Exploring the Regulatory Effect of Hydroxytyrosol on Ovarian Inflammaging Through Autophagy-Targeted Mechanisms: A Bioinformatics Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ovarian Aging Data Acquisition and Differential Analysis
2.2. HT Target Prediction and Autophagy-Related Gene Databases
2.3. KEGG and GO Enrichment Analyses
2.4. PPI Network Construction and Hub Gene Screening
2.5. Annotation of Hub Genes Using AutophagyNet and Prediction of LIR and FIR Motifs
2.6. Molecular Docking and Molecular Dynamics Simulation
3. Results
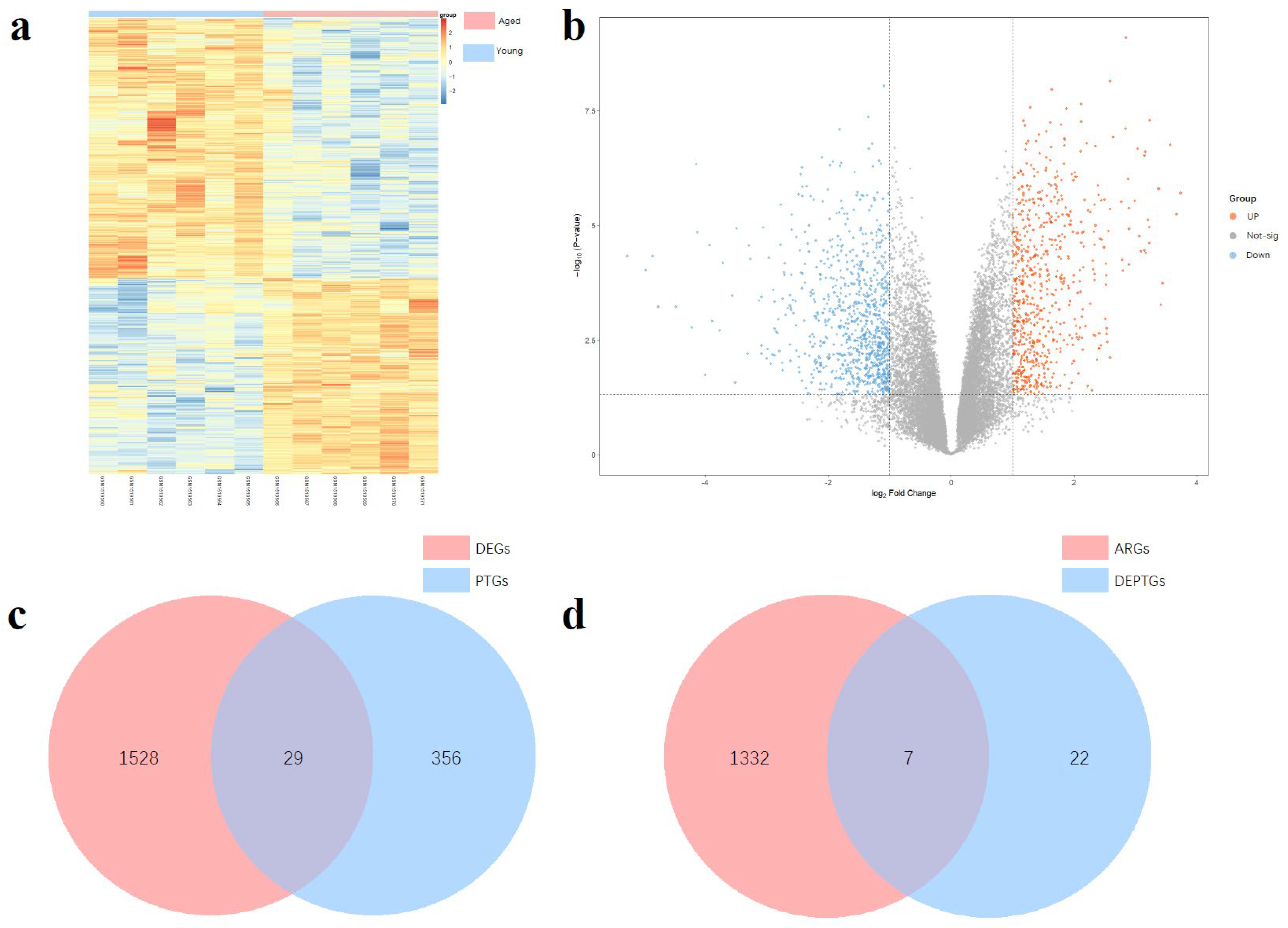
3.1. Differential Expressed Genes Analysis
3.2. Screening of Autophagy-Related DEPTGs
3.3. KEGG and GO Enrichment Analysis
3.4. PPI Network and Hub Gene Results
3.5. Functional Annotation Using AutophagyNet and Prediction of LIR and FIR Motifs
- SARs containing only LIR motifs (indicating involvement solely in LC3-dependent selective autophagy);
- SARs containing only FIR motifs (indicating involvement solely in non-canonical selective autophagy (FIP200 dependent));
- SARs containing both LIR and FIR motifs (indicating potential participation in both LC3-dependent and non-canonical selective autophagy (FIP200 dependent)).
