Impact of Kiwifruit Consumption on Cholesterol Metabolism in Rat Liver: A Gene Expression Analysis in Induced Hypercholesterolemia
Abstract
1. Introduction
2. Materials and Methods
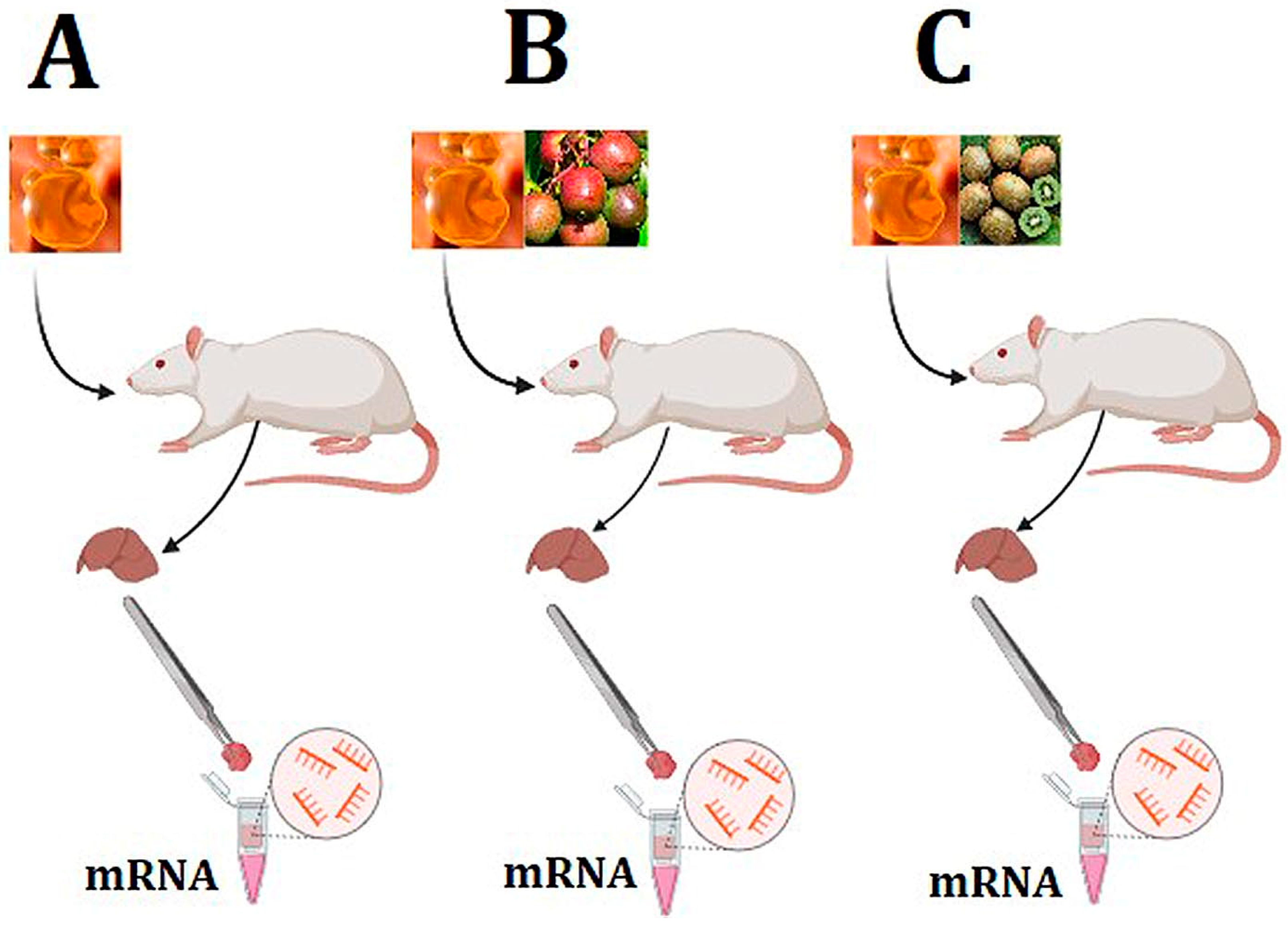
2.1. Study Design
2.2. Sample Collection and RNA Extraction
2.3. Microarray Analysis
2.4. Real-Time qPCR Analysis
2.5. Pathway, Gene Ontology, and Literature Analysis
2.6. MiRNA and Transcription Factor Prediction
3. Results
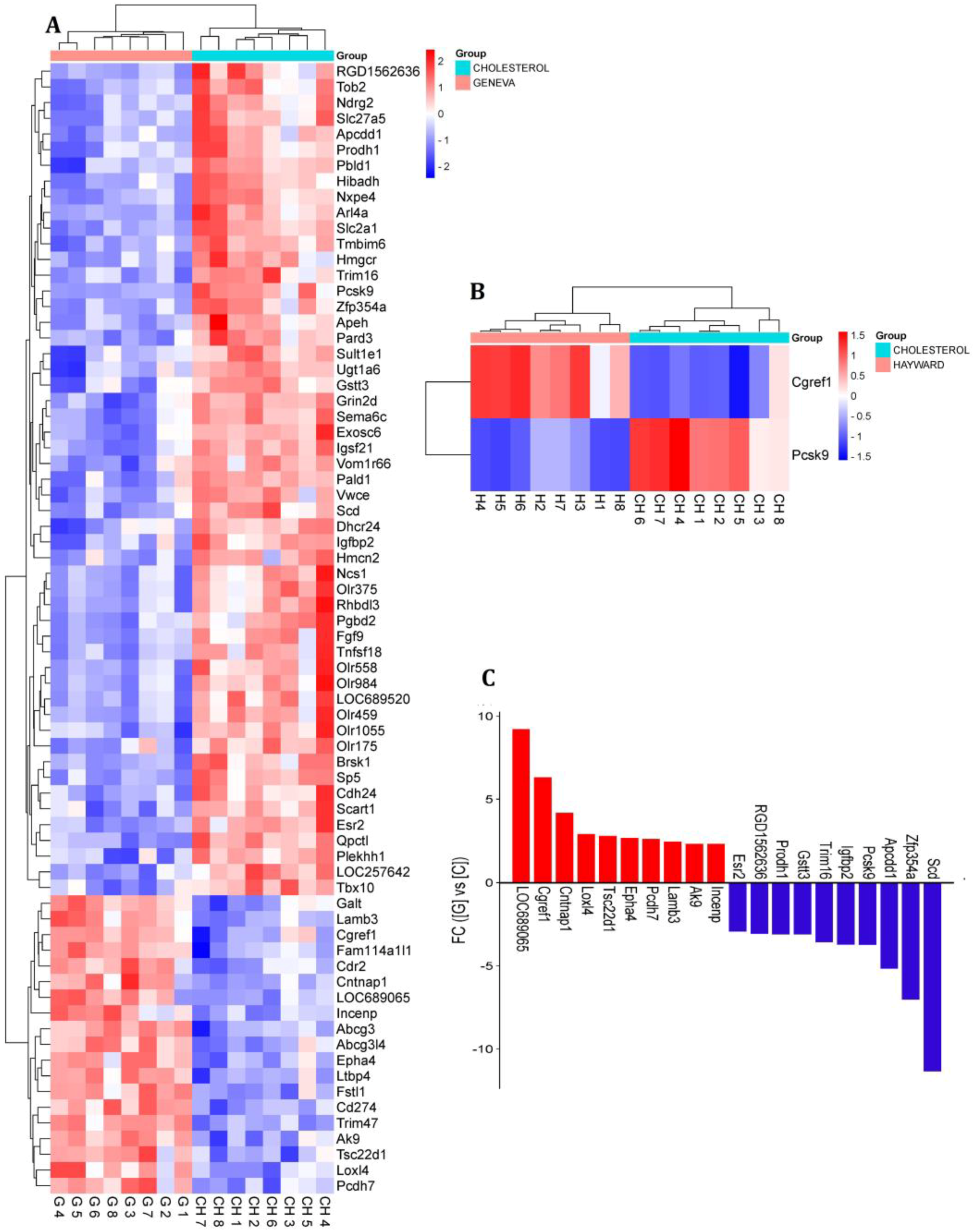
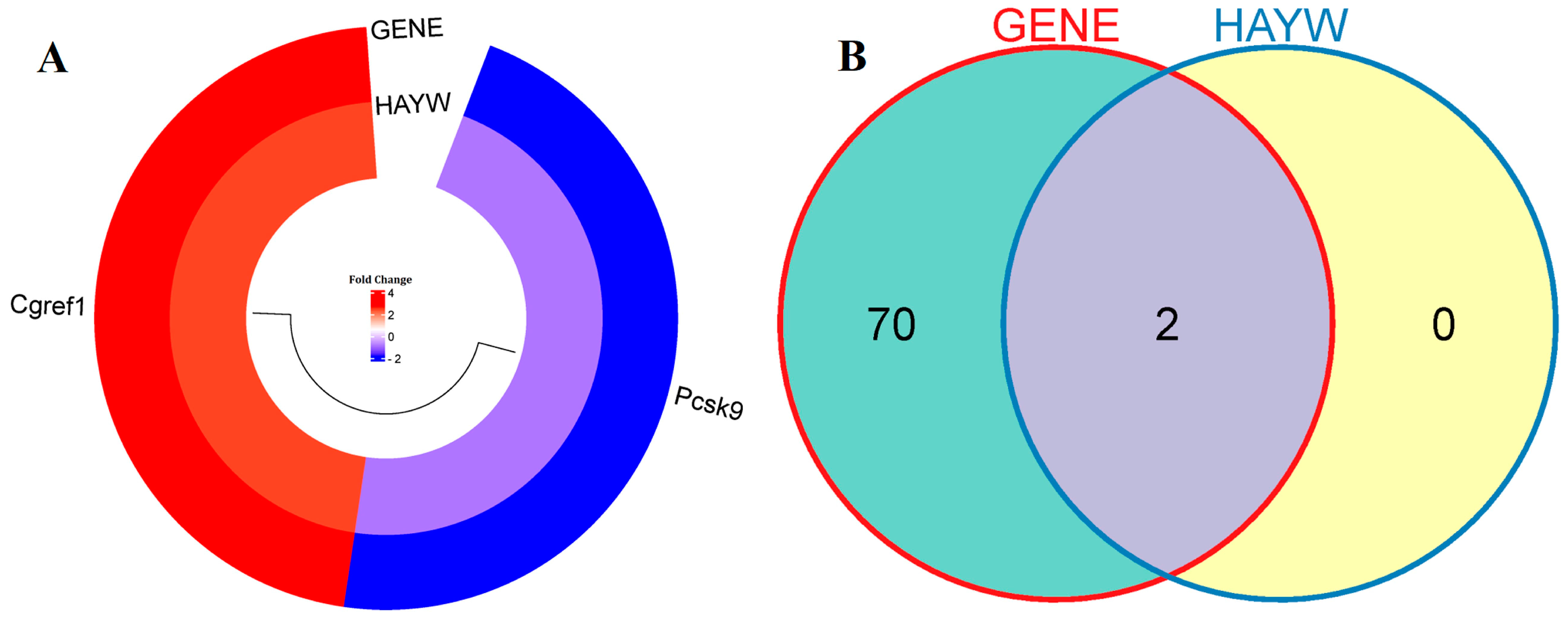
3.1. Identification of Differentially Expressed Genes
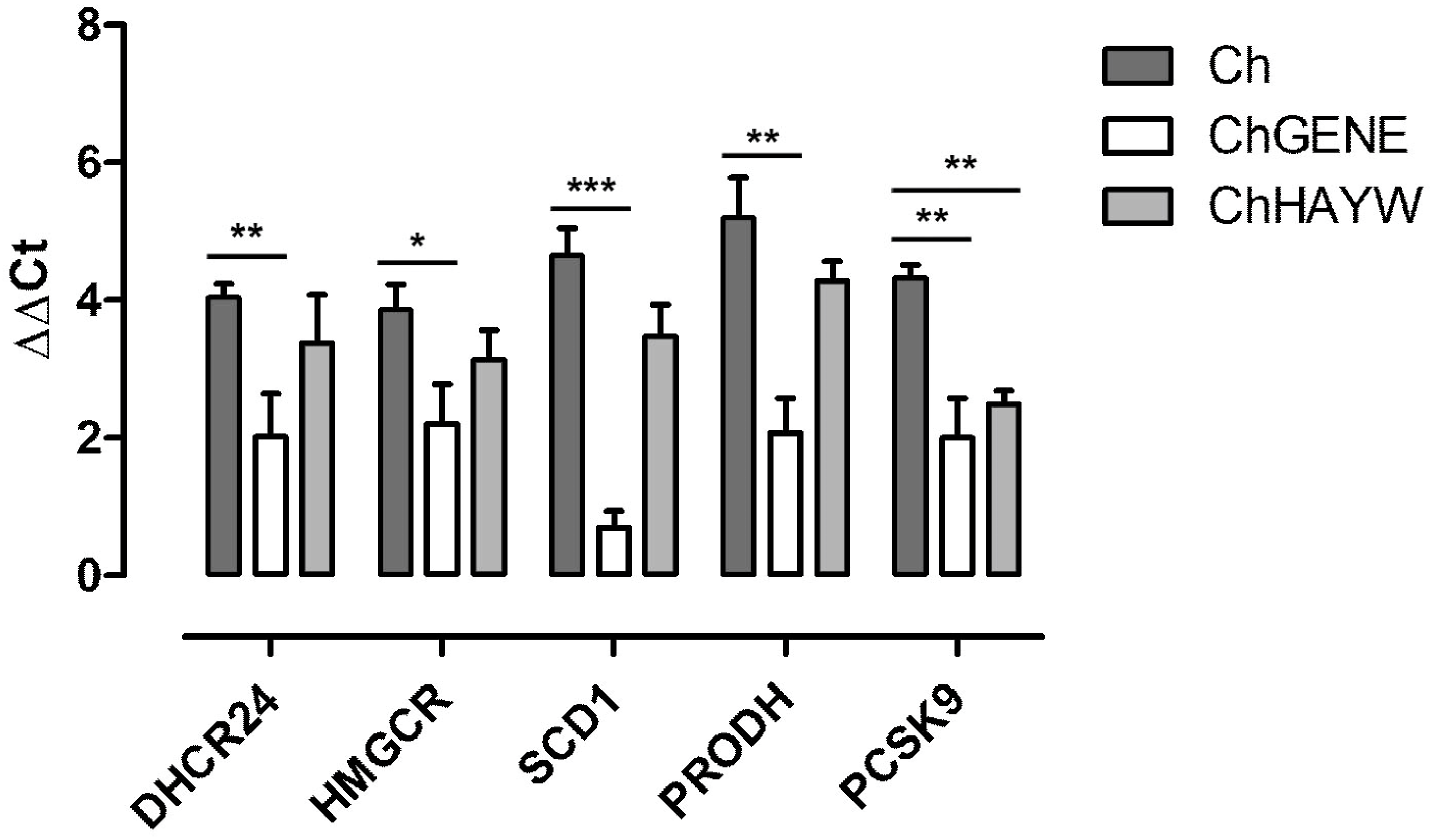
3.2. Real-Time qPCR Validation
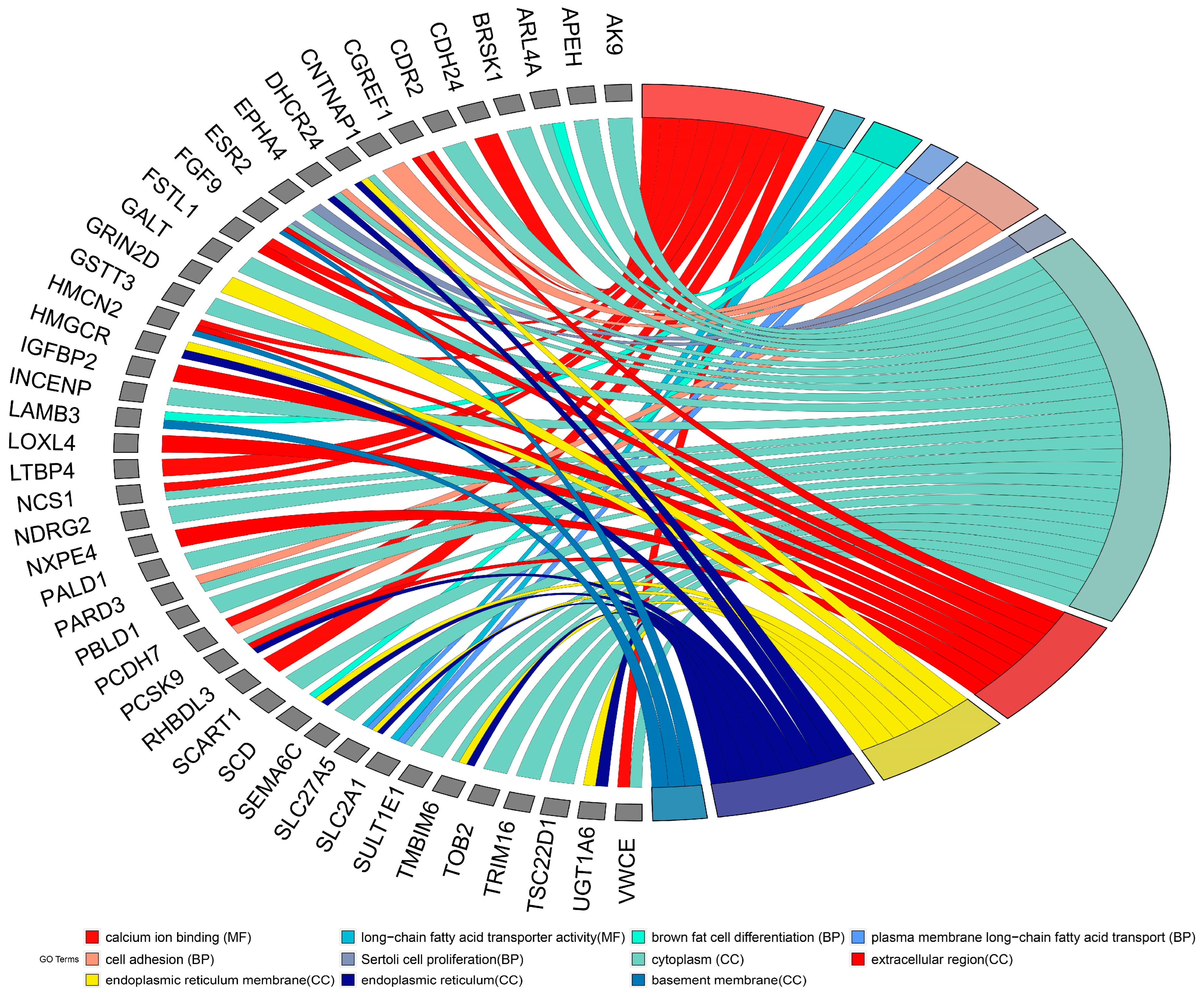
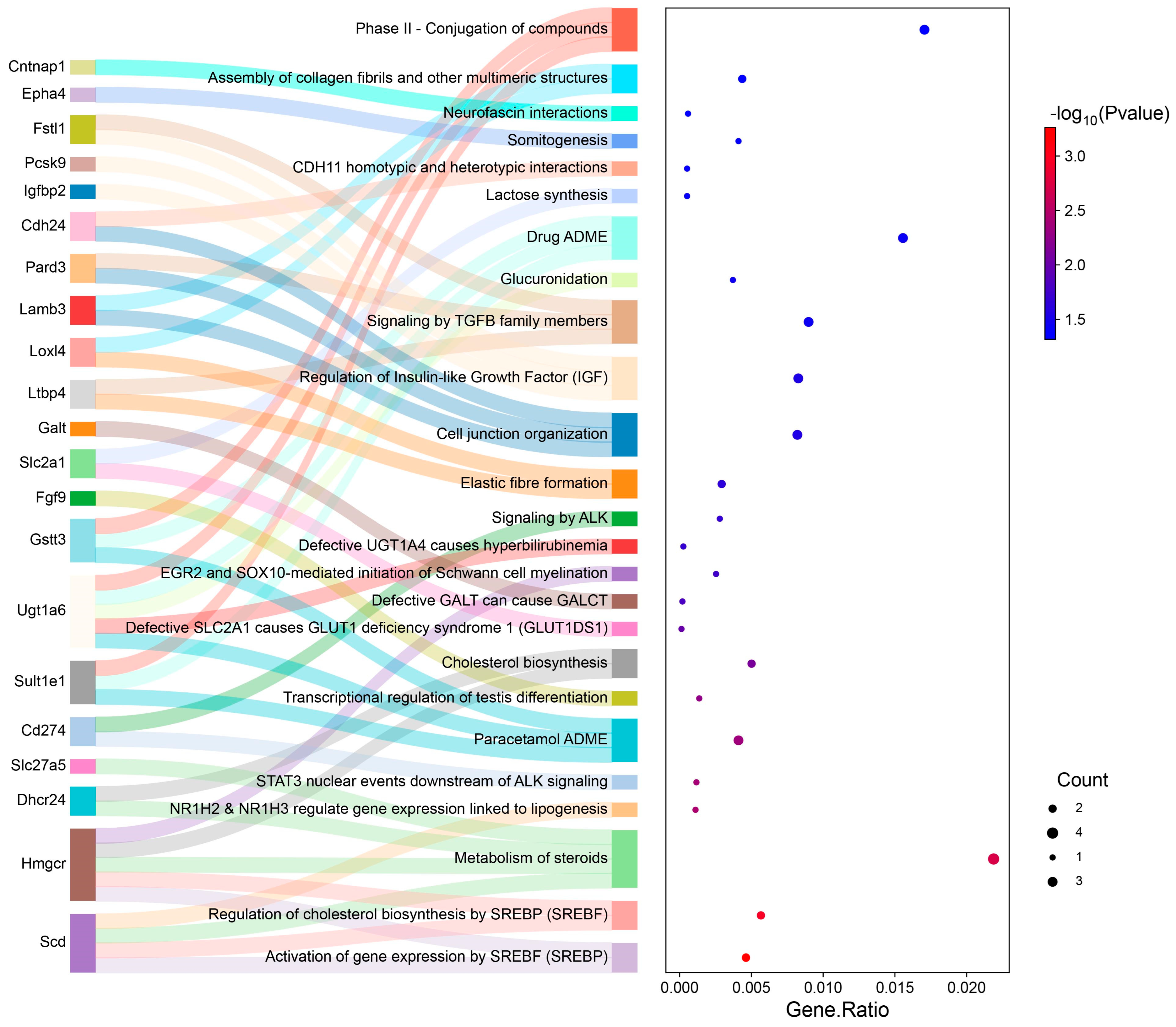
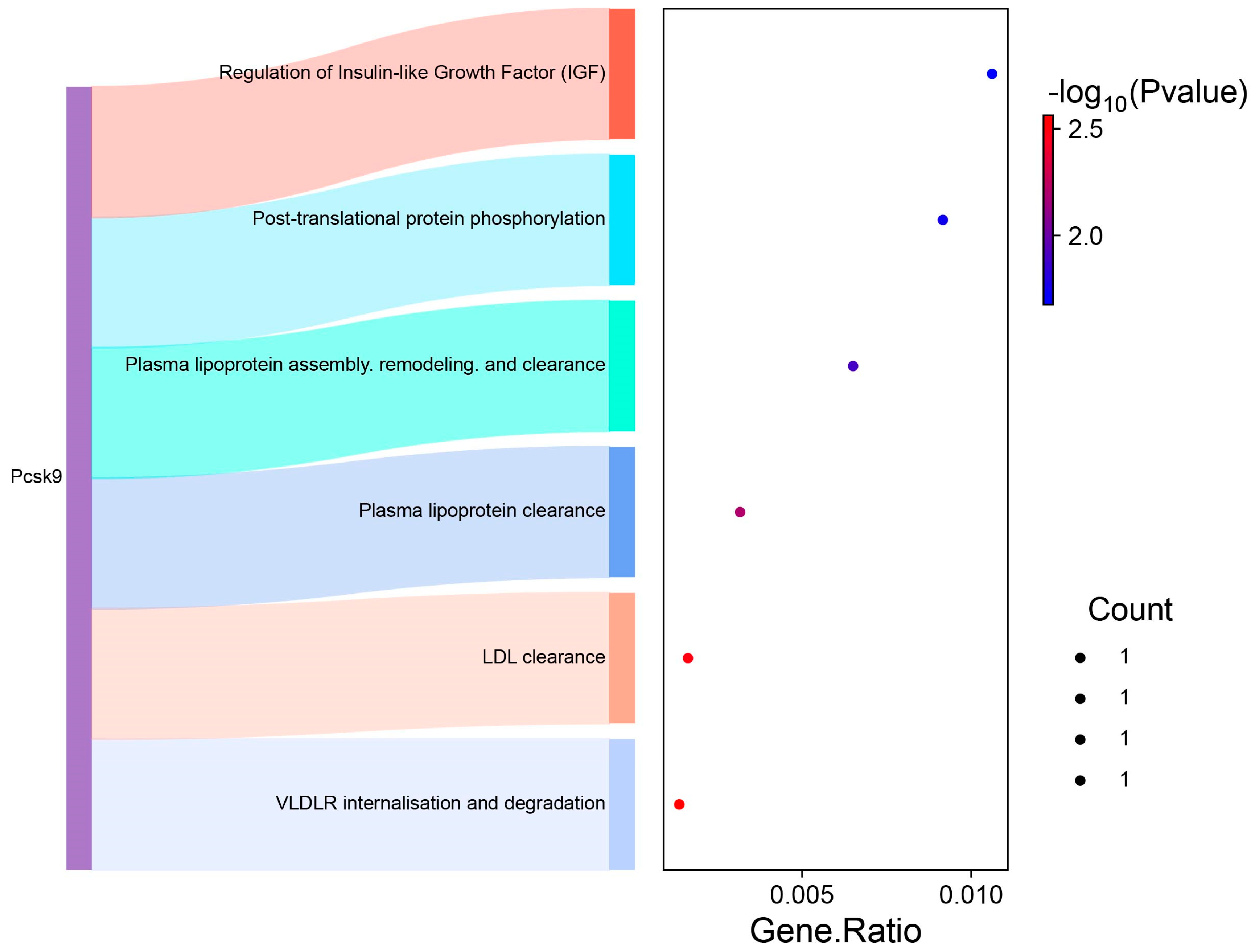
3.3. GO and Pathway Analysis
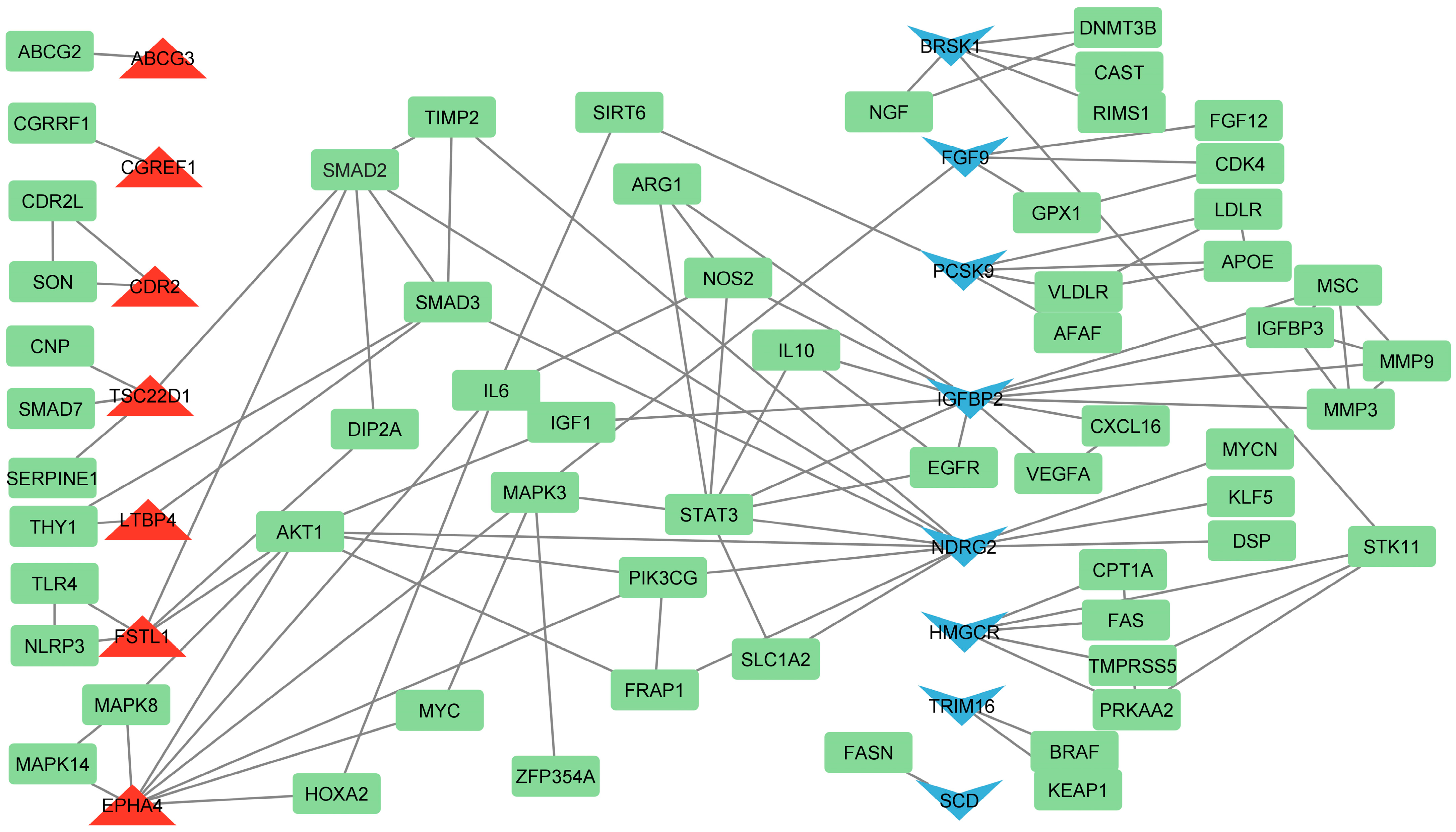
3.4. Identification of Hub Genes Based on Literature Analysis
3.5. Prediction of miRNA and Transcription Factors
4. Discussion
4.1. The Effect of ChGENE/ChHAYW Common Genes in Hypercholesterolemia
4.2. The Effect of Geneva Kiwifruit Supplementation
4.2.1. Reduced Biosynthesis of Cholesterol
4.2.2. Lipid Transport
4.2.3. Anti-Inflammatory and Regenerative Effects
4.2.4. Altered Glucose and Fatty Acid Metabolism
4.2.5. Role of Olfactory Receptors
4.3. The Prediction of miRNA and TF Interaction with Hub Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Berlanga, A.; Guiu-Jurado, E.; Porras, J.A.; Auguet, T. Molecular Pathways in Non-Alcoholic Fatty Liver Disease. Clin. Exp. Gastroenterol. 2014, 7, 221–239. [Google Scholar] [CrossRef] [PubMed]
- Zhou, E.; Ge, X.; Nakashima, H.; Li, R.; van der Zande, H.J.P.; Liu, C.; Li, Z.; Müller, C.; Bracher, F.; Mohammed, Y.; et al. Inhibition of DHCR24 Activates LXRα to Ameliorate Hepatic Steatosis and Inflammation. EMBO Mol. Med. 2023, 15, e16845. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Coker, O.O.; Chu, E.S.; Fu, K.; Lau, H.C.H.; Wang, Y.-X.; Chan, A.W.H.; Wei, H.; Yang, X.; Sung, J.J.Y.; et al. Dietary Cholesterol Drives Fatty Liver-Associated Liver Cancer by Modulating Gut Microbiota and Metabolites. Gut 2021, 70, 761–774. [Google Scholar] [CrossRef] [PubMed]
- Ahn, A.-L.; Choi, J.-K.; Kim, M.-N.; Kim, S.-A.; Oh, E.-J.; Kweon, H.-J.; Cho, D.-Y. Non-Alcoholic Fatty Liver Disease and Chronic Kidney Disease in Koreans Aged 50 Years or Older. Korean J. Fam. Med. 2013, 34, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Kim, B.; Seo, H.S.; Lee, Y.J.; Kim, H.H.; Son, H.-H.; Choi, M.H. Cholesterol-Induced Non-Alcoholic Fatty Liver Disease and Atherosclerosis Aggravated by Systemic Inflammation. PLoS ONE 2014, 9, e97841. [Google Scholar] [CrossRef]
