Association between Gut Microbiota and Breast Cancer: Diet as a Potential Modulating Factor
Abstract
:1. Introduction
2. Methods
2.1. Participants and Sample Collection
2.2. Diet and Healthy Eating Index Assessment
2.3. Microbiome Data
2.3.1. Bacterial DNA Extraction and Next-Generation Sequencing
2.3.2. Bioinformatics and Statistical Analysis
3. Results
3.1. Participant Characteristics
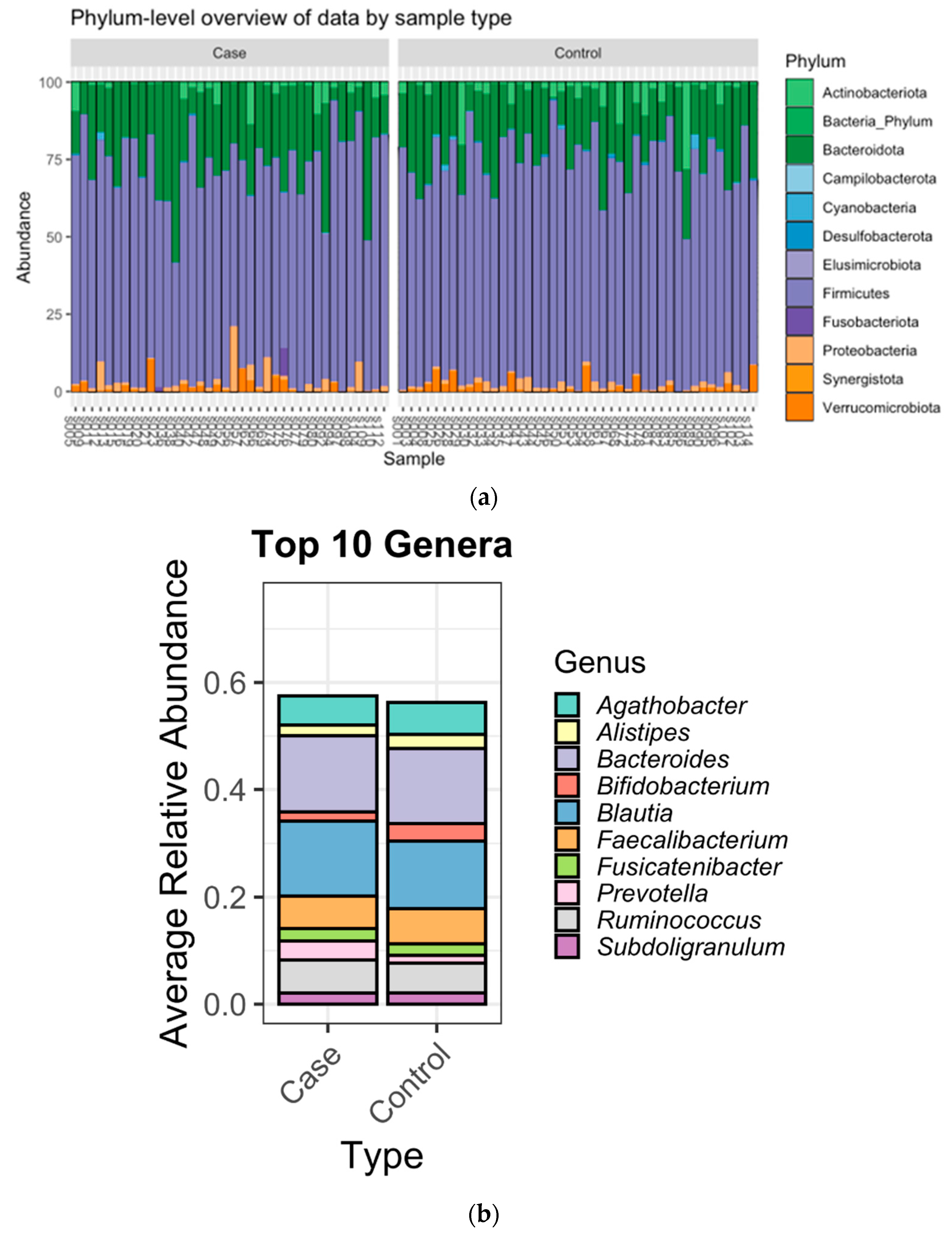
3.2. Microbiome Composition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Giaquinto, A.N.; Sung, H.; Miller, K.D.; Kramer, J.L.; Newman, L.A.; Minihan, A.; Jemal, A.; Siegel, R.L. Breast Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 524–541. [Google Scholar] [CrossRef] [PubMed]
- Britt, K.L.; Cuzick, J.; Phillips, K.A. Key steps for effective breast cancer prevention. Nat. Rev. Cancer 2020, 20, 417–436. [Google Scholar] [CrossRef] [PubMed]
- Uhomoibhi, T.O.; Okobi, T.J.; Okobi, O.E.; Koko, J.O.; Uhomoibhi, O.; Igbinosun, O.E.; Ehibor, U.D.; Boms, M.G.; Abdulgaffar, R.A.; Hammed, B.L.; et al. High-Fat Diet as a Risk Factor for Breast Cancer: A Meta-Analysis. Cureus 2022, 14, e32309. [Google Scholar] [CrossRef] [PubMed]
- Kazemi, A.; Barati-Boldaji, R.; Soltani, S.; Mohammadipoor, N.; Esmaeilinezhad, Z.; Clark, C.C.T.; Babajafari, S.; Akbarzadeh, M. Intake of Various Food Groups and Risk of Breast Cancer: A Systematic Review and Dose-Response Meta-Analysis of Prospective Studies. Adv. Nutr. 2021, 12, 809–849. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Q.; Zhang, Y.; Xie, Q.; Tan, X. Physical Activity and Risk of Breast Cancer: A Meta-Analysis of 38 Cohort Studies in 45 Study Reports. Value Health 2019, 22, 104–128. [Google Scholar] [CrossRef]
- Sun, Q.; Xie, W.; Wang, Y.; Chong, F.; Song, M.; Li, T.; Xu, L.; Song, C. Alcohol Consumption by Beverage Type and Risk of Breast Cancer: A Dose-Response Meta-Analysis of Prospective Cohort Studies. Alcohol Alcohol. 2020, 55, 246–253. [Google Scholar] [CrossRef]
- He, Y.; Si, Y.; Li, X.; Hong, J.; Yu, C.; He, N. The relationship between tobacco and breast cancer incidence: A systematic review and meta-analysis of observational studies. Front. Oncol. 2022, 12, 961970. [Google Scholar] [CrossRef]
- Sarhangi, N.; Hajjari, S.; Heydari, S.F.; Ganjizadeh, M.; Rouhollah, F.; Hasanzad, M. Breast cancer in the era of precision medicine. Mol. Biol. Rep. 2022, 49, 10023–10037. [Google Scholar] [CrossRef]