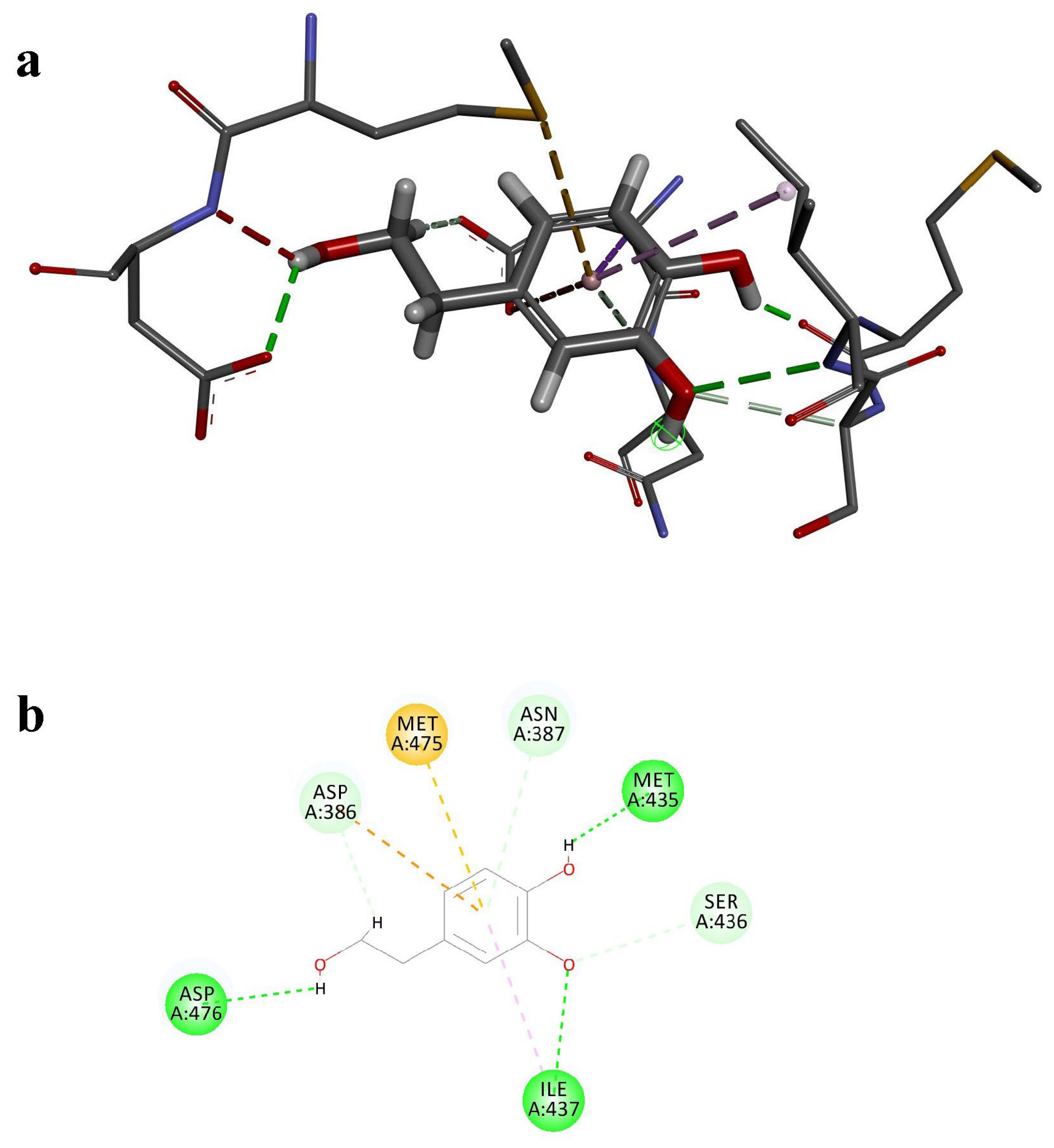
3.6. Molecular Docking
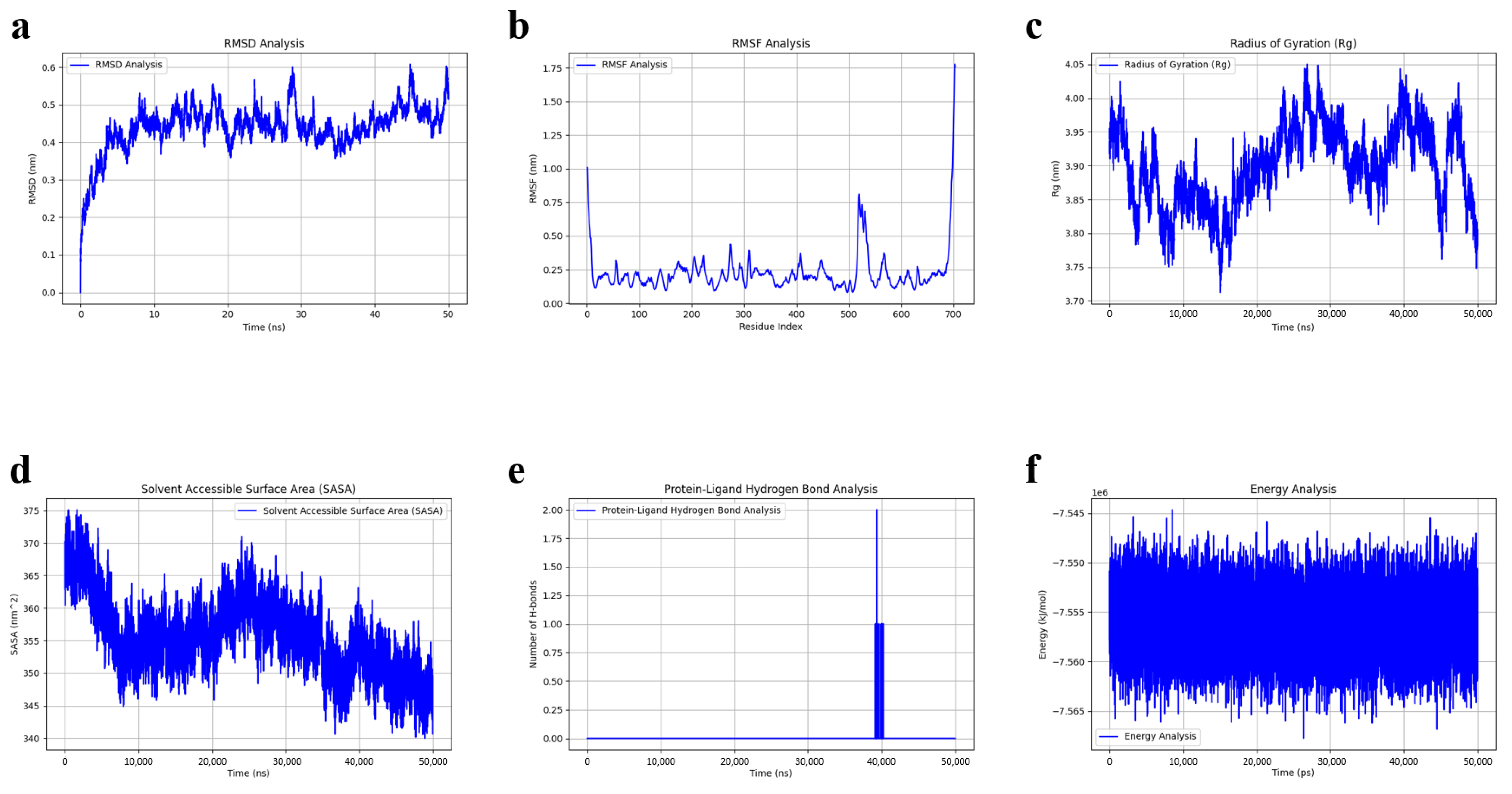
3.7. Molecular Dynamics Simulation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AR-DEPTGs | autophagy-related differentially expressed potential target genes |
| ARGs | autophagy-related gene |
| CRP | C-reactive protein |
| CTD | Comparative Toxicogenomics Database |
| DEGs | differentially expressed genes |
| DEPTGs | differentially expressed potential target genes |
| DGIdb | Drug-Gene Interaction Database |
| ERK/MAPK | extracellular signal-regulated kinase/ mitogen-activated protein kinase |
| FDR | false discovery rate |
| FIP200 | focal adhesion kinase family interacting protein of 200 kD |
| FIR | FIP200-interacting region |
| GEO | Gene Expression Omnibus |
| GO | Gene Ontology |
| HADb | Human Autophagy Database |
| HT | hydroxytyrosol |
| IL-6 | interleukin-6 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LC3 | microtubule-associated protein 1 light chain 3 |
| LIR | LC3-interacting region |
| logFC | log fold change |
| MD | molecular dynamics |
| NF-κB | nuclear factor kappa-light-chain-enhancer of activated B cells |
| NLRP3 | NOD-like receptor family pyrin domain containing 3 |
| NPT | constant pressure and temperature ensemble |
| NVT | constant volume and temperature ensemble |
| PPI | protein-protein interaction |
| PTGs | predicted target genes |
| Rg | radius of gyration |
| RMSD | root mean square deviation |
| RMSF | root mean square fluctuation |
| ROS | reactive oxygen species |
| SARs | selective autophagy receptors |
| SASA | solvent accessible surface area |
| TCMSP | Traditional Chinese Medicine Systems Pharmacology Database |
| TGF-β | transforming growth factor-beta |
| TNF-α | tumor necrosis factor-alpha |
References
- Broekmans, F.J.; Soules, M.R.; Fauser, B.C. Ovarian Aging: Mechanisms and Clinical Consequences. Endocr. Rev. 2009, 30, 465–493. [Google Scholar] [CrossRef] [PubMed]
- Seals, D.R.; Justice, J.N.; LaRocca, T.J. Physiological geroscience: Targeting function to increase healthspan and achieve optimal longevity. J. Physiol. 2016, 594, 2001–2024. [Google Scholar] [CrossRef]
- Franceschi, C.; Bonafè, M.; Valensin, S.; Olivieri, F.; De Luca, M.; Ottaviani, E.; De Benedictis, G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci. 2000, 908, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Khosro, S.; Alireza, S.; Omid, A.; Forough, S. Night work and inflammatory markers. Indian. J. Occup. Environ. Med. 2011, 15, 38–41. [Google Scholar] [CrossRef]