- Maxfield, F.R.; Tabas, I. Role of Cholesterol and Lipid Organization in Disease. Nature 2005, 438, 612–621. [Google Scholar] [CrossRef]
- Collado, A.; Domingo, E.; Piqueras, L.; Sanz, M.-J. Primary Hypercholesterolemia and Development of Cardiovascular Disorders: Cellular and Molecular Mechanisms Involved in Low-Grade Systemic Inflammation and Endothelial Dysfunction. Int. J. Biochem. Cell Biol. 2021, 139, 106066. [Google Scholar] [CrossRef]
- Sekhar, C.C.; Jindal, P.; Karna, V.G.; Aggarwala, M.; Sekhar, S. Cholestrol Emboli Syndrome: Acute Renal Insufficiency After a Procedure or a Thrombolytic Therapy or Anticoagulant Therapy. Indian J. Surg. 2013, 75, 432–435. [Google Scholar] [CrossRef]
- Ding, X.; Zhang, W.; Li, S.; Yang, H. The Role of Cholesterol Metabolism in Cancer. Am. J. Cancer Res. 2019, 9, 219–227. [Google Scholar]
- Havel, P.J. Dietary Fructose: Implications for Dysregulation of Energy Homeostasis and Lipid/Carbohydrate Metabolism. Nutr. Rev. 2005, 63, 133–157. [Google Scholar] [CrossRef]
- Harasym, J.; Oledzki, R. Effect of Fruit and Vegetable Antioxidants on Total Antioxidant Capacity of Blood Plasma. Nutrition 2014, 30, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Chesoniene, L.; Daubaras, R.; Viskelis, P. Biochemical Composition of Berries of Some Kolomikta Kiwi (Actinidia Kolomikta) Cultivars and Detection of Harvest Maturity. In Proceedings of the XI Eucarpia Symposium on Fruit Breeding and Genetics Angers, France, 11–15 September 2003; pp. 305–308. [Google Scholar]
- Shehata, M.M.S.M.; Soltan, S.S.A. Effects of Bioactive Component of Kiwi Fruit and Avocado (Fruit and Seed) on Hypercholesterolemic Rats. World J. Dairy Food Sci. 2013, 8, 82–93. [Google Scholar]
- Wang, Y.; Li, H.; Ren, Y.; Wang, Y.; Ren, Y.; Wang, X.; Yue, T.; Wang, Z.; Gao, Z. Preparation, Model Construction and Efficacy Lipid-Lowering Evaluation of Kiwifruit Juice Fermented by Probiotics. Food Biosci. 2022, 47, 101710. [Google Scholar] [CrossRef]
- Qu, L.; Liu, Q.; Zhang, Q.; Liu, D.; Zhang, C.; Fan, D.; Deng, J.; Yang, H. Kiwifruit Seed Oil Ameliorates Inflammation and Hepatic Fat Metabolism in High-Fat Diet-Induced Obese Mice. J. Funct. Foods 2019, 52, 715–723. [Google Scholar] [CrossRef]
- Leontowicz, H.; Leontowicz, M.; Latocha, P.; Jesion, I.; Park, Y.-S.; Katrich, E.; Barasch, D.; Nemirovski, A.; Gorinstein, S. Bioactivity and Nutritional Properties of Hardy Kiwi Fruit Actinidia arguta in Comparison with Actinidia deliciosa ‘Hayward’ and Actinidia eriantha ‘Bidan’. Food Chem. 2016, 196, 281–291. [Google Scholar] [CrossRef]
