A Probiotic Bacillus amyloliquefaciens D-1 Strain Is Responsible for Zearalenone Detoxifying in Coix Semen
Abstract
:1. Introduction
2. Results
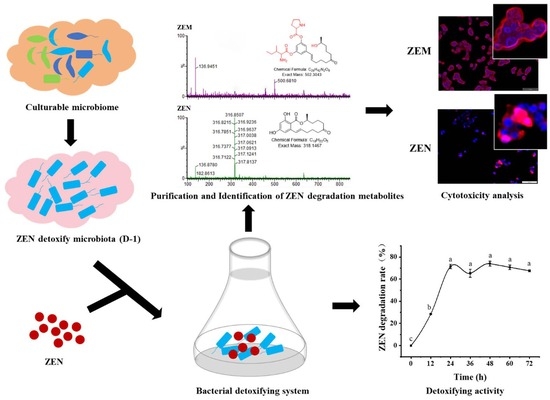
2.1. A Highly Active Bacillus Strain Catabolizes ZEN into ZEM
2.2. Analysis of the New ZEN-Derived Bacterial Metabolite Using UPLC-Q/TOF MS/MS
2.3. ZEM Has Decreased the Leakage of LDH, the Content of Intracellular MDA, and the Activity of Intracellular SOC of ZEM on TM4 and HepG2 Cell
2.4. Decreased Apoptosis of ZEM on TM4 and HepG2 Cell
2.5. D-1 Strain with No Adverse Impact on Cell
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Soil, Chemicals, Media, Kits and Cell
5.2. Construction of Culturable Microbiome from Rhizosphere Soil of Pseudostellaria heterophylla
5.3. Screening of ZEN-Degrading Strain from the Culturable Microbiome
5.4. Taxonomic Characterization of the Isolated Bacterial Strain
5.5. The Degradation Activity Analysis of D-1 Strain Versus Time, Temperature, pH, and Inoculation Amount
5.6. Assay of D-1 ZEN-Degrading Activity in Coix Semen Contaminated with ZEN
5.7. Analysis of the ZEN-Degradation of D-1 Is via Enzyme Conversion
5.8. UPLC- qTOF-MS/MS Analysis
5.9. Cytotoxicity Analysis of ZEN and Degradation Products to TM4 and HepG2 Cell
5.9.1. Cell Culture and Experiment Design
5.9.2. MTT Assay for Detecting Cell Viability
5.9.3. Fluorescence-Staining Assay
5.9.4. Detection of Lactate Dehydrogenase (LDH), Intracellular Malondialdehyde (MDA) Content and Superoxide Dismutase (SOD) Activity
5.9.5. Detection of Intracellular Reactive Oxygen Species (ROS) Content, Mitochondrial-Membrane Potential, and Cell Apoptosis using Flow Cytometry
5.9.6. Detection of Apoptosis-Related Genes Using qRT-PCR
5.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, M.; Yin, L.; Hu, H.; Selvaraj, J.N.; Zhou, Y.; Zhang, G. Expression, functional analysis and mutation of a novel neutral zearalenone-degrading enzyme. Int. J. Biol. Macromol. 2018, 118, 1284–1292. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, W.; Wu, H.; Zhang, W.; Mu, W. Identification of a potent enzyme for the detoxification of zearalenone. J. Agric. Food Chem. 2020, 68, 376–383. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.J.; Shen, H.H.; Zhang, X.F.; Yang, X.L.; Qiu, F.; Ou-Yang, Z.; Yang, M.H. Analysis of zearalenone and α-zearalenol in 100 foods and medicinal plants determined by HPLC-FLD and positive confirmation by LC-MS-MS. J. Sci. Food Agric. Ecosyst. Environ. 2013, 93, 1584–1590. [Google Scholar] [CrossRef] [PubMed]
- Borzekowski, A.; Drewitz, T.; Keller, J.; Pfeifer, D.; Kunte, H.J.; Koch, M.; Rohn, S.; Maul, R. Biosynthesis and characterization of zearalenone-14-sulfate, zearalenone-14-glucoside and zearalenone-16-glucoside using common fungal strains. Toxins 2018, 10, 104. [Google Scholar] [CrossRef]