- Eslami, S.Z.; Majidzadeh, A.K.; Halvaei, S.; Babapirali, F.; Esmaeili, R. Microbiome and Breast Cancer: New Role for an Ancient Population. Front. Oncol. 2020, 10, 120. [Google Scholar] [CrossRef]
- Shreiner, A.B.; Kao, J.Y.; Young, V.B. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015, 31, 69–75. [Google Scholar] [CrossRef]
- Bodai, B.I.; Nakata, T.E. Breast Cancer: Lifestyle, the Human Gut Microbiota/Microbiome, and Survivorship. Perm. J. 2020, 24, 19.129. [Google Scholar] [CrossRef]
- Wu, A.H.; Tseng, C.; Vigen, C.; Yu, Y.; Cozen, W.; Garcia, A.A.; Spicer, D. Gut microbiome associations with breast cancer risk factors and tumor characteristics: A pilot study. Breast Cancer Res. Treat. 2020, 182, 451–463. [Google Scholar] [CrossRef]
- Jaye, K.; Chang, D.; Li, C.G.; Bhuyan, D.J. Gut Metabolites and Breast Cancer: The Continuum of Dysbiosis, Breast Cancer Risk, and Potential Breast Cancer Therapy. Int. J. Mol. Sci. 2022, 23, 9490. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef]
- Feng, T.Y.; Azar, F.N.; Dreger, S.A.; Rosean, C.B.; McGinty, M.T.; Putelo, A.M.; Kolli, S.H.; Carey, M.A.; Greenfield, S.; Fowler, W.J.; et al. Reciprocal Interactions Between the Gut Microbiome and Mammary Tissue Mast Cells Promote Metastatic Dissemination of HR+ Breast Tumors. Cancer Immunol. Res. 2022, 10, 1309–1325. [Google Scholar] [CrossRef] [PubMed]
- Laborda-Illanes, A.; Sanchez-Alcoholado, L.; Boutriq, S.; Plaza-Andrades, I.; Peralta-Linero, J.; Alba, E.; Gonzalez-Gonzalez, A.; Queipo-Ortuno, M.I. A New Paradigm in the Relationship between Melatonin and Breast Cancer: Gut Microbiota Identified as a Potential Regulatory Agent. Cancers 2021, 13, 3141. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.F.; Reina-Perez, I.; Astorga, J.M.; Rodriguez-Carrillo, A.; Plaza-Diaz, J.; Fontana, L. Breast Cancer and Its Relationship with the Microbiota. Int. J. Environ. Res. Public Health 2018, 15, 1747. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef]
- Mahmood, R.; Voisin, A.; Olof, H.; Khorasaniha, R.; Lawal, S.A.; Armstrong, H.K. Host Microbiomes Influence the Effects of Diet on Inflammation and Cancer. Cancers 2023, 15, 521. [Google Scholar] [CrossRef] [PubMed]
- Diet History Questionnaire, Version 3.0; National Institutes of Health, Applied Research Program; National Cancer Institute: Bethesda, MD, USA, 2020.
- Reedy, J.; Lerman, J.L.; Krebs-Smith, S.M.; Kirkpatrick, S.I.; Pannucci, T.E.; Wilson, M.M.; Subar, A.F.; Kahle, L.L.; Tooze, J.A. Evaluation of the Healthy Eating Index-2015. J. Acad. Nutr. Diet. 2018, 118, 1622–1633. [Google Scholar] [CrossRef] [PubMed]