- Leontowicz, M.; Leontowicz, H.; Jesion, I.; Bielecki, W.; Najman, K.; Latocha, P.; Park, Y.-S.; Gorinstein, S. Actinidia Arguta Supplementation Protects Aorta and Liver in Rats with Induced Hypercholesterolemia. Nutr. Res. 2016, 36, 1231–1242. [Google Scholar] [CrossRef]
- Ciecierska, A.; Motyl, T.; Sadkowski, T. Transcriptomic Profile of Primary Culture of Skeletal Muscle Cells Isolated from Semitendinosus Muscle of Beef and Dairy Bulls. Int. J. Mol. Sci. 2020, 21, 4794. [Google Scholar] [CrossRef]
- Szcześniak, K.A.; Ciecierska, A.; Ostaszewski, P.; Sadkowski, T. Characterisation of Equine Satellite Cell Transcriptomic Profile Response to β -Hydroxy- β -Methylbutyrate (HMB). Br. J. Nutr. 2016, 116, 1315–1325. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Lopez, D. PCSK9: An Enigmatic Protease. Biochim. Biophys. Acta BBA-Mol. Cell Biol. Lipids 2008, 1781, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Mahley, R.W.; Huang, Y. Apolipoprotein E Sets the Stage: Response to Injury Triggers Neuropathology. Neuron 2012, 76, 871–885. [Google Scholar] [CrossRef] [PubMed]
- Seidah, N.G.; Prat, A. The Biology and Therapeutic Targeting of the Proprotein Convertases. Nat. Rev. Drug Discov. 2012, 11, 367–383. [Google Scholar] [CrossRef] [PubMed]
- Miranda, M.X.; van Tits, L.J.; Lohmann, C.; Arsiwala, T.; Winnik, S.; Tailleux, A.; Stein, S.; Gomes, A.P.; Suri, V.; Ellis, J.L.; et al. The Sirt1 Activator SRT3025 Provides Atheroprotection in Apoe−/− Mice by Reducing Hepatic Pcsk9 Secretion and Enhancing Ldlr Expression. Eur. Heart J. 2015, 36, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Poirier, S.; Mayer, G.; Benjannet, S.; Bergeron, E.; Marcinkiewicz, J.; Nassoury, N.; Mayer, H.; Nimpf, J.; Prat, A.; Seidah, N.G. The Proprotein Convertase PCSK9 Induces the Degradation of Low Density Lipoprotein Receptor (LDLR) and Its Closest Family Members VLDLR and ApoER2. J. Biol. Chem. 2008, 283, 2363–2372. [Google Scholar] [CrossRef]
- Gerrard, S.D.; Biase, F.H.; Yonke, J.A.; Yadav, R.; Shafron, A.J.; Sunny, N.E.; Gerrard, D.E.; El-Kadi, S.W. Non-Alcoholic Fatty Liver Disease Induced by Feeding Medium-Chain Fatty Acids Upregulates Cholesterol and Lipid Homeostatic Genes in Skeletal Muscle of Neonatal Pigs. Metabolites 2024, 14, 384. [Google Scholar] [CrossRef]
- Deng, W.; Wang, L.; Xiong, Y.; Li, J.; Wang, Y.; Shi, T.; Ma, D. The Novel Secretory Protein CGREF1 Inhibits the Activation of AP-1 Transcriptional Activity and Cell Proliferation. Int. J. Biochem. Cell Biol. 2015, 65, 32–39. [Google Scholar] [CrossRef]
- Guo, C.; Zhang, L.; Zhao, M.; Ai, Y.; Liao, W.; Wan, L.; Liu, Q.; Li, S.; Zeng, J.; Ma, X.; et al. Targeting Lipid Metabolism with Natural Products: A Novel Strategy for Gastrointestinal Cancer Therapy. Phytother. Res. PTR 2023, 37, 2036–2050. [Google Scholar] [CrossRef]