- Franceschi, C.; Campisi, J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69 (Suppl. S1), S4–S9. [Google Scholar] [CrossRef]
- Vilser, C.; Hueller, H.; Nowicki, M.; Hmeidan, F.A.; Blumenauer, V.; Spanel-Borowski, K. The variable expression of lectin-like oxidized low-density lipoprotein receptor (LOX-1) and signs of autophagy and apoptosis in freshly harvested human granulosa cells depend on gonadotropin dose, age, and body weight. Fertil. Steril. 2010, 93, 2706–2715. [Google Scholar] [CrossRef]
- Deretic, V.; Kroemer, G. Autophagy in metabolism and quality control: Opposing, complementary or interlinked functions? Autophagy 2022, 18, 283–292. [Google Scholar] [CrossRef]
- Kobayashi, H.; Imanaka, S. Exploring potential pathways from oxidative stress to ovarian aging. J. Obs. Gynaecol. Res. 2025, 51, e16166. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhang, D.; Hu, D.; Zhou, X.; Zhou, Y. The role of mitochondria in NLRP3 inflammasome activation. Mol. Immunol. 2018, 103, 115–124. [Google Scholar] [CrossRef]
- Salminen, A.; Kaarniranta, K.; Kauppinen, A. Inflammaging: Disturbed interplay between autophagy and inflammasomes. Aging 2012, 4, 166–175. [Google Scholar] [CrossRef]
- Kaushik, S.; Tasset, I.; Arias, E.; Pampliega, O.; Wong, E.; Martinez-Vicente, M.; Cuervo, A.M. Autophagy and the hallmarks of aging. Ageing Res. Rev. 2021, 72, 101468. [Google Scholar] [CrossRef]
- Popelka, H.; Klionsky, D.J. The RB1CC1 Claw-binding motif: A new piece in the puzzle of autophagy regulation. Autophagy 2022, 18, 237–239. [Google Scholar] [CrossRef] [PubMed]
- Turco, E.; Witt, M.; Abert, C.; Bock-Bierbaum, T.; Su, M.Y.; Trapannone, R.; Sztacho, M.; Danieli, A.; Shi, X.; Zaffagnini, G.; et al. FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates. Mol. Cell 2019, 74, 330–346.e11. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Liu, J.; Fu, T.; Wu, P.; Peng, C.; Gong, X.; Wang, Y.; Zhang, M.; Li, Y.; Wang, Y.; et al. Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation. Nat. Commun. 2021, 12, 1570. [Google Scholar] [CrossRef]
- Priem, D.; Huyghe, J.; Bertrand, M.J. LC3-independent autophagy is vital to prevent TNF cytotoxicity. Autophagy 2023, 19, 2585–2589. [Google Scholar] [CrossRef] [PubMed]