- Ware, J., Jr.; Kosinski, M.; Keller, S.D. A 12-Item Short-Form Health Survey: Construction of scales and preliminary tests of reliability and validity. Med. Care 1996, 34, 220–233. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Whittaker, R.H. Vegetation of the Siskiyou Mountains, Oregon and California. Ecol. Monogr. 1960, 30, 279–338. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Technol. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Simpson, E.H. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Gotelli, N.J.; Colwell, R.K. Quantifying biodiversity: Procedures and pitfalls in the measurement and comparison of species richness. Ecol. Lett. 2001, 4, 379–391. [Google Scholar] [CrossRef]
- Pielou, E.C. The measurement of diversity in different types of biological collections. J. Theor. Biol. 1966, 13, 131–144. [Google Scholar] [CrossRef]
- Schroeder, P.J.; Jenkins, D.G. How robust are popular beta diversity indices to sampling error? Ecosphere 2018, 9, e02100. [Google Scholar] [CrossRef]
- Legendre, P.; De Cáceres, M. Beta diversity as the variance of community data: Dissimilarity coefficients and partitioning. Ecol. Lett. 2013, 16, 951–963. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Guo, W.; Fensom, G.K.; Reeves, G.K.; Key, T.J. Physical activity and breast cancer risk: Results from the UK Biobank prospective cohort. Br. J. Cancer 2020, 122, 726–732. [Google Scholar] [CrossRef]
- Heath, A.K.; Muller, D.C.; van den Brandt, P.A.; Papadimitriou, N.; Critselis, E.; Gunter, M.; Vineis, P.; Weiderpass, E.; Fagherazzi, G.; Boeing, H.; et al. Nutrient-wide association study of 92 foods and nutrients and breast cancer risk. Breast Cancer Res. 2020, 22, 5. [Google Scholar] [CrossRef]
- Zhu, G.L.; Xu, C.; Yang, K.B.; Tang, S.Q.; Tang, L.L.; Chen, L.; Li, W.F.; Mao, Y.P.; Ma, J. Causal relationship between genetically predicted depression and cancer risk: A two-sample bi-directional mendelian randomization. BMC Cancer 2022, 22, 353. [Google Scholar] [CrossRef] [PubMed]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 2015, 26, 26191. [Google Scholar] [CrossRef] [PubMed]
- Bobin-Dubigeon, C.; Luu, H.T.; Leuillet, S.; Lavergne, S.N.; Carton, T.; Le Vacon, F.; Michel, C.; Nazih, H.; Bard, J.M. Faecal Microbiota Composition Varies between Patients with Breast Cancer and Healthy Women: A Comparative Case-Control Study. Nutrients 2021, 13, 2705. [Google Scholar] [CrossRef]
