 2019, 11, 1421. [Google Scholar] [CrossRef] [Green Version]
- Łykowska-Szuber, L.; Rychter, A.M.; Dudek, M.; Ratajczak, A.E.; Szymczak-Tomczak, A.; Zawada, A.; Eder, P.; Lesiak, M.; Dobrowolska, A.; Krela-Kaźmierczak, I. What links an increased cardiovascular risk and inflammatory bowel disease? A narrative review. Nutrients 2021, 13, 2661. [Google Scholar] [CrossRef]
- Peters, V.; Bolte, L.; Schuttert, E.; Andreu-Sánchez, S.; Dijkstra, G.; Weersma, R.; Campmans-Kuijpers, M. Western and carnivorous dietary patterns are associated with greater likelihood of IBD-development in a large prospective population-based cohort. J. Crohn’s Colitis 2021, jjab219. [Google Scholar] [CrossRef] [PubMed]
- Papada, E.; Amerikanou, C.; Forbes, A.; Kaliora, A.C. Adherence to Mediterranean diet in Crohn’s disease. Eur. J. Nutr. 2020, 59, 1115–1121. [Google Scholar] [CrossRef]
- Chicco, F.; Magrì, S.; Cingolani, A.; Paduano, D.; Pesenti, M.; Zara, F.; Tumbarello, F.; Urru, E.; Melis, A.; Casula, L.; et al. Multidimensional impact of Mediterranean diet on IBD patients. Inflamm. Bowel Dis. 2021, 27, 1–9. [Google Scholar] [CrossRef] [PubMed]
- de Silva, P.S.A.; Luben, R.; Shrestha, S.S.; Khaw, K.T.; Hart, A.R. Dietary arachidonic and oleic acid intake in ulcerative colitis etiology: A prospective cohort study using 7-day food diaries. Eur. J. Gastroenterol. Hepatol. 2014, 26, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Morvaridi, M.; Jafarirad, S.; Seyedian, S.S.; Alavinejad, P.; Cheraghian, B. The effects of extra virgin olive oil and canola oil on inflammatory markers and gastrointestinal symptoms in patients with ulcerative colitis. Eur. J. Clin. Nutr. 2020, 74, 891–899. [Google Scholar] [CrossRef]
- Vrdoljak, J.; Kumric, M.; Vilovic, M.; Martinovic, D.; Tomic, I.J.; Krnic, M.; Ticinovic Kurir, T.; Bozic, J. Effects of olive oil and its components on intestinal inflammation and inflammatory bowel disease. Nutrients 2022, 14, 757. [Google Scholar] [CrossRef]
- Vrdoljak, J.; Vilović, M.; Živković, P.M.; Tadin Hadjina, I.; Rušić, D.; Bukić, J.; Borovac, J.A.; Božić, J. Mediterranean diet adherence and dietary attitudes in patients with inflammatory bowel disease. Nutrients 2020, 12, 3429. [Google Scholar] [CrossRef]
- Fiorindi, C.; Dinu, M.; Gavazzi, E.; Scaringi, S.; Ficari, F.; Nannoni, A.; Sofi, F.; Giudici, F. Adherence to Mediterranean diet in patients with inflammatory bowel disease. Clin. Nutr. ESPEN 2021, 46, 416–423. [Google Scholar] [CrossRef]
- Godny, L.; Reshef, L.; Pfeffer-Gik, T.; Goren, I.; Yanai, H.; Tulchinsky, H.; Gophna, U.; Dotan, I. Adherence to the Mediterranean diet is associated with decreased fecal calprotectin in patients with ulcerative colitis after pouch surgery. Eur. J. Nutr. 2020, 59, 3183–3190. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [Green Version]