- Nutrition and Allergies EFSA Panel on Dietetic Products. Scientific Opinion on the Substantiation of Health Claims Related to Polyphenols in Olive and Protection of LDL Particles from Oxidative Damage (ID 1313, 1333, 1638, 1639, 1696, 2865) Pursuant to Article 13(1) of Regulation (EC) No 1924/2006; European Food Safety Authority (EFSA): Parma, Italy, 2011; p. 2033. [Google Scholar]
- Visioli, F.; Galli, C.; Bornet, F.; Mattei, A.; Patelli, R.; Galli, G.; Caruso, D. Olive oil phenolics are dose-dependently absorbed in humans. FEBS Lett. 2000, 468, 159–160. [Google Scholar] [CrossRef]
- Robles-Almazan, M.; Pulido-Moran, M.; Moreno-Fernandez, J.; Ramirez-Tortosa, C.; Rodriguez-Garcia, C.; Quiles, J.L.; Ramirez-Tortosa, M. Hydroxytyrosol: Bioavailability, toxicity, and clinical applications. Food Res. Int. 2018, 105, 654–667. [Google Scholar] [CrossRef]
- Wen, X.; Tang, S.; Wan, F.; Zhong, R.; Chen, L.; Zhang, H. The PI3K/Akt-Nrf2 Signaling Pathway and Mitophagy Synergistically Mediate Hydroxytyrosol to Alleviate Intestinal Oxidative Damage. Int. J. Biol. Sci. 2024, 20, 4258–4276. [Google Scholar] [CrossRef]
- Schaffer, S.; Podstawa, M.; Visioli, F.; Bogani, P.; Müller, W.E.; Eckert, G.P. Hydroxytyrosol-rich olive mill wastewater extract protects brain cells in vitro and ex vivo. J. Agric. Food Chem. 2007, 55, 5043–5049. [Google Scholar] [CrossRef]
- Velotti, F.; Bernini, R. Hydroxytyrosol Interference with Inflammaging via Modulation of Inflammation and Autophagy. Nutrients 2023, 15, 1774. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Liu, Z.; Feng, Z.; Hao, J.; Shen, W.; Li, X.; Sun, L.; Sharman, E.; Wang, Y.; Wertz, K.; et al. Hydroxytyrosol protects against oxidative damage by simultaneous activation of mitochondrial biogenesis and phase II detoxifying enzyme systems in retinal pigment epithelial cells. J. Nutr. Biochem. 2010, 21, 1089–1098. [Google Scholar] [CrossRef]
- de Pablos, R.M.; Espinosa-Oliva, A.M.; Hornedo-Ortega, R.; Cano, M.; Arguelles, S. Hydroxytyrosol protects from aging process via AMPK and autophagy; a review of its effects on cancer, metabolic syndrome, osteoporosis, immune-mediated and neurodegenerative diseases. Pharmacol. Res. 2019, 143, 58–72. [Google Scholar] [CrossRef] [PubMed]
- Miao, F. Hydroxytyrosol alleviates dextran sodium sulfate-induced colitis by inhibiting NLRP3 inflammasome activation and modulating gut microbiota in vivo. Nutrition 2022, 97, 111579. [Google Scholar] [CrossRef]