- Kriszt, R.; Krifaton, C.; Szoboszlay, S.; Cserháti, M.; Kriszt, B.; Kukolya, J.; Czéh, Á.; Fehér-Tóth, S.; Török, L.; Szőke, Z.; et al. A new zearalenone biodegradation strategy using non-pathogenic Rhodococcus pyridinivorans K408 strain. PloS ONE 2012, 7, e43608. [Google Scholar] [CrossRef]
- Takahashi-Ando, N.; Ohsato, S.; Shibata, T.; Hamamoto, H.; Yamaguchi, I.; Kimura, M. Metabolism of zearalenone by genetically modified organisms expressing the detoxification gene from Clonostachys rosea. Appl. Environ. Microbiol. 2004, 70, 3239–3245. [Google Scholar] [CrossRef] [PubMed]
- Abd Alla, E.S. Zearalenone: Incidence, toxigenic fungi and chemical decontamination in Egyptian cereals. Die Nahr. 1997, 41, 362–365. [Google Scholar] [CrossRef]
- Kabak, B.; Dobson, A.D.; Var, I. Strategies to prevent mycotoxin contamination of food and animal feed: A review. Crit. Rev. Food Sci. Nutr. 2006, 46, 593–619. [Google Scholar] [CrossRef]
- Krifaton, C.; Kriszt, B.; Risa, A.; Szoboszlay, S.; Cserháti, M.; Harkai, P.; Eldridge, M.; Wang, J.; Kukolya, J. Application of a yeast estrogen reporter system for screening zearalenone degrading microbes. J. Hazard. Mater. 2013, 244, 429–435. [Google Scholar] [CrossRef]
- Guzel, E.; Arlier, S.; Guzeloglu-Kayisli, O.; Tabak, M.S.; Ekiz, T.; Semerci, N.; Larsen, K.; Schatz, F.; Lockwood, C.J.; Kayisli, U.A. Endoplasmic reticulum stress and homeostasis in reproductive physiology and pathology. Int. J. Mol. Sci. 2017, 18, 792. [Google Scholar] [CrossRef]
- Long, M.; Yang, S.; Zhang, W.; Zhang, Y.; Li, P.; Guo, Y.; Wang, Y.; Chen, X.; He, J. The influence of selenium yeast on hematological, biochemical and reproductive hormone level changes in Kunming mice following acute exposure to zearalenone. Biol. Trace Elem. Res. 2016, 174, 362–368. [Google Scholar] [CrossRef]
- Hussein, H.S.; Brasel, J.M. Toxicity, metabolism, and impact of mycotoxins on humans and animals. Toxicology 2001, 167, 101–134. [Google Scholar] [CrossRef] [PubMed]
- Altalhi, A.D.; El-Deeb, B. Localization of zearalenone detoxification gene(s) in pzea-1 plasmid of Pseudomonas putida zea-1 and expressed in Escherichia coli. J. Hazard. Mater. 2009, 161, 1166–1172. [Google Scholar] [CrossRef]
- Bueno, D.J.; Di Marco, L.; Oliver, G.; Bardón, A. In vitro binding of zearalenone to different adsorbents. J. Food Prot. 2005, 68, 613–615. [Google Scholar] [CrossRef]
- Ryu, D.; Hanna, M.A.; Bullerman, L.B. Stability of zearalenone during extrusion of corn grits. J. Food Prot. 1999, 62, 1482–1484. [Google Scholar] [CrossRef]
- Binder, E.M. Managing the risk of mycotoxins in modern feed production. Anim. Feed. Sci. Technol. 2007, 133, 149–166. [Google Scholar] [CrossRef]
- He, W.-J.; Yuan, Q.-S.; Zhang, Y.-B.; Guo, M.-W.; Gong, A.-D.; Zhang, J.-B.; Wu, A.-B.; Huang, T.; Qu, B.; Li, H.-P.; et al. Aerobic de-epoxydation of Trichothecene mycotoxins by a soil bacterial consortium isolated using in situ soil enrichment. Toxins 2016, 8, 277. [Google Scholar] [CrossRef]