- Eswaran, S.; Muir, J.; Chey, W.D. Fiber and functional gastrointestinal disorders. Off. J. Am. Coll. Gastroenterol. ACG 2013, 108, 718–727. [Google Scholar] [CrossRef] [PubMed]
- Kaser, A.; Lee, A.-H.; Franke, A.; Glickman, J.N.; Zeissig, S.; Tilg, H.; Nieuwenhuis, E.E.S.; Higgins, D.E.; Schreiber, S.; Glimcher, L.H.; et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 2008, 134, 743–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Region Name | OMIM Number | Location | Disease | Marker (LOD Score), rs Number (p-Value) | Candidate Genes |
|---|---|---|---|---|---|
| IBD1 | 266,600 | 16q12 | CD | D1S3669 (2.65) | NOD2 |
| IBD2 | 601,458 | 12p13.2-q24.1 | CD, UC | D12S83 (5.47) | VDR, IFNG |
| IBD3 | 604,519 | 6p21.3 | CD, UC | D6S289 and D6S276 (2.07) D6S461 (4.2) | HLA-B, HLA-DRB1, TNF, LTA |
| IBD4 | 606,675 | 14q11-q12 | CD | D14S261 (3.0) | RNASE2, RNASE3 |
| IBD5 | 606,348 | 5q31-q33 | CD | 5q31-q33 (3.9) | IL4, IL5, IL13, IRF1, CSF2, SLC22A4, SLC22A5 |
| IBD6 | 606,674 | 19p13 | CD, UC | D19S591 (4.6) | ICAM1, C3, TBXA2R, LTB4H |
| IBD7 | 605,225 | 1p36 | CD, UC | D1S1597 (3.01) | TNFRSF1B, TNFRSF4 CASP9 |
| IBD8 | 606,668 | 16p | CD, UC | D16S408 (>2.5) | IL27, SULT1A1, SULT1A2 |
| IBD9 | 608,448 | 3p26 | CD, UC | D3S1297 (3.69) | |
| IBD10 | 611,081 | 2q37.1 | CD | ATG16L1 | |
| IBD11 | 191,390 | 7q22 | CD, UC | D7S669 (3.08) | MUC3A |
| IBD12 | 612,241 | 3p21.3 | CD, UC | D3S2432 (1.68) | MST1, BSN, GNAI2 |
| IBD13 | 612,244 | 7q21.1 | CD, UC | D7S669 (3.08) | ABCB1 |
| IBD14 | 612,245 | 7q32.1 | CD, UC | IRF5 | |
| IBD15 | 612,255 | 10q21 | CD, UC | rs224136 (<10 × 10−10) | SIRT1 |
| IBD16 | 612,259 | 9q32 | CD, UC | D9S2157 (1.41) | TNFSF15 |
| IBD17 | 612,261 | 1p31.3 | CD, UC | rs11209026 (<10−9) | IL23R |
| IBD18 | 612,262 | 5p13.1 | CD, UC | rs1373692 (2.1 × 10−12) | PTGER4 |
| IBD19 | 612,278 | 5q33.1 | CD | IRGM | |
| IBD20 | 612,288 | 10q23-q24 | CD, UC | D10S547 and D10S20 (2.30) | DLG5 |
| IBD21 | 612,354 | 18p11 | CD, UC | rs2542151 (3.16 × 10−8) | PTPN2 |
| IBD22 | 612,380 | 17q21.2 | CD | rs744166 (6.82 × 10−12) rs2872507 (5.00 × 10−9) | STAT3, ORMDL3 |
| IBD23 | 612,381 | 1q32.1 | CD, UC | rs11584383 (1.43 × 10−11) | IL10 |
| IBD24 | 612,566 | 20q13 | CD, UC | rs2315008 (6.30 × 10−8) rs4809330 (6.95 × 10−8) | TNFRSF6B |
| IBD25 | 612,567 | 21q22.1 | CD, UC | rs2836878 (6.01 × 10−8) | IL10RB |
| IBD26 | 612,639 | 12q15 | UC | rs1558744 (2.5 × 10−12) | |
| IBD27 | 612,796 | 13q13.3 | CD | rs20411 (LOD 3.98) | |
| IBD28 | 613,148 | 11q23.3 | CD, UC | IL10RA |
| Crohn’s Disease | Ulcerative Colitis |
|---|---|
| Th1 | Th2 |
| IFNγ, IL-2, TNFα | IL-4 |
| IL-2 | IL-5 |
| TNFα | IL-13 |
| Il-17A | |
| Th17 lymphocyte and regulatory T (Treg) lymphocyte imbalance | |
| Il-23 polymorphism | |
| Factors | Risk of Crohn’s Disease | Risk of Ulcerative Colitis |
|---|---|---|
| Smoking | ↑ | ↓ |
| Appendectomy over the age of 20 | Conflicting data | ↓ |