- Tatone, C.; Amicarelli, F.; Carbone, M.C.; Monteleone, P.; Caserta, D.; Marci, R.; Artini, P.G.; Piomboni, P.; Focarelli, R. Cellular and molecular aspects of ovarian follicle ageing. Hum. Reprod. Update 2008, 14, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.B.; Zhuang, H.Z.; Ji, X.J.; Dong, L.; Duan, M.L. Hydroxytyrosol suppresses LPS-induced intrahepatic inflammatory responses via inhibition of ERK signaling pathway activation in acute liver injury. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 6455–6462. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef]
- Dunkel, M.; Günther, S.; Ahmed, J.; Wittig, B.; Preissner, R. SuperPred: Drug classification and target prediction. Nucleic Acids Res. 2008, 36, W55–W59. [Google Scholar] [CrossRef]
- Liu, T.; Hwang, L.; Burley, S.K.; Nitsche, C.I.; Southan, C.; Walters, W.P.; Gilson, M.K. BindingDB in 2024: A FAIR knowledgebase of protein-small molecule binding data. Nucleic Acids Res. 2025, 53, D1633–D1644. [Google Scholar] [CrossRef]
- Davis, A.P.; Wiegers, T.C.; Sciaky, D.; Barkalow, F.; Strong, M.; Wyatt, B.; Wiegers, J.; McMorran, R.; Abrar, S.; Mattingly, C.J. Comparative Toxicogenomics Database’s 20th anniversary: Update 2025. Nucleic Acids Res. 2025, 53, D1328–D1334. [Google Scholar] [CrossRef]
- Freshour, S.L.; Kiwala, S.; Cotto, K.C.; Coffman, A.C.; McMichael, J.F.; Song, J.J.; Griffith, M.; Griffith, O.L.; Wagner, A.H. Integration of the Drug-Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021, 49, D1144–D1151. [Google Scholar] [CrossRef] [PubMed]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y.; et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.N.; Dong, J.; Zhang, L.; Ouyang, D.; Cheng, Y.; Chen, A.F.; Lu, A.P.; Cao, D.S. HAMdb: A database of human autophagy modulators with specific pathway and disease information. J. Cheminform. 2018, 10, 34. [Google Scholar] [CrossRef]
- Csabai, L.; Balázs, B.; Dénes, T.; Sowmya, P.; László, F.-N.; Matthew, M.; Dávid, F.; Dezső, M.; Márton, Ö.; Themis, H.; et al. AutophagyNet: High-resolution data source for the analysis of autophagy and its regulation. Autophagy 2024, 20, 188–201. [Google Scholar] [CrossRef] [PubMed]
- Csabai, L.; Fazekas, D.; Kadlecsik, T.; Szalay-Bekő, M.; Bohár, B.; Madgwick, M.; Módos, D.; Ölbei, M.; Gul, L.; Sudhakar, P.; et al. SignaLink3: A multi-layered resource to uncover tissue-specific signaling networks. Nucleic Acids Res. 2021, 50, D701–D709. [Google Scholar] [CrossRef]
