Association of the Gut Microbiota with the Host’s Health through an Analysis of Biochemical Markers, Dietary Estimation, and Microbial Composition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Anthropometric and Biochemical Measurements
2.3. Dietary Estimation
2.4. Fecal Sample Collection, DNA Extraction, and Metagenomic Data
2.5. Statistical Analysis
3. Results
3.1. Characteristics of the Study Population
3.2. Dietary Intake Characteristics
3.3. Microbiota Composition: Biochemical Markers
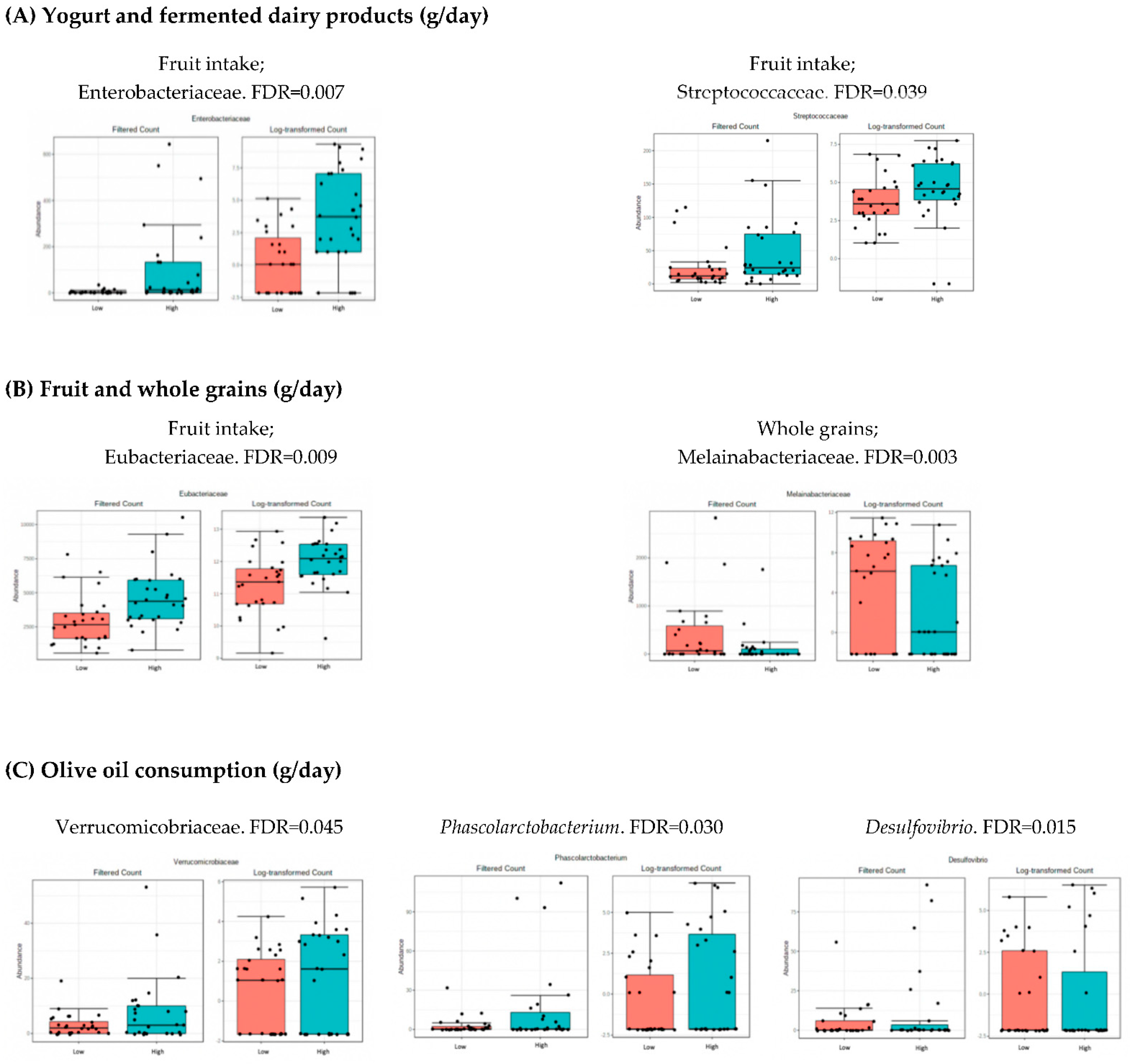
3.4. Microbiota Composition: Nutritional Markers
4. Discussion
4.1. Analysis of Microbiota with Metabolic and Hepatic Health
4.2. Analysis of Microbiota and Dietary Intake
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of Diet on the Gut Microbiome and Implications for Human Health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gebrayel, P.; Nicco, C.; al Khodor, S.; Bilinski, J.; Caselli, E.; Comelli, E.M.; Egert, M.; Giaroni, C.; Karpinski, T.M.; Loniewski, I.; et al. Microbiota Medicine: Towards Clinical Revolution. J. Transl. Med. 2022, 20, 111. [Google Scholar] [CrossRef] [PubMed]
- Dabke, K.; Hendrick, G.; Devkota, S. The Gut Microbiome and Metabolic Syndrome. J. Clin. Investig. 2019, 129, 4050–4057. [Google Scholar] [CrossRef]
- Tiffany, C.R.; Bäumler, A.J. Dysbiosis: From Fiction to Function. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 317, G602–G608. [Google Scholar] [CrossRef]