- Cánovas, A.; Quintanilla, R.; Gallardo, D.; Díaz, I.; Noguera, J.L.; Ramírez, O.; Pena, R.N. Functional and Association Studies on the Pig HMGCR Gene, a Cholesterol-Synthesis Limiting Enzyme. Anim. Int. J. Anim. Biosci. 2010, 4, 224–233. [Google Scholar] [CrossRef]
- Hu, N.; Chen, C.; Wang, J.; Huang, J.; Yao, D.; Li, C. Atorvastatin Ester Regulates Lipid Metabolism in Hyperlipidemia Rats via the PPAR-Signaling Pathway and HMGCR Expression in the Liver. Int. J. Mol. Sci. 2021, 22, 11107. [Google Scholar] [CrossRef]
- Bai, X.; Mai, M.; Yao, K.; Zhang, M.; Huang, Y.; Zhang, W.; Guo, X.; Xu, Y.; Zhang, Y.; Qurban, A.; et al. The Role of DHCR24 in the Pathogenesis of AD: Re-Cognition of the Relationship between Cholesterol and AD Pathogenesis. Acta Neuropathol. Commun. 2022, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Kozan, D.; Young, E.D.; Hensley, M.; Shen, M.-C.; Wen, J.; Moll, T.; Anderson, J.L.; Kozan, H.; Rawls, J.F.; et al. A High-Cholesterol Zebrafish Diet Promotes Hypercholesterolemia and Fasting-Associated Liver Steatosis. J. Lipid Res. 2024, 65, 100637. [Google Scholar] [CrossRef] [PubMed]
- Doege, H.; Baillie, R.A.; Ortegon, A.M.; Tsang, B.; Wu, Q.; Punreddy, S.; Hirsch, D.; Watson, N.; Gimeno, R.E.; Stahl, A. Targeted Deletion of FATP5 Reveals Multiple Functions in Liver Metabolism: Alterations in Hepatic Lipid Homeostasis. Gastroenterology 2006, 130, 1245–1258. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Zhang, G.; Zheng, Y.; Yang, Y.; Chen, C.; Xia, J.; Liang, L.; Lei, C.; Hu, Y.; Cai, X.; et al. SLC27A5 Deficiency Activates NRF2/TXNRD1 Pathway by Increased Lipid Peroxidation in HCC. Cell Death Differ. 2020, 27, 1086–1104. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qiao, Y.; Sun, H.; Chang, H.; Zhao, H.; Zhang, S.; Shan, C. Decreased SLC27A5 Suppresses Lipid Synthesis and Tyrosine Metabolism to Activate the Cell Cycle in Hepatocellular Carcinoma. Biomedicines 2022, 10, 234. [Google Scholar] [CrossRef]
- Enooku, K.; Tsutsumi, T.; Kondo, M.; Fujiwara, N.; Sasako, T.; Shibahara, J.; Kado, A.; Okushin, K.; Fujinaga, H.; Nakagomi, R.; et al. Hepatic FATP5 Expression Is Associated with Histological Progression and Loss of Hepatic Fat in NAFLD Patients. J. Gastroenterol. 2020, 55, 227–243. [Google Scholar] [CrossRef]
- Lorkowski, S.; Cullen, P. ABCG Subfamily of Human ATP-Binding Cassette Proteins. Pure Appl. Chem. 2002, 74, 2057–2081. [Google Scholar] [CrossRef]