- Shier, W.T.; Shier, A.C.; Xie, W.; Mirocha, C.J. Structure-activity relationships for human estrogenic activity in zearalenone mycotoxins. Toxicon Off. J. Int. Soc. Toxinology 2001, 39, 1435–1438. [Google Scholar] [CrossRef]
- El-Sharkawy, S.; Abul-Hajj, Y. Microbial transformation of zearalenone, i. Formation of zearalenone-4-o-β-glucoside. J. Nat. Prod. 2004, 50, 520–521. [Google Scholar] [CrossRef]
- Kamimura, H. Conversion of zearalenone to zearalenone glycoside by Rhizopus sp. Appl. Environ. Microbiol. 1986, 52, 515–519. [Google Scholar] [CrossRef]
- El-Sharkaway, S.H.; Selim, M.I.; Afifi, M.S.; Halaweish, F.T. Microbial transformation of zearalenone to a zearalenone sulfate. Appl. Environ. Microbiol. 1991, 57, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Gareis, M.; Bauer, J.; Thiem, J.; Plank, G.; Grabley, S.; Gedek, B. Cleavage of zearalenone-glycoside, a “masked” mycotoxin, during digestion in swine. Zentralblatt Veterinarmedizin. Reihe B. J. Vet. Medicine. Ser. B 1990, 37, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Plasencia, J.; Mirocha, C.J. Isolation and characterization of zearalenone sulfate produced by Fusarium spp. Appl. Environ. Microbiol. 1991, 57, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Berthiller, F.; Schuhmacher, R.; Adam, G.; Krska, R. Formation, determination and significance of masked and other conjugated mycotoxins. Anal. Bioanal. Chem. 2009, 395, 1243–1252. [Google Scholar] [CrossRef]
- Vekiru, E.; Hametner, C.; Mitterbauer, R.; Rechthaler, J.; Adam, G.; Schatzmayr, G.; Krska, R.; Schuhmacher, R. Cleavage of zearalenone by trichosporon mycotoxinivorans to a novel nonestrogenic metabolite. Appl. Environ. Microbiol. 2010, 76, 2353–2359. [Google Scholar] [CrossRef]
- Kakeya, H.; Takahashi-Ando, N.; Kimura, M.; Onose, R.; Yamaguchi, I.; Osada, H. Biotransformation of the mycotoxin, zearalenone, to a non-estrogenic compound by a fungal strain of Clonostachys sp. Biosci. Biotechnol. Biochem. 2002, 66, 2723–2726. [Google Scholar] [CrossRef] [PubMed]
- El-Sharkawy, S.; Abul-Hajj, Y.J. Microbial cleavage of zearalenone. Xenobiotica 1988, 18, 365–371. [Google Scholar] [CrossRef]
- Wang, N.; Wu, W.; Pan, J.; Long, M.J.M. Detoxification strategies for zearalenone using microorganisms: A review. Microorganisms 2019, 7, 208. [Google Scholar] [CrossRef]
- Siahmoshteh, F.; Siciliano, I.; Banani, H.; Hamidi-Esfahani, Z.; Razzaghi-Abyaneh, M.; Gullino, M.L.; Spadaro, D. Efficacy of Bacillus subtilis and Bacillus amyloliquefaciens in the control of Aspergillus parasiticus growth and aflatoxins production on pistachio. Int. J. Food Microbiol. 2017, 254, 47–53. [Google Scholar] [CrossRef]
- Chang, X.; Wu, Z.; Wu, S.; Dai, Y.; Sun, C. Degradation of ochratoxin a by Bacillus amyloliquefaciens asag1. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2015, 32, 564–571. [Google Scholar] [CrossRef]