- Aron-Wisnewsky, J.; Warmbrunn, M.v.; Nieuwdorp, M.; Clément, K. Nonalcoholic Fatty Liver Disease: Modulating Gut Microbiota to Improve Severity? Gastroenterology 2020, 158, 1881–1898. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.C.; Hoffmann, C.; Mota, J.F. The Human Gut Microbiota: Metabolism and Perspective in Obesity. Gut Microbes 2018, 9, 308–325. [Google Scholar] [CrossRef] [Green Version]
- Makki, K.; Deehan, E.C.; Walter, J.; Bäckhed, F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar] [CrossRef] [Green Version]
- Satokari, R. High Intake of Sugar and the Balance between Pro- and Anti-Inflammatory Gut Bacteria. Nutrients 2020, 12, 1348. [Google Scholar] [CrossRef] [PubMed]
- Rosés, C.; Cuevas-Sierra, A.; Quintana, S.; Riezu-Boj, J.I.; Martínez, J.A.; Milagro, F.I.; Barceló, A. Gut Microbiota Bacterial Species Associated with Mediterranean Diet-Related Food Groups in a Northern Spanish Population. Nutrients 2021, 13, 636. [Google Scholar] [CrossRef]
- World Medical Association. World Medical Association Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects. J. Am. Med. Assoc. 2013, 310, 2191–2194. [Google Scholar] [CrossRef]
- WHO. A Healthy Lifestyle—WHO Recommendations. Available online: https://www.who.int/europe/news-room/fact-sheets/item/a-healthy-lifestyle—Who-recommendations (accessed on 2 November 2022).