- Byrd, D.A.; Vogtmann, E.; Wu, Z.; Han, Y.; Wan, Y.; Clegg-Lamptey, J.N.; Yarney, J.; Wiafe-Addai, B.; Wiafe, S.; Awuah, B.; et al. Associations of fecal microbial profiles with breast cancer and nonmalignant breast disease in the Ghana Breast Health Study. Int. J. Cancer 2021, 148, 2712–2723. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.F.; Ou-Yang, F.; Li, C.L.; Chen, F.M.; Chuang, C.H.; Kan, J.Y.; Wu, C.C.; Shih, S.L.; Shiau, J.P.; Kao, L.C.; et al. Comprehensive profiles and diagnostic value of menopausal-specific gut microbiota in premenopausal breast cancer. Exp. Mol. Med. 2021, 53, 1636–1646. [Google Scholar] [CrossRef] [PubMed]
- Goedert, J.J.; Jones, G.; Hua, X.; Xu, X.; Yu, G.; Flores, R.; Falk, R.T.; Gail, M.H.; Shi, J.; Ravel, J.; et al. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: A population-based case-control pilot study. J. Natl. Cancer Inst. 2015, 107, djv147. [Google Scholar] [CrossRef]
- Ruo, S.W.; Alkayyali, T.; Win, M.; Tara, A.; Joseph, C.; Kannan, A.; Srivastava, K.; Ochuba, O.; Sandhu, J.K.; Went, T.R.; et al. Role of Gut Microbiota Dysbiosis in Breast Cancer and Novel Approaches in Prevention, Diagnosis, and Treatment. Cureus 2021, 13, e17472. [Google Scholar] [CrossRef]
- Parida, S.; Sharma, D. Microbial Alterations and Risk Factors of Breast Cancer: Connections and Mechanistic Insights. Cells 2020, 9, 1091. [Google Scholar] [CrossRef]
- Kelly, J.R.; Kennedy, P.J.; Cryan, J.F.; Dinan, T.G.; Clarke, G.; Hyland, N.P. Breaking down the barriers: The gut microbiome, intestinal permeability and stress-related psychiatric disorders. Front. Cell. Neurosci. 2015, 9, 392. [Google Scholar] [CrossRef]
- Han, K.M.; Ham, B.J. How Inflammation Affects the Brain in Depression: A Review of Functional and Structural MRI Studies. J. Clin. Neurol. 2021, 17, 503–515. [Google Scholar] [CrossRef]
- Asfaw, M.S.; Dagne, W.K. Physical activity can improve diabetes patients’ glucose control; A systematic review and meta-analysis. Heliyon 2022, 8, e12267. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Fuhler, G.M.; Bn, N.; Jose, T.; Bruno, M.J.; Peppelenbosch, M.P.; Konstantinov, S.R. Pancreatic cyst fluid harbors a unique microbiome. Microbiome 2017, 5, 147. [Google Scholar] [CrossRef] [PubMed]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed]
- al-Waiz, M.; Mikov, M.; Mitchell, S.C.; Smith, R.L. The exogenous origin of trimethylamine in the mouse. Metabolism 1992, 41, 135–136. [Google Scholar] [CrossRef] [PubMed]