- Hairul Islam, V.I.; Prakash Babu, N.; Pandikumar, P.; Ignacimuthu, S. Isolation and characterization of putative probiotic bacterial strain, Bacillus amyloliquefaciens, from North East Himalayan soil based on in vitro and in vivo functional properties. Probiotics Antimicrob. Proteins 2011, 3, 175–185. [Google Scholar] [CrossRef]
- Larsen, N.; Thorsen, L.; Kpikpi, E.N.; Stuer-Lauridsen, B.; Cantor, M.D.; Nielsen, B.; Brockmann, E.; Derkx, P.M.F.; Jespersen, L. Characterization of Bacillus spp. Strains for use as probiotic additives in pig feed. Appl. Microbiol. Biotechnol. 2014, 98, 1105–1118. [Google Scholar] [CrossRef]
- Ahmed, S.T.; Islam, M.M.; Mun, H.-S.; Sim, H.-J.; Kim, Y.-J.; Yang, C.-J. Effects of Bacillus amyloliquefaciens as a probiotic strain on growth performance, cecal microflora, and fecal noxious gas emissions of broiler chickens. Poult. Sci. 2014, 93, 1963–1971. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Cheng, K.-C.; Liu, J.-R. Isolation and characterization of a Bacillus amyloliquefaciens strain with zearalenone removal ability and its probiotic potential. PloS ONE 2017, 12, e0182220. [Google Scholar] [CrossRef]
- Woldemariam, Y.K.; Wan, Z.; Yu, Q.; Li, H.; Wei, X.; Liu, Y.; Wang, J.; Sun, B. Prebiotic, probiotic, antimicrobial, and functional food applications of Bacillus amyloliquefaciens. J. Agric. Food Chem. 2020, 68, 14709–14727. [Google Scholar] [CrossRef]
- Deng, T.; Yuan, Q.S.; Zhou, T.; Guo, L.P.; Jiang, W.K.; Zhou, S.H.; Yang, C.G.; Kang, C.Z. Screening of zearalenone-degrading bacteria and analysis of degradation conditions. China J. Chin. Mater. Medica 2021, 46, 5240–5246. [Google Scholar]
- Qu, R.; Jiang, C.; Wu, W.; Pang, B.; Lei, S.; Lian, Z.; Shao, D.; Jin, M.; Shi, J. Conversion of don to 3-epi-don in vitro and toxicity reduction of don in vivo by Lactobacillus rhamnosus. Food Funct. 2019, 10, 2785–2796. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowski, S.; Bozko, M.; Zarod, M.; Witecka, A.; Kocdemir, K.; Jagielski, A.K.; Drozak, J. Recharacterization of the mammalian cytosolic type 2 (r)-β-hydroxybutyrate dehydrogenase as 4-oxo-l-proline reductase (ec 1.1.1.104). J. Biol. Chem. 2022, 298, 101708–101724. [Google Scholar] [CrossRef] [PubMed]
- Ali, R.; Clark, L.D.; Zahm, J.A.; Lemoff, A.; Ramesh, K.; Rosenbaum, D.M.; Rosen, M.K. Improved strategy for isoleucine (1)h/(13)c methyl labeling in Pichia pastoris. J. Biomol. NMR 2019, 73, 687–697. [Google Scholar] [CrossRef]
- Poppenberger, B.; Berthiller, F.; Bachmann, H.; Lucyshyn, D.; Peterbauer, C.; Mitterbauer, R.; Schuhmacher, R.; Krska, R.; Glössl, J.; Adam, G. Heterologous expression of arabidopsis udp-glucosyltransferases in Saccharomyces cerevisiae for production of zearalenone-4-o-glucoside. Appl. Enviromental Microbiolgy 2006, 72, 4404–4410. [Google Scholar] [CrossRef]
- Poppenberger, B.; Berthiller, F.; Lucyshyn, D.; Schuhmacher, R.; Krska, R.; Adam, G. First results of gen-au: Cloning of deoxynivalenol- and zearalenone-inactivating udp-glucosyltransferase genes from Arabidopsis thaliana and expression in yeast for production of mycotoxin-glucosides. Mycotoxin Res. 2005, 21, 108–111. [Google Scholar] [CrossRef]