- Shatenstein, B.; Nadon, S.; Godin, C.; Ferland, G. Development and Validation of a Food Frequency Questionnaire. Can. J. Diet. Pract. Res. 2005, 66, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Aitchison, J. Principal Component Analysis of Compositional Data. Biometrika 1983, 70, 57–65. [Google Scholar] [CrossRef]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for Comprehensive Statistical, Functional, and Meta-Analysis of Microbiome Data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef] [PubMed]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A Web-Based Tool for Comprehensive Statistical, Visual and Meta-Analysis of Microbiome Data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef] [PubMed]
- Schoeler, M.; Caesar, R. Dietary Lipids, Gut Microbiota and Lipid Metabolism. Rev. Endocr. Metab. Disord. 2019, 20, 461–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mithieux, G. Gut Microbiota and Host Metabolism: What Relationship. Neuroendocrinology 2018, 106, 352–356. [Google Scholar] [CrossRef]
- Dubinkina, V.B.; Tyakht, A.v.; Odintsova, V.Y.; Yarygin, K.S.; Kovarsky, B.A.; Pavlenko, A.v.; Ischenko, D.S.; Popenko, A.S.; Alexeev, D.G.; Taraskina, A.Y.; et al. Links of Gut Microbiota Composition with Alcohol Dependence Syndrome and Alcoholic Liver Disease. Microbiome 2017, 5, 141. [Google Scholar] [CrossRef] [Green Version]
- Lala, V.; Zubair, M.; Minter, D.A. Liver Function Tests; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Abdelmalek, M.F. Nonalcoholic Fatty Liver Disease: Another Leap Forward. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 85–86. [Google Scholar] [CrossRef]
- Azad, M.A.K.; Sarker, M.; Li, T.; Yin, J. Probiotic Species in the Modulation of Gut Microbiota: An Overview. Biomed. Res. Int. 2018, 2018, 9478630. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Li, Y.; Wen, Z.; Liu, W.; Meng, L.; Huang, H. Oscillospira—A Candidate for the next-Generation Probiotics. Gut Microbes 2021, 13, 1987783. [Google Scholar] [CrossRef]
- Han, W.; Huang, C.; Ji, Y.; Zhou, L.; Chen, J.; Hou, J. Alterations in the Gut Microbiota and Hepatitis-B-Virus Infection in Southern Chinese Patients with Coexisting Non-Alcoholic Fatty Liver Disease and Type-2 Diabetes Mellitus. Front. Med. 2021, 8, 2608. [Google Scholar] [CrossRef] [PubMed]
- Kashtanova, D.A.; Klimenko, N.S.; Tkacheva, O.N.; Strazhesko, I.D.; Metelskaya, V.A.; Gomyranova, N.V.; Boytsov, S.A. Subfractional Spectrum of Serum Lipoproteins and Gut Microbiota Composition in Healthy Individuals. Microorganisms 2021, 9, 1461. [Google Scholar] [CrossRef]
- Waters, J.L.; Ley, R.E. The Human Gut Bacteria Christensenellaceae Are Widespread, Heritable, and Associated with Health. BMC Biol. 2019, 17, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González, S.; Fernández-Navarro, T.; Arboleya, S.; de Los Reyes-Gavilán, C.G.; Salazar, N.; Gueimonde, M. Fermented Dairy Foods: Impact on Intestinal Microbiota and Health-Linked Biomarkers. Front. Microbiol. 2019, 10, 1046. [Google Scholar] [CrossRef] [PubMed]