- Genoni, A.; Christophersen, C.T.; Lo, J.; Coghlan, M.; Boyce, M.C.; Bird, A.R.; Lyons-Wall, P.; Devine, A. Long-term Paleolithic diet is associated with lower resistant starch intake, different gut microbiota composition and increased serum TMAO concentrations. Eur. J. Nutr. 2020, 59, 1845–1858. [Google Scholar] [CrossRef] [PubMed]
- Rawat, P.S.; Li, Y.; Zhang, W.; Meng, X.; Liu, W. Hungatella hathewayi, an Efficient Glycosaminoglycan-Degrading Firmicutes from Human Gut and Its Chondroitin ABC Exolyase with High Activity and Broad Substrate Specificity. Appl. Environ. Microbiol. 2022, 88, e0154622. [Google Scholar] [CrossRef]
- Xia, X.; Wu, W.K.K.; Wong, S.H.; Liu, D.; Kwong, T.N.Y.; Nakatsu, G.; Yan, P.S.; Chuang, Y.-M.; Chan, M.W.-Y.; Coker, O.O.; et al. Bacteria pathogens drive host colonic epithelial cell promoter hypermethylation of tumor suppressor genes in colorectal cancer. Microbiome 2020, 8, 108. [Google Scholar] [CrossRef]
- Kelly, T.N.; Bazzano, L.A.; Ajami, N.J.; He, H.; Zhao, J.; Petrosino, J.F.; Correa, A.; He, J. Gut Microbiome Associates With Lifetime Cardiovascular Disease Risk Profile Among Bogalusa Heart Study Participants. Circ. Res. 2016, 119, 956–964. [Google Scholar] [CrossRef]
- Xu, A.A.; Kennedy, L.K.; Hoffman, K.; White, D.L.; Kanwal, F.; El-Serag, H.B.; Petrosino, J.F.; Jiao, L. Dietary Fatty Acid Intake and the Colonic Gut Microbiota in Humans. Nutrients 2022, 14, 2722. [Google Scholar] [CrossRef]
- Al-Qadami, G.H.; Secombe, K.R.; Subramaniam, C.B.; Wardill, H.R.; Bowen, J.M. Gut Microbiota-Derived Short-Chain Fatty Acids: Impact on Cancer Treatment Response and Toxicities. Microorganisms 2022, 10, 2048. [Google Scholar] [CrossRef]
- He, Y.; Fu, L.; Li, Y.; Wang, W.; Gong, M.; Zhang, J.; Dong, X.; Huang, J.; Wang, Q.; Mackay, C.R.; et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8(+) T cell immunity. Cell Metab. 2021, 33, 988–1000. [Google Scholar] [CrossRef] [PubMed]
- Rangan, P.; Mondino, A. Microbial short-chain fatty acids: A strategy to tune adoptive T cell therapy. J. Immunother. Cancer 2022, 10, e004147. [Google Scholar] [CrossRef] [PubMed]


| Variable | BCa Cases (N = 42) | Controls (N = 44) | p-Value |
|---|---|---|---|
| Demographic Characteristics | |||
| Age at enrollment, mean (SD), yr | 60.3 (11.3) | 58.5 (12.6) | 0.49 |
| Race, No. (%) | |||
| White | 40 (95.2) | 41 (93.2) | 0.68 |
| Non-White | 2 (4.8) | 3 (6.8) | |