- Moradi, S.; Tavilani, H.; Saidijam, M.; Hashemnia, M.; Vaisi-Raygani, A. Kiwifruit Supplementation Increases the Gene Expression of ATP-Binding Cassette Transporter A1 and Liver X Receptor α in Liver and Intestine of Hamsters Fed with High-Fat Diet. Mediterr. J. Nutr. Metab. 2021, 14, 343–352. [Google Scholar] [CrossRef]
- Wang, N.; Westerterp, M. ABC Transporters, Cholesterol Efflux, and Implications for Cardiovascular Diseases. Adv. Exp. Med. Biol. 2020, 1276, 67–83. [Google Scholar] [CrossRef]
- Dean, M.; Moitra, K.; Allikmets, R. The Human ATP-Binding Cassette (ABC) Transporter Superfamily. Hum. Mutat. 2022, 43, 1162–1182. [Google Scholar] [CrossRef]
- Inaguma, S.; Lasota, J.; Wang, Z.; Felisiak-Golabek, A.; Ikeda, H.; Miettinen, M. Clinicopathologic Profile, Immunophenotype, and Genotype of CD274 (PD-L1)-Positive Colorectal Carcinomas. Mod. Pathol. 2017, 30, 278–285. [Google Scholar] [CrossRef]
- Fukai, J.; Yokote, H.; Yamanaka, R.; Arao, T.; Nishio, K.; Itakura, T. EphA4 Promotes Cell Proliferation and Migration through a Novel EphA4-FGFR1 Signaling Pathway in the Human Glioma U251 Cell Line. Mol. Cancer Ther. 2008, 7, 2768–2778. [Google Scholar] [CrossRef] [PubMed]
- Aoki, M.; Yamashita, T.; Tohyama, M. EphA Receptors Direct the Differentiation of Mammalian Neural Precursor Cells through a Mitogen-Activated Protein Kinase-Dependent Pathway*. J. Biol. Chem. 2004, 279, 32643–32650. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Pan, Y.; Liu, L.; Li, H.; Li, Y.; Jiang, J.; Xiang, J.; Zhang, J.; Chu, W. Effects of High Fat Diet on Lipid Accumulation, Oxidative Stress and Autophagy in the Liver of Chinese Softshell Turtle (Pelodiscus sinensis). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2020, 240, 110331. [Google Scholar] [CrossRef] [PubMed]
- Serviddio, G.; Bellanti, F.; Villani, R.; Tamborra, R.; Zerbinati, C.; Blonda, M.; Ciacciarelli, M.; Poli, G.; Vendemiale, G.; Iuliano, L. Effects of Dietary Fatty Acids and Cholesterol Excess on Liver Injury: A Lipidomic Approach. Redox Biol. 2016, 9, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K. FIBROBLAST CELL SHAPE AND ADHESION IN VITRO IS ALTERED BY OVEREXPRESSION OF THE 7A AND 7B ISOFORMS OF PROTOCADHERIN 7, BUT NOT THE 7C ISOFORM. Cell. Mol. Biol. Lett. 2003, 8, 735–741. [Google Scholar]
- Rashid, D.; Puettmann, P.; Roy, E.; Bradley, R.S. Neural Crest Development in Xenopus Requires Protocadherin 7 at the Lateral Neural Crest Border. Mech. Dev. 2018, 149, 41–52. [Google Scholar] [CrossRef]
- Hintermann, E.; Christen, U. The Many Roles of Cell Adhesion Molecules in Hepatic Fibrosis. Cells 2019, 8, 1503. [Google Scholar] [CrossRef]
- Springer, T.A. Adhesion Receptors of the Immune System. Nature 1990, 346, 425–434. [Google Scholar] [CrossRef]