- Ormazabal, V.; Nair, S.; Elfeky, O.; Aguayo, C.; Salomon, C.; Zuñiga, F.A. Association between Insulin Resistance and the Development of Cardiovascular Disease. Cardiovasc. Diabetol. 2018, 17, 122. [Google Scholar] [CrossRef] [Green Version]
- Yuan, X.; Chen, R.; Zhang, Y.; Lin, X.; Yang, X.; McCormick, K.L. Gut Microbiota of Chinese Obese Children and Adolescents with and without Insulin Resistance. Front. Endocrinol. 2021, 12, 163. [Google Scholar] [CrossRef]
- Cuevas-Sierra, A.; Ramos-Lopez, O.; Riezu-Boj, J.I.; Milagro, F.I.; Martinez, J.A. Diet, Gut Microbiota, and Obesity: Links with Host Genetics and Epigenetics and Potential Applications. Adv. Nutr. 2019, 10, S17–S30. [Google Scholar] [CrossRef] [Green Version]
- Loo, Y.T.; Howell, K.; Chan, M.; Zhang, P.; Ng, K. Modulation of the Human Gut Microbiota by Phenolics and Phenolic Fiber-Rich Foods. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1268–1298. [Google Scholar] [CrossRef]
- Redondo-Useros, N.; Gheorghe, A.; Díaz-Prieto, L.E.; Villavisencio, B.; Marcos, A.; Nova, E. Associations of Probiotic Fermented Milk (PFM) and Yogurt Consumption with Bifidobacterium and Lactobacillus Components of the Gut Microbiota in Healthy Adults. Nutrients 2019, 11, 651. [Google Scholar] [CrossRef] [Green Version]
- Papier, K.; Hartman, L.; Tong, T.Y.N.; Key, T.J.; Knuppel, A. Higher Meat Intake Is Associated with Higher Inflammatory Markers, Mostly Due to Adiposity: Results from UK Biobank. J. Nutr. 2022, 152, 183–189. [Google Scholar] [CrossRef]
- Yubero-Serrano, E.; Lopez-Moreno, J.; Gomez-Delgado, F.; Lopez-Miranda, J. Extra Virgin Olive Oil: More than a Healthy Fat. Eur. J. Clin. Nutr. 2019, 72, 8–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia Muciniphila in Overweight and Obese Human Volunteers: A Proof-of-Concept Exploratory Study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Cândido, T.L.N.; Bressan, J.; Alfenas, R.d.C.G. Dysbiosis and Metabolic Endotoxemia Induced by High-Fat Diet. Nutr. Hosp. 2018, 35, 1432–1440. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, Y.; Nagai, F.; Morotomi, M. Characterization of Phascolarctobacterium succinatutens sp. nov., an asaccharolytic, succinate-utilizing bacterium isolated from human feces. Appl. Environ. Microbiol. 2012, 78, 511–518. [Google Scholar] [CrossRef] [PubMed]



| Variables | All Participants (n = 60) | High Group (n = 30) | Low Group (n = 30) |
|---|---|---|---|
| Age (y) | 47.3 ± 11.2 | 55.8 ± 5.8 | 38.3 ± 9.3 |
| Weight (kg) | 69.9 ± 14.1 | 79.8 ± 11.6 | 59.0 ± 5.0 |
| BMI (kg/cm2) | 24.6 ± 3.9 | 27.5 ± 2.8 | 21.4 ± 1.6 |
| Glucose (mg/dL) | 94.5 ± 11.3 | 101.3 ± 11.1 | 86.3 ± 3.9 |
| Total cholesterol (mg/dL) | 219.4 ± 35.4 | 247.5 ± 21.7 | 192.8 ± 21.0 |
| HDL (mg/dL) | 64.3 ± 15.6 | 78.0 ± 11.9 | 52.7 ± 6.8 |
| Triglycerides (mg/dL) | 79.0 ± 36.6 | 102.2 ± 36.1 | 53.1 ± 9.0 |
| Insulin (µIU/mL) | 8.4 ± 4.3 | 11.4 ± 3.7 | 5.2 ± 1.6 |
| AST (µ/L) | 22.0 ± 14.3 | 27.3 ± 17.7 | 16.2 ± 2.4 |
| ALT (µ/L) | 22.0 ± 20.6 | 30.2 ± 25.1 | 12.8 ± 2.4 |