- Varadi, M.; Bertoni, D.; Magana, P.; Paramval, U.; Pidruchna, I.; Radhakrishnan, M.; Tsenkov, M.; Nair, S.; Mirdita, M.; Yeo, J.; et al. AlphaFold Protein Structure Database in 2024: Providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 2023, 52, D368–D375. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Taherbhoy, A.M.; Tait, S.W.; Kaiser, S.E.; Williams, A.H.; Deng, A.; Nourse, A.; Hammel, M.; Kurinov, I.; Rock, C.O.; Green, D.R.; et al. Atg8 transfer from Atg7 to Atg3: A distinctive E1-E2 architecture and mechanism in the autophagy pathway. Mol. Cell 2011, 44, 451–461. [Google Scholar] [CrossRef]
- Kochetkova, E.Y.; Blinova, G.I.; Bystrova, O.A.; Martynova, M.G.; Pospelov, V.A.; Pospelova, T.V. Targeted elimination of senescent Ras-transformed cells by suppression of MEK/ERK pathway. Aging 2017, 9, 2352–2375. [Google Scholar] [CrossRef]
- Hu, P.; Lai, D.; Lu, P.; Gao, J.; He, H. ERK and Akt signaling pathways are involved in advanced glycation end product-induced autophagy in rat vascular smooth muscle cells. Int. J. Mol. Med. 2012, 29, 613–618. [Google Scholar] [CrossRef]
- Pokharel, S.M.; Shil, N.K.; Bose, S. Autophagy, TGF-β, and SMAD-2/3 Signaling Regulates Interferon-β Response in Respiratory Syncytial Virus Infected Macrophages. Front. Cell Infect. Microbiol. 2016, 6, 174. [Google Scholar] [CrossRef] [PubMed]
- Stavropoulos, A.; Divolis, G.; Manioudaki, M.; Gavriil, A.; Kloukina, I.; Perrea, D.N.; Sountoulidis, A.; Ford, E.; Doulou, A.; Apostolidou, A.; et al. Coordinated activation of TGF-β and BMP pathways promotes autophagy and limits liver injury after acetaminophen intoxication. Sci. Signal. 2022, 15, eabn4395. [Google Scholar] [CrossRef] [PubMed]
- Yeager, M.P.; Pioli, P.A.; Guyre, P.M. Cortisol exerts bi-phasic regulation of inflammation in humans. Dose Response 2011, 9, 332–347. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.Y.; Lee, C.-H.; Li, C.-y. Abstract 7476: Obtusifolin attenuates LPS-induced inflammation and NLRP3 inflammasome activation by suppressing the MAPK/NF-κB signaling pathway and ROS production in BV2 cells. Cancer Res. 2024, 84, 7476. [Google Scholar] [CrossRef]
- Wang, Y.; Han, D.; Huang, Y.; Dai, Y.; Wang, Y.; Liu, M.; Wang, N.; Yin, T.; Du, W.; He, K.; et al. Oral administration of punicalagin attenuates imiquimod-induced psoriasis by reducing ROS generation and inflammation via MAPK/ERK and NF-κB signaling pathways. Phytother. Res. 2024, 38, 713–726. [Google Scholar] [CrossRef]