- Montefort, S.; Holgate, S.T. Adhesion Molecules and Their Role in Inflammation. Respir. Med. 1991, 85, 91–99. [Google Scholar] [CrossRef]
- Wu, X.; Zou, X.; Chang, Q.; Zhang, Y.; Li, Y.; Zhang, L.; Huang, J.; Liang, B. The Evolutionary Pattern and the Regulation of Stearoyl-CoA Desaturase Genes. BioMed Res. Int. 2013, 2013, e856521. [Google Scholar] [CrossRef] [PubMed]
- Ntambi, J.M. The Regulation of Stearoyl-CoA Desaturase (SCD). Prog. Lipid Res. 1995, 34, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Tsukamoto, H. Stearoyl-CoA Desaturase and Tumorigenesis. Chem. Biol. Interact. 2020, 316, 108917. [Google Scholar] [CrossRef]
- Li, J.; Savransky, V.; Nanayakkara, A.; Smith, P.L.; O’Donnell, C.P.; Polotsky, V.Y. Hyperlipidemia and Lipid Peroxidation Are Dependent on the Severity of Chronic Intermittent Hypoxia. J. Appl. Physiol. 2007, 102, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Major, C.A.; Ryan, K.; Bennett, A.J.; Lock, A.L.; Bauman, D.E.; Salter, A.M. Inhibition of Stearoyl CoA Desaturase Activity Induces Hypercholesterolemia in the Cholesterol-Fed Hamster. J. Lipid Res. 2008, 49, 1456–1465. [Google Scholar] [CrossRef]
- Pai, J.T.; Guryev, O.; Brown, M.S.; Goldstein, J.L. Differential Stimulation of Cholesterol and Unsaturated Fatty Acid Biosynthesis in Cells Expressing Individual Nuclear Sterol Regulatory Element-Binding Proteins. J. Biol. Chem. 1998, 273, 26138–26148. [Google Scholar] [CrossRef]
- Pallag, G.; Nazarian, S.; Ravasz, D.; Bui, D.; Komlódi, T.; Doerrier, C.; Gnaiger, E.; Seyfried, T.N.; Chinopoulos, C. Proline Oxidation Supports Mitochondrial ATP Production When Complex I Is Inhibited. Int. J. Mol. Sci. 2022, 23, 5111. [Google Scholar] [CrossRef]
- Fungfuang, W.; Srisuksai, K.; Santativongchai, P.; Charoenlappanit, S.; Phaonakrop, N.; Roytrakul, S.; Tulayakul, P.; Parunyakul, K. Targeted Proteomic Analysis Reveals That Crocodile Oil from Crocodylus Siamensis May Enhance Hepatic Energy Metabolism in Rats. Exp. Anim. 2023, 72, 425. [Google Scholar] [CrossRef]
- Lettieri Barbato, D.; Aquilano, K.; Baldelli, S.; Cannata, S.M.; Bernardini, S.; Rotilio, G.; Ciriolo, M.R. Proline Oxidase–Adipose Triglyceride Lipase Pathway Restrains Adipose Cell Death and Tissue Inflammation. Cell Death Differ. 2014, 21, 113–123. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, G.; Chen, Y.; Xu, W.; Liu, Y.; Ji, G.; Xu, H. Can Proline Dehydrogenase—A Key Enzyme Involved in Proline Metabolism—Be a Novel Target for Cancer Therapy? Front. Oncol. 2023, 13, 1254439. [Google Scholar] [CrossRef]