| Variables | All (n = 60) | High Group (n = 30) | Low Group (n = 30) | Variables | All (n = 60) | High Group (n = 30) | Low Group (n = 30) |
|---|---|---|---|---|---|---|---|
| Energy intake (kcal/day) | 2381 ± 796 | 3118 ± 801 | 1819 ± 299 | Yogurt (g/day) | 75.4 ± 78.2 | 129.9 ± 72.4 | 18.0 ± 21.8 |
| Carbohydrate intake (%) | 38.2 ± 7.7 | 43.8 ± 4.2 | 31.9 ± 5.3 | Fermented dairy (g/day) | 93.0 ± 79.8 | 149.6 ± 72.2 | 34.3 ± 22.7 |
| Protein intake (%) | 18.3 ± 3.4 | 21.1 ± 3.0 | 15.9 ± 2.0 | Meat (g/day) | 169.4 ± 75.6 | 263.8 ± 166.0 | 115.1 ± 34.6 |
| Fat intake (%) | 41.3 ± 6.8 | 46.8 ± 5.1 | 36.3 ± 3.2 | Cold meat (g/day) | 7.5 ± 9.2 | 12.8 ± 10.1 | 1.7 ± 1.7 |
| Fiber intake (g/day) | 28.9 ± 11.8 | 39.9 ± 9.6 | 20.1 ± 4.7 | Olive oil (g/day) | 27.6 ± 27.2 | 45.8 ± 30.5 | 12.6 ± 5.6 |
| Vegetables (g/day) | 432.6 ± 206.8 | 632.5 ± 194.1 | 281.3 ± 90.6 | Soda (g/day) | 13.2 ± 29.5 | 26.5 ± 36.5 | 0.0 ± 0.0 |
| Fruit (g/day) | 301.8 ± 204.7 | 527.6 ± 343.2 | 156.6 ± 65.3 | Soda light (g/day) | 20.3 ± 42.2 | 45.9 ± 59.2 | 0.0 ± 0.0 |
| Legumes (g/day) | 23.8 ± 12.6 | 32.2 ± 12.0 | 14.9 ± 4.7 | Total cholesterol (mg/day) | 526.5 ± 210.7 | 727.4 ± 237.0 | 379.5 ± 88.0 |
| Cereals (g/day) | 166.3 ± 98.5 | 241.0 ± 83.4 | 97.1 ± 36.9 | Trans fat (g/day) | 0.8 ± 0.5 | 1.1 ± 0.4 | 0.4 ± 0.1 |
| Whole grains (g/day) | 35.8 ± 39.9 | 64.8 ± 36.8 | 5.8 ± 8.5 | Monounsaturated fat (g/day) | 48.8 ± 22.0 | 69.2 ± 22.7 | 33.4 ± 7.6 |
| Dairy intake (g/day) | 301.3 ± 187.5 | 442.3 ± 142.3 | 154.8 ± 80.1 | Polyunsaturated fat (g/day) | 16.8 ± 6.5 | 24.3 ± 7.6 | 11.6 ± 2.7 |
| Saturated fat (g/day) | 30.4 ± 11.9 | 41.3 ± 8.7 | 21.4 ± 5.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villaseñor-Aranguren, M.; Rosés, C.; Riezu-Boj, J.I.; López-Yoldi, M.; Ramos-Lopez, O.; Barceló, A.M.; Milagro, F.I. Association of the Gut Microbiota with the Host’s Health through an Analysis of Biochemical Markers, Dietary Estimation, and Microbial Composition. Nutrients 2022, 14, 4966. https://doi.org/10.3390/nu14234966
Villaseñor-Aranguren M, Rosés C, Riezu-Boj JI, López-Yoldi M, Ramos-Lopez O, Barceló AM, Milagro FI. Association of the Gut Microbiota with the Host’s Health through an Analysis of Biochemical Markers, Dietary Estimation, and Microbial Composition. Nutrients. 2022; 14(23):4966. https://doi.org/10.3390/nu14234966
Chicago/Turabian StyleVillaseñor-Aranguren, Maite, Carles Rosés, José Ignacio Riezu-Boj, Miguel López-Yoldi, Omar Ramos-Lopez, Anna M. Barceló, and Fermín I. Milagro. 2022. "Association of the Gut Microbiota with the Host’s Health through an Analysis of Biochemical Markers, Dietary Estimation, and Microbial Composition" Nutrients 14, no. 23: 4966. https://doi.org/10.3390/nu14234966
APA StyleVillaseñor-Aranguren, M., Rosés, C., Riezu-Boj, J. I., López-Yoldi, M., Ramos-Lopez, O., Barceló, A. M., & Milagro, F. I. (2022). Association of the Gut Microbiota with the Host’s Health through an Analysis of Biochemical Markers, Dietary Estimation, and Microbial Composition. Nutrients, 14(23), 4966. https://doi.org/10.3390/nu14234966