- Brereton, C.F.; Sutton, C.E.; Lalor, S.J.; Lavelle, E.C.; Mills, K.H. Inhibition of ERK MAPK suppresses IL-23- and IL-1-driven IL-17 production and attenuates autoimmune disease. J. Immunol. 2009, 183, 1715–1723. [Google Scholar] [CrossRef]
- Wu, Z.; Yu, Y.; Niu, L.; Fei, A.; Pan, S. IGF-1 protects tubular epithelial cells during injury via activation of ERK/MAPK signaling pathway. Sci. Rep. 2016, 6, 28066. [Google Scholar] [CrossRef]
- Wang, C.; Jin, A.; Huang, W.; Tsang, L.L.; Cai, Z.; Zhou, X.; Chen, H.; Chan, H.C. Up-regulation of Bcl-2 by CD147 Through ERK Activation Results in Abnormal Cell Survival in Human Endometriosis. J. Clin. Endocrinol. Metab. 2015, 100, E955–E963. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, L.; Li, X.; Xu, S. EphA10 drives tumor progression and immune evasion by regulating the MAPK/ERK cascade in lung adenocarcinoma. Int. Immunopharmacol. 2022, 110, 109031. [Google Scholar] [CrossRef]
- Wu, N.; Ji, J.; Gou, X.; Hu, P.; Cheng, Y.; Liu, Y.; Wang, Y.; Zhang, Q.; Zuo, L. DENV-2 NS1 promotes AMPK-LKB1 interaction to activate AMPK/ERK/mTOR signaling pathway to induce autophagy. Virol. J. 2023, 20, 231. [Google Scholar] [CrossRef]
- Li, T.; Zhang, L. Autophagy-related biomarkers in hepatocellular carcinoma and their relationship with immune infiltration. Discov. Oncol. 2024, 15, 299. [Google Scholar] [CrossRef] [PubMed]
- Dolcetti, F.J.C.; Falomir-Lockhart, E.; Acuña, F.; Herrera, M.L.; Cervellini, S.; Barbeito, C.G.; Grassi, D.; Arevalo, M.A.; Bellini, M.J. IGF1 gene therapy in middle-aged female rats delays reproductive senescence through its effects on hypothalamic GnRH and kisspeptin neurons. Aging 2022, 14, 8615–8632. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Sun, H.; Gao, X.; Zhang, C.; Sun, Y.; Wang, H.; Zhang, H.; Shi, Y.; He, X. A comprehensive analysis of age-related metabolomics and transcriptomics reveals metabolic alterations in rat bone marrow mesenchymal stem cells. Aging 2022, 14, 1014–1032. [Google Scholar] [CrossRef]
- Tracz-Gaszewska, Z.; Dobrzyn, P. Stearoyl-CoA Desaturase 1 as a Therapeutic Target for the Treatment of Cancer. Cancers 2019, 11, 948. [Google Scholar] [CrossRef]
- Bai, Y.; Onuma, H.; Bai, X.; Medvedev, A.V.; Misukonis, M.; Weinberg, J.B.; Cao, W.; Robidoux, J.; Floering, L.M.; Daniel, K.W.; et al. Persistent nuclear factor-kappa B activation in Ucp2-/- mice leads to enhanced nitric oxide and inflammatory cytokine production. J. Biol. Chem. 2005, 280, 19062–19069. [Google Scholar] [CrossRef] [PubMed]