| BMI (kg/m2) at enrollment, mean (SD) continuous variable | 28.1 (6.2) | 25.1 (5.6) | 0.02 |
| Family history of BCa, No. (%) | 0.95 | ||
| Yes | 15 (35.7) | 16 (36.4) | |
| No | 27 (64.3) | 28 (63.6) | |
| Ever full-term live birth, No. (%) | 0.97 | ||
| Yes | 30 (71.4) | 28 (71.8) | |
| No | 12 (28.6) | 11 (28.2) | |
| Missing | 0 | 5 | |
| Menopausal status, No. (%) | 0.90 | ||
| Premenopausal | 10 (23.8) | 11 (25.0) | |
| Post/peri-menopausal | 32 (76.2) | 33 (75.0) | |
| Age at menarche, No. (%) | 0.83 | ||
| ≤11 years old | 10 (23.8) | 9 (20.5) | |
| 12–14 years old | 26 (61.9) | 30 (68.2) | |
| ≥15 years old | 6 (14.3) | 5 (11.4) | |
| Marital Status, No. (%) | 0.25 | ||
| Married/Living as married | 27 (64.3) | 29 (65.9) | |
| Divorced/Separated/Widowed | 13 (31.0) | 9 (20.5) | |
| Single, never married | 2 (4.8) | 6 (13.6) | |
| Employment Status, No. (%) | 0.13 | ||
| Employed/self-employed | 20 (47.6) | 28 (63.6) | |
| Unemployed/Disabled/Retired/Homemaker | 22 (52.4) | 16 (36.4) |
| Variable | BCa Cases (N = 42) | Controls (N = 44) | p-Value |
|---|---|---|---|
| Life Style Characteristics | |||
| Total dietary energy intake (kcal/day) | 1664.9 (752.8) | 1521.5 (590.8) | 0.33 |
| Total energy expended from recreational physical activity (MET-minutes/week) | 2093.9 (1247.1) | 2728.4 (1274.4) | 0.02 |
| Ever Smoked 100 cigarettes in life | 0.95 | ||
| Yes | 15 (35.7) | 16 (36.4) | |
| No | 27 (64.3) | 28 (63.6) | |
| Alcohol, No. (%) | 0.45 | ||
| Current drinkers | 36 (85.7) | 34 (77.3) | |
| Past drinkers | 3 (7.1) | 7 (15.9) | |
| Never drinkers | 3 (7.1) | 3 (6.8) | |
| Missing | |||
| Sleep Quality Score, No. (%) | 0.58 | ||
| 11–14 | 13 (31.0) | 10 (22.7) | |
| 15–16 | 15 (35.7) | 15 (34.1) | |
| 17–18 | 14 (33.3) | 19 (43.2) | |
| Ever used hormone therapy, No. (%) | 0.29 | ||
| Yes | 24 (57.1) | 30 (68.2) | |
| No | 18 (42.9) | 14 (31.8) | |
| Regular (at least once a week) Probiotic product use, No. (%) | 0.52 | ||
| Yes | 21 (50.0) | 19 (43.2) | |
| No | 21 (50.0) | 25 (56.8) | |
| Feeling downhearted and blue in the past year, No. (%) | 0.02 | ||
| All/most of the time | 8 (19.1) | 2 (4.6) | |
| Some of the time | 19 (45.2) | 13 (29.6) | |
| A little of the time | 11 (26.2) | 19 (43.2) | |
| None of the time | 4 (9.5) | 10 (22.7) | |
| Feeling downhearted and blue around age 25, No. (%) | 0.01 | ||
| All/most of the time | 2 (5.0) | 4 (9.1) | |
| Some of the time | 9 (22.5) | 12 (27.3) | |
| A little of the time | 17 (42.5) | 15 (34.1) | |