- Van Wilpe, S.; Koornstra, R.; Den Brok, M.; De Groot, J.W.; Blank, C.; De Vries, J.; Gerritsen, W.; Mehra, N. Lactate dehydrogenase: A marker of diminished antitumor immunity. Oncoimmunology 2020, 9, 1731942–1731952. [Google Scholar] [CrossRef]
- Abbès, S.; Ouanes, Z.; Salah-Abbès, J.B.; Houas, Z.; Oueslati, R.; Bacha, H.; Othman, O. The protective effect of hydrated sodium calcium aluminosilicate against haematological, biochemical and pathological changes induced by zearalenone in mice. Toxicon Off. J. Int. Soc. Toxinology 2006, 47, 567–574. [Google Scholar] [CrossRef]
- Jiang, S.Z.; Yang, Z.B.; Yang, W.R.; Gao, J.; Liu, F.X.; Broomhead, J.; Chi, F. Effects of purified zearalenone on growth performance, organ size, serum metabolites, and oxidative stress in postweaning gilts1. J. Anim. Sci. 2011, 89, 3008–3015. [Google Scholar] [CrossRef]
- Hou, Y.-J.; Zhao, Y.-Y.; Xiong, B.; Cui, X.-S.; Kim, N.-H.; Xu, Y.-X.; Sun, S.-C. Mycotoxin-containing diet causes oxidative stress in the mouse. PLoS ONE 2013, 8, e60374. [Google Scholar] [CrossRef] [PubMed]
- Hassen, W.; Ayed-Boussema, I.; Oscoz, A.A.; Lopez Ade, C.; Bacha, H. The role of oxidative stress in zearalenone-mediated toxicity in hep g2 cells: Oxidative DNA damage, gluthatione depletion and stress proteins induction. Toxicology 2007, 232, 294–302. [Google Scholar] [CrossRef]
- Chen, F.; Li, Q.; Zhang, Z.; Lin, P.; Lei, L.; Wang, A.; Jin, Y. Endoplasmic reticulum stress cooperates in zearalenone-induced cell death of raw 264.7 macrophages. Int. J. Mol. Sci. 2015, 16, 19780. [Google Scholar] [CrossRef]
- Ben Salem, I.; Boussabbeh, M.; Prola, A.; Guilbert, A.; Bacha, H.; Lemaire, C.; Abid-Essefi, S. Crocin protects human embryonic kidney cells (hek293) from α- and β-zearalenol-induced er stress and apoptosis. Environ. Sci. Pollut. Res. 2016, 23, 15504–15514. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Shu, C.-W.; Xu, W.; Shiau, C.-W.; Grant, D.; Vasile, S.; Cosford, N.D.P.; Reed, J.C. Chemical biology investigation of cell death pathways activated by endoplasmic reticulum stress reveals cytoprotective modulators of ask1*. J. Biol. Chem. 2009, 284, 1593–1603. [Google Scholar] [CrossRef] [PubMed]
- Pfaffenbach, K.T.; Lee, A.S. The critical role of grp78 in physiologic and pathologic stress. Curr. Opin. Cell Biol. 2011, 23, 150–156. [Google Scholar] [CrossRef]
- Barati, M.T.; Powell, D.W.; Kechavarzi, B.D.; Isaacs, S.M.; Zheng, S.; Epstein, P.N.; Cai, L.; Coventry, S.; Rane, M.J.; Klein, J.B. Differential expression of endoplasmic reticulum stress-response proteins in different renal tubule subtypes of ove26 diabetic mice. Cell Stress Chaperones 2016, 21, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.-Q.; Chen, Z.; Chen, L.-X. Endoplasmic reticulum stress: A novel mechanism and therapeutic target for cardiovascular diseases. Acta Pharmacol. Sin. 2016, 37, 425–443. [Google Scholar] [CrossRef]