| Oral contraceptives | ↑ | |
| Antibiotics and non-steroidal anti-inflammatory drugs (NSAIDs) | ↑ | |
| Water and air pollution | ↑ | |
| Stress | ↑ | |
| Being breastfed | ↓ | |
| Decreased Abundance | Increased Abundance |
|---|---|
| Firmicutes | Ruminococcus gnavu |
| Faecalibacterium prausnitzii | Proteobacteria |
| Lactobacillus | Actinobacteria |
| Roseburia faecis | Escherichia coli |
| Clostridium XIVa | Desulfovibrio |
| Clostridium IV | Akkermansia muciniphila |
| Faecalibacterium prausnitzii | Escherichia |
| Bacteroidetes | Fusobacterium |
| Verrucomicrobia | |
| Anaerostipes | |
| Methanobrevibacter | |
| Faecalibacterium | |
| Peptostreptococcaceae | |
| Collinsella | |
| Christensenellaceae |
| Dietary Pattern | Risk of Crohn’s Disease | Risk of Ulcerative Colitis |
|---|---|---|
| High intake of saturated fatty acid | No data | ↑ |
| Fast food intake | ↑ | ↑ |
| Unsaturated fatty acids | Mixed results | |
| Sucrose | ↑ | ↑ |
| Vitamin C | ↓ | No data |
| Fiber | Mixed results | |
| Western diet pattern | ↑ | ↑ |
| Mediterranean diet pattern | ↓ | ↓ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krela-Kaźmierczak, I.; Zakerska-Banaszak, O.; Skrzypczak-Zielińska, M.; Łykowska-Szuber, L.; Szymczak-Tomczak, A.; Zawada, A.; Rychter, A.M.; Ratajczak, A.E.; Skoracka, K.; Skrzypczak, D.; et al. Where Do We Stand in the Behavioral Pathogenesis of Inflammatory Bowel Disease? The Western Dietary Pattern and Microbiota—A Narrative Review. Nutrients 2022, 14, 2520. https://doi.org/10.3390/nu14122520
Krela-Kaźmierczak I, Zakerska-Banaszak O, Skrzypczak-Zielińska M, Łykowska-Szuber L, Szymczak-Tomczak A, Zawada A, Rychter AM, Ratajczak AE, Skoracka K, Skrzypczak D, et al. Where Do We Stand in the Behavioral Pathogenesis of Inflammatory Bowel Disease? The Western Dietary Pattern and Microbiota—A Narrative Review. Nutrients. 2022; 14(12):2520. https://doi.org/10.3390/nu14122520
Chicago/Turabian StyleKrela-Kaźmierczak, Iwona, Oliwia Zakerska-Banaszak, Marzena Skrzypczak-Zielińska, Liliana Łykowska-Szuber, Aleksandra Szymczak-Tomczak, Agnieszka Zawada, Anna Maria Rychter, Alicja Ewa Ratajczak, Kinga Skoracka, Dorota Skrzypczak, and et al. 2022. "Where Do We Stand in the Behavioral Pathogenesis of Inflammatory Bowel Disease? The Western Dietary Pattern and Microbiota—A Narrative Review" Nutrients 14, no. 12: 2520. https://doi.org/10.3390/nu14122520
APA StyleKrela-Kaźmierczak, I., Zakerska-Banaszak, O., Skrzypczak-Zielińska, M., Łykowska-Szuber, L., Szymczak-Tomczak, A., Zawada, A., Rychter, A. M., Ratajczak, A. E., Skoracka, K., Skrzypczak, D., Marcinkowska, E., Słomski, R., & Dobrowolska, A. (2022). Where Do We Stand in the Behavioral Pathogenesis of Inflammatory Bowel Disease? The Western Dietary Pattern and Microbiota—A Narrative Review. Nutrients, 14(12), 2520. https://doi.org/10.3390/nu14122520