| None of the time | 12 (30.0) | 13 (27.3) | |
| Don’t know | 2 | 0 |
| Alpha Diversity | Cases | Controls | p-Value (Unadjusted) ANOVA | p-Value (Adjusted BMI) ANOVA | Wilcoxon Rank Sum Test with Continuity Correction p-Value |
|---|---|---|---|---|---|
| Phylum Level | |||||
| Observed | 198.57 (52.14) | 228.73 (68.94) | 0.025 | 0.025 | 0.045 |
| Shannon | 3.91 (0.40) | 4.13 (0.42) | 0.013 | 0.012 | 0.013 |
| Inverse Simpson | 25.97 (11.18) | 34.22 (15.47) | 0.006 | 0.005 | 0.007 |
| Pielou | 0.74 (0.06) | 0.77 (0.04) | 0.030 | 0.028 | 0.009 |
| Case-Control | Sample Size | Permutations | pseudo_F | p-Value | q-Value |
|---|---|---|---|---|---|
| Jaccard-significance | 86 | 999 | 1.261412 | 0.04 | 0.04 |
| Bray-Curtis-significance | 86 | 999 | 1.121184 | 0.23 | 0.23 |
| Healthy Eating Index 2015 | Min-Max Score | Case-Control Status | Acidaminococus | Hungatella | Tyzzerella | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases (n = 39) | Controls (n = 44) | p Value | Positive (n = 14) | Negative (n = 69) | p Value | Positive (n = 18) | Negative (n = 65) | p Value | Positive (n = 24) | Negative (n = 59) | p Value | ||
| Adequacy | |||||||||||||
| Total vegetable score | 0–5 | 5.00 (0.60) | 4.90 (1.13) | 0.25 | 5.00 (1.40) | 5.00 (0.59) | 0.989 | 5.00 (0.00) | 4.88 (1.20) | 0.024 | 5.00 (0.63) | 5.00 (1.19) | 0.282 |
| Greens and beans score | 0–5 | 5.00 (0.00) | 5.00 (0.00) | 0.83 | 5.00 (3.05) | 5.00 (0) | 0.055 | 5.00 (0.00) | 5.00 (0.00) | 0.73 | 5.00 (0.00) | 5.00 (0.00) | 0.941 |
| Total fruits score | 0–5 | 5.00 (2.61) | 5.00 (0.59) | 0.23 | 5.00 (4.10) | 5.00 (1.35) | 0.06 | 5.00 (1.35) | 5.00 (2.01) | 0.749 | 5.00 (1.05) | 5.00 (2.01) | 0.417 |
| Whole fruits score | 0–5 | 5.00 (1.40) | 5.00 (0.00) | 0.10 | 4.50 (3.31) | 5.00 (0.00) | 0.005 | 5.00 (0.00) | 5.00 (0.00) | 0.573 | 5.00 (0.55) | 5.00 (0.00) | 0.795 |
| Whole grains score | 0–10 | 2.68 (2.32) | 3.45 (2.96) | 0.25 | 2.76 (3.89) | 3.15 (2.41) | 0.507 | 2.60 (2.33) | 3.15 (2.43) | 0.973 | 2.31 (3.12) | 3.15 (2.48) | 0.563 |
| Dairy score | 0–10 | 5.88 (4.52) | 6.97 (4.70) | 0.27 | 5.73 (5.49) | 6.60 (4.25) | 0.22 | 4.99 (4.22) | 6.60 (4.31) | 0.029 | 5.39 (4.99) | 7.01 (3.72) | 0.084 |
| Total protein food score | 0–5 | 5.00 (0.72) | 5.00 (0.00) | 0.29 | 5.00 (1.02) | 5.00 (0.00) | 0.511 | 5.00 (1.02) | 5.00 (0.00) | 0.207 | 5.00 (0.33) | 5.00 (0.00) | 0.702 |
| Seafood and plant proteins score | 0–5 | 5.00 (0.21) | 5.00 (0.00) | 0.35 | 5.00 (0.15) | 5.00 (0.00) | 0.679 | 5.00 (0.93) | 5.00 (0.00) | 0.1 | 5.00 (0.55) | 5.00 (0.00) | 0.58 |