- Nakka, V.P.; Prakash, B.P.; Vemuganti, R. Crosstalk between endoplasmic reticulum stress, oxidative stress, and autophagy: Potential therapeutic targets for acute cns injuries. Mol. Neurobiol. 2016, 53, 532–544. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Jin, H.; Lan, J.; Zhang, R.; Ren, H.; Zhang, X.; Yu, G. Detoxification of zearalenone by three strains of Lactobacillus plantarum from fermented food in vitro. Food Control 2015, 54, 158–164. [Google Scholar] [CrossRef]
- Tinyiro, S.E.; Wokadala, C.; Xu, D.; Yao, W. Adsorption and degradation of zearalenone by bacillus strains. Folia Microbiol. 2011, 56, 321–327. [Google Scholar] [CrossRef]
- Huang, W.; Chang, J.; Wang, P.; Liu, C.; Yin, Q.; Zhu, Q.; Lu, F.; Gao, T. Effect of the combined compound probiotics with mycotoxin–degradation enzyme on detoxifying aflatoxin b1 and zearalenone. J. Toxicol. Sci. 2018, 43, 377–385. [Google Scholar] [CrossRef]
- Armando, M.R.; Pizzolitto, R.P.; Dogi, C.A.; Cristofolini, A.; Merkis, C.; Poloni, V.; Dalcero, A.M.; Cavaglieri, L.R. Adsorption of ochratoxin a and zearalenone by potential probiotic Saccharomyces cerevisiae strains and its relation with cell wall thickness. J. Appl. Microbiol. 2012, 113, 256–264. [Google Scholar] [CrossRef]
- Sun, X.; He, X.; Xue, K.S.; Li, Y.; Xu, D.; Qian, H. Biological detoxification of zearalenone by Aspergillus niger strain fs10. Food Chem. Toxicol. 2014, 72, 76–82. [Google Scholar] [CrossRef]
- Wang, N.; Li, P.; Pan, J.; Wang, M.; Long, M.; Zang, J.; Yang, S. Bacillus velezensis a2 fermentation exerts a protective effect on renal injury induced by zearalenone in mice. Sci. Rep. 2018, 8, 13646–13659. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Wang, Y.; Wang, K.; Wei, H.; Shen, L. Isolation and characterization of the bacillus cereus bc7 strain, which is capable of zearalenone removal and intestinal flora modulation in mice. Toxicon Off. J. Int. Soc. Toxinology 2018, 155, 9–20. [Google Scholar] [CrossRef]
- Hsu, T.-C.; Yi, P.-J.; Lee, T.-Y.; Liu, J.-R. Probiotic characteristics and zearalenone-removal ability of a Bacillus licheniformis strain. PLoS ONE 2018, 13, 1–18. [Google Scholar] [CrossRef] [PubMed]
- El-Nezami, H.; Polychronaki, N.; Lee, Y.K.; Haskard, C.; Juvonen, R.; Salminen, S.; Mykkänen, H. Chemical moieties and interactions involved in the binding of zearalenone to the surface of Lactobacillus rhamnosus strains gg. J. Agric. Food Chem. 2004, 52, 4577–4581. [Google Scholar] [CrossRef] [PubMed]
- Yiannikouris, A.; François, J.; Poughon, L.; Dussap, C.G.; Bertin, G.; Jeminet, G.; Jouany, J.P. Alkali extraction of beta-d-glucans from Saccharomyces cerevisiae cell wall and study of their adsorptive properties toward zearalenone. J. Agric. Food Chem. 2004, 52, 3666–3673. [Google Scholar] [CrossRef]