- Rao, G.; Peng, X.; Li, X.; An, K.; He, H.; Fu, X.; Li, S.; An, Z. Unmasking the Enigma of Lipid Metabolism in Metabolic Dysfunction-Associated Steatotic Liver Disease: From Mechanism to the Clinic. Front. Med. 2023, 10, 1294267. [Google Scholar] [CrossRef] [PubMed]
- Daci, A.; Bozalija, A.; Jashari, F.; Krasniqi, S. Individualizing Treatment Approaches for Epileptic Patients with Glucose Transporter Type1 (GLUT-1) Deficiency. Int. J. Mol. Sci. 2018, 19, 122. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Zhang, L.; Zhang, M.; Huang, J.; Zhang, J.; Chang, Y. FGF9 Alleviates the Fatty Liver Phenotype by Regulating Hepatic Lipid Metabolism. Front. Pharmacol. 2022, 13, 850128. [Google Scholar] [CrossRef] [PubMed]
- Menco, B.P.M. Ultrastructural Aspects of Olfactory Signaling. Chem. Senses 1997, 22, 295–311. [Google Scholar] [CrossRef] [PubMed]
- Michaloski, J.S.; Galante, P.A.F.; Nagai, M.H.; Armelin-Correa, L.; Chien, M.-S.; Matsunami, H.; Malnic, B. Common Promoter Elements in Odorant and Vomeronasal Receptor Genes. PLoS ONE 2011, 6, e29065. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Hwang, S.H.; Jia, Y.; Choi, J.; Kim, Y.-J.; Choi, D.; Pathiraja, D.; Choi, I.-G.; Koo, S.-H.; Lee, S.-J. Olfactory Receptor 544 Reduces Adiposity by Steering Fuel Preference toward Fats. J. Clin. Investig. 2017, 127, 4118–4123. [Google Scholar] [CrossRef]
- Zhang, S.; Li, L.; Li, H. Role of Ectopic Olfactory Receptors in Glucose and Lipid Metabolism. Br. J. Pharmacol. 2021, 178, 4792–4807. [Google Scholar] [CrossRef]
- Gwinn, D.M.; Shackelford, D.B.; Egan, D.F.; Mihaylova, M.M.; Mery, A.; Vasquez, D.S.; Turk, B.E.; Shaw, R.J. AMPK Phosphorylation of Raptor Mediates a Metabolic Checkpoint. Mol. Cell 2008, 30, 214–226. [Google Scholar] [CrossRef]
- Lin, S.-C.; Hardie, D.G. AMPK: Sensing Glucose as Well as Cellular Energy Status. Cell Metab. 2018, 27, 299–313. [Google Scholar] [CrossRef]
- Leem, J.; Shim, H.; Cho, H.; Park, J.-H. Octanoic Acid Potentiates Glucose-Stimulated Insulin Secretion and Expression of Glucokinase through the Olfactory Receptor in Pancreatic β-Cells. Biochem. Biophys. Res. Commun. 2018, 503, 278–284. [Google Scholar] [CrossRef]
- Tang, Z.F.; McMillen, D.R. Design Principles for the Analysis and Construction of Robustly Homeostatic Biological Networks. J. Theor. Biol. 2016, 408, 274–289. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, J.; Zhao, S.; Chen, L.; Zhang, M.; Ma, Y.; Yao, D. Regulation of Bta-miRNA29d-3p on Lipid Accumulation via GPAM in Bovine Mammary Epithelial Cells. Agriculture 2023, 13, 501. [Google Scholar] [CrossRef]