| Fatty acids score | 0–10 | 5.5 (4.90) | 5.22 (7.21) | 0.65 | 4.52 (5.80) | 5.26 (4.41) | 0.591 | 6.20 (6.68) | 5.06 (4.65) | 0.694 | 5.24 (5.64) | 5.24 (4.51) | 0.259 |
| Moderation | |||||||||||||
| Sodium score | 0–10 | 5.25 (2.44) | 5.25 (2.12) | 0.99 | 4.50 (2.18) | 5.40 (2.26) | 0.18 | 5.76 (2.69) | 5.11 (2.13) | 0.28 | 5.49 (2.06) | 5.15 (2.34) | 0.55 |
| Refined grains | 0–10 | 9.96 (1.56) | 10.00 (1.25) | 0.28 | 10.00 (2.49) | 10.00 (1.26) | 0.178 | 9.56 (1.60) | 10.00 (1.18) | 0.326 | 10.00 (0.54) | 9.96 (1.56) | 0.126 |
| Saturated fat | 0–10 | 6.27 (3.34) | 6.32 (5.44) | 0.87 | 6.44 (4.61) | 6.23 (4.84) | 0.883 | 6.88 (4.91) | 5.85 (4.26) | 0.232 | 10.00 (4.07) | 5.55 (4.48) | 0.116 |
| Added sugar | 0–10 | 9.19 (2.41) | 9.52 (2.36) | 0.68 | 8.95 (2.93) | 9.54 (2.29) | 0.243 | 9.14 (2.93) | 9.50 (2.37) | 0.502 | 9.49 (2.02) | 9.30 (2.44) | 0.946 |
| Total HEI-2015 Score | 0–100 | 70.27 (13.08) | 75.77 (15.37) | 0.17 | 68.80 (32.20) | 72.73 (14.39) | 0.45 | 70.55 (10.16) | 72.73 (15.19) | 0.48 | 75.11 (13.50) | 71.28 (15.19) | 0.14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Altinok Dindar, D.; Chun, B.; Palma, A.; Cheney, J.; Krieger, M.; Kasschau, K.; Stagaman, K.; Mitri, Z.I.; Goodyear, S.M.; Shannon, J.; et al. Association between Gut Microbiota and Breast Cancer: Diet as a Potential Modulating Factor. Nutrients 2023, 15, 4628. https://doi.org/10.3390/nu15214628
Altinok Dindar D, Chun B, Palma A, Cheney J, Krieger M, Kasschau K, Stagaman K, Mitri ZI, Goodyear SM, Shannon J, et al. Association between Gut Microbiota and Breast Cancer: Diet as a Potential Modulating Factor. Nutrients. 2023; 15(21):4628. https://doi.org/10.3390/nu15214628
Chicago/Turabian StyleAltinok Dindar, Duygu, Brie Chun, Amy Palma, John Cheney, Madeline Krieger, Kristin Kasschau, Keaton Stagaman, Zahi I. Mitri, Shaun M. Goodyear, Jackilen Shannon, and et al. 2023. "Association between Gut Microbiota and Breast Cancer: Diet as a Potential Modulating Factor" Nutrients 15, no. 21: 4628. https://doi.org/10.3390/nu15214628
APA StyleAltinok Dindar, D., Chun, B., Palma, A., Cheney, J., Krieger, M., Kasschau, K., Stagaman, K., Mitri, Z. I., Goodyear, S. M., Shannon, J., Karstens, L., Sharpton, T., & Zhang, Z. (2023). Association between Gut Microbiota and Breast Cancer: Diet as a Potential Modulating Factor. Nutrients, 15(21), 4628. https://doi.org/10.3390/nu15214628