- Forte, M.; Bianchi, F.; Cotugno, M.; Marchitti, S.; Stanzione, R.; Maglione, V.; Sciarretta, S.; Valenti, V.; Carnevale, R.; Versaci, F.; et al. An interplay between UCP2 and ROS protects cells from high-salt-induced injury through autophagy stimulation. Cell Death Dis. 2021, 12, 919. [Google Scholar] [CrossRef]
- Lv, Y.; Liang, Q.; Li, Y.; Liu, X.; Zhang, D.; Li, X. Study of the binding mechanism between hydroxytyrosol and bovine serum albumin using multispectral and molecular docking. Food Hydrocoll. 2022, 122, 107072. [Google Scholar] [CrossRef]
- Gu, M.; Shi, J.; Zhang, B.; Wang, X.; Wang, X.; Tian, B. Interaction of soy protein isolate with hydroxytyrosol based on an alkaline method: Implications for structural and functional properties. Food Chem. 2024, 446, 138813. [Google Scholar] [CrossRef]
- Navarro, M.; Morales, F.J. In vitro investigation on the antiglycative and carbonyl trapping activities of hydroxytyrosol. Eur. Food Res. Technol. 2016, 242, 1101–1110. [Google Scholar] [CrossRef]


| Hub Genes | Direct Regulators |
|---|---|
| TNF * | BCL2 |
| IGF1 * | No result |
| UCP2 * | No result |
| PPARA | BCL2 |
| SCD * | MAP1LC3B ATG7 RB1CC1 |
| NR1H4 | No result |
| CSF1R | No result |
| PCK1 | SQSTM1 |
| TGFB2 | RB1CC1 |
| ALOX5 | No result |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, X.; Guo, X.; Cai, M.; Xu, M. Exploring the Regulatory Effect of Hydroxytyrosol on Ovarian Inflammaging Through Autophagy-Targeted Mechanisms: A Bioinformatics Approach. Nutrients 2025, 17, 1421. https://doi.org/10.3390/nu17091421
An X, Guo X, Cai M, Xu M. Exploring the Regulatory Effect of Hydroxytyrosol on Ovarian Inflammaging Through Autophagy-Targeted Mechanisms: A Bioinformatics Approach. Nutrients. 2025; 17(9):1421. https://doi.org/10.3390/nu17091421
Chicago/Turabian StyleAn, Xiaoyang, Xiaoyu Guo, Meng Cai, and Meihong Xu. 2025. "Exploring the Regulatory Effect of Hydroxytyrosol on Ovarian Inflammaging Through Autophagy-Targeted Mechanisms: A Bioinformatics Approach" Nutrients 17, no. 9: 1421. https://doi.org/10.3390/nu17091421
APA StyleAn, X., Guo, X., Cai, M., & Xu, M. (2025). Exploring the Regulatory Effect of Hydroxytyrosol on Ovarian Inflammaging Through Autophagy-Targeted Mechanisms: A Bioinformatics Approach. Nutrients, 17(9), 1421. https://doi.org/10.3390/nu17091421