- Dalgaard, L.T.; Sørensen, A.E.; Hardikar, A.A.; Joglekar, M.V. The microRNA-29 Family: Role in Metabolism and Metabolic Disease. Am. J. Physiol.-Cell Physiol. 2022, 323, C367–C377. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, G.; Yuan, J.; Li, N.; Yan, B.; Yan, J.; Sheng, Y. Hsa-miR-34a-5p Ameliorates Hepatic Ischemia/Reperfusion Injury Via Targeting HNF4α. Turk. J. Gastroenterol. 2022, 33, 596. [Google Scholar] [CrossRef]
- Ruiz-Roso, M.B.; Gil-Zamorano, J.; López de Las Hazas, M.C.; Tomé-Carneiro, J.; Crespo, M.C.; Latasa, M.J.; Briand, O.; Sánchez-López, D.; Ortiz, A.I.; Visioli, F. Intestinal Lipid Metabolism Genes Regulated by miRNAs. Front. Genet. 2020, 11, 707. [Google Scholar] [CrossRef] [PubMed]
- Da Dalt, L.; Bonacina, F. Metabolic Impact of Extrahepatic PCSK9 Modulation: Extrahepatic PCSK9 Modulation. Eur. Atheroscler. J. 2022, 1, 41–47. [Google Scholar] [CrossRef]

| Gene Symbol | Accession Number | Forward Primer | Reverse Primer | Annealing Temp. (°C) | Annealing Time (s) | PCR Product Length |
|---|---|---|---|---|---|---|
| DHCR24 | NM_001080148 | 5′AAGGGGTTGGAGTTCGTTC3′ | 5′TGAGACAGTGAGCCATCCA3′ | 58 | 15 | 243 |
| HMGCR | NM_013134 | 5′GGACTGAAACACGGGCATT3′ | 5′AACACGGCACGGAAAGAAC3′ | 58 | 15 | 197 |
| PCSK9 | NM_199253 | 5′GTGTGTGTGGCACGAATCC3′ | 5′AAGTTCCCCCAGGCAGAGT3′ | 58 | 15 | 224 |
| PRODH | NM_001135778 | 5′CGCAGGTTCAATGTGGAT3′ | 5′GGCATTGGTGGCTTCATA3′ | 58 | 15 | 222 |
| SCD1 | NM_139192 | 5′CACACTGGTGCCCTGGTA3′ | 5′GGGAAGGCGTGATGGTAG3′ | 58 | 15 | 225 |
| GAPDH | NM_017008 | 5′CCCACACTGTGCCCATCTAT3′ | 5′AAGGGTGTAAAACGCAGCTC3′ | 60 | 15 | 198 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gorji, A.E.; Ciecierska, A.; Leontowicz, H.; Roudbari, Z.; Sadkowski, T. Impact of Kiwifruit Consumption on Cholesterol Metabolism in Rat Liver: A Gene Expression Analysis in Induced Hypercholesterolemia. Nutrients 2024, 16, 3999. https://doi.org/10.3390/nu16233999
Gorji AE, Ciecierska A, Leontowicz H, Roudbari Z, Sadkowski T. Impact of Kiwifruit Consumption on Cholesterol Metabolism in Rat Liver: A Gene Expression Analysis in Induced Hypercholesterolemia. Nutrients. 2024; 16(23):3999. https://doi.org/10.3390/nu16233999
Chicago/Turabian StyleGorji, Abdolvahab Ebrahimpour, Anna Ciecierska, Hanna Leontowicz, Zahra Roudbari, and Tomasz Sadkowski. 2024. "Impact of Kiwifruit Consumption on Cholesterol Metabolism in Rat Liver: A Gene Expression Analysis in Induced Hypercholesterolemia" Nutrients 16, no. 23: 3999. https://doi.org/10.3390/nu16233999
APA StyleGorji, A. E., Ciecierska, A., Leontowicz, H., Roudbari, Z., & Sadkowski, T. (2024). Impact of Kiwifruit Consumption on Cholesterol Metabolism in Rat Liver: A Gene Expression Analysis in Induced Hypercholesterolemia. Nutrients, 16(23), 3999. https://doi.org/10.3390/nu16233999