- Takahashi-Ando, N.; Kimura, M.; Kakeya, H.; Osada, H.; Yamaguchi, I. A novel Lactonohydrolase responsible for the detoxification of zearalenone: Enzyme purification and gene cloning. Biochem. J. 2002, 365, 1–6. [Google Scholar] [CrossRef]
- Ben Salem, I.; Prola, A.; Boussabbeh, M.; Guilbert, A.; Bacha, H.; Abid-Essefi, S.; Lemaire, C. Crocin and quercetin protect hct116 and hek293 cells from zearalenone-induced apoptosis by reducing endoplasmic reticulum stress. Cell Stress Chaperones 2015, 20, 927–938. [Google Scholar] [CrossRef]
- Zheng, W.; Wang, B.; Si, M.; Zou, H.; Song, R.; Gu, J.; Yuan, Y.; Liu, X.; Zhu, G.; Bai, J.; et al. Zearalenone altered the cytoskeletal structure via er stress- autophagy- oxidative stress pathway in mouse tm4 sertoli cells. Sci. Rep. 2018, 8, 3320–3333. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Jiang, T.; Lin, P.; Chen, H.; Wang, L.; Wang, N.; Zhao, F.; Tang, K.; Zhou, D.; Wang, A.; et al. Apoptosis inducing factor gene depletion inhibits zearalenone-induced cell death in a goat leydig cell line. Reprod. Toxicol. 2017, 67, 129–139. [Google Scholar] [CrossRef] [PubMed]
- He, W.J.; Zhang, L.M.; Yi, S.Y.; Tang, X.L.; Yuan, Q.S.; Guo, M.W.; Wu, A.B.; Qu, B.; Li, H.P.; Liao, Y.C. An aldo-keto reductase is responsible for fusarium toxin-degrading activity in a soil Sphingomonas strain. Sci. Rep. 2017, 7, 9549–9561. [Google Scholar] [CrossRef]
- Yang, S.B.; Zheng, H.C.; Xu, J.Y.; Zhao, X.Y.; Shu, W.J.; Li, X.M.; Song, H.; Ma, Y.H. New biotransformation mode of zearalenone identified in Bacillus subtilis y816 revealing a novel zen conjugate. J. Agric. Food Chem. 2021, 69, 7409–7419. [Google Scholar] [CrossRef]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of cell viability by the mtt assay. Cold Spring Harb. Protoc. 2018, 2018, 469–471. [Google Scholar] [CrossRef]
- Wang, H.; Lv, D.; Jiang, S.; Hou, Q.; Zhang, L.; Li, S.; Zhu, X.; Xu, X.; Wen, J.; Zeng, C.; et al. Complement induces podocyte pyroptosis in membranous nephropathy by mediating mitochondrial dysfunction. Cell Death Dis. 2022, 13, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, H.; Yang, R.; Ji, D.; Xia, X. Gsk872 and necrostatin-1 protect retinal ganglion cells against necroptosis through inhibition of rip1/rip3/mlkl pathway in glutamate-induced retinal excitotoxic model of glaucoma. J. Neuroinflamm. 2022, 19, 262–283. [Google Scholar] [CrossRef] [PubMed]
- Karaman, E.F.; Zeybel, M.; Ozden, S. Evaluation of the epigenetic alterations and gene expression levels of hepg2 cells exposed to zearalenone and α-zearalenol. Toxicol. Lett. 2020, 326, 52–60. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, T.; Chen, Y.; Zhang, J.; Gao, Y.; Yang, C.; Jiang, W.; Ou, X.; Wang, Y.; Guo, L.; Zhou, T.; et al. A Probiotic Bacillus amyloliquefaciens D-1 Strain Is Responsible for Zearalenone Detoxifying in Coix Semen. Toxins 2023, 15, 674. https://doi.org/10.3390/toxins15120674
Deng T, Chen Y, Zhang J, Gao Y, Yang C, Jiang W, Ou X, Wang Y, Guo L, Zhou T, et al. A Probiotic Bacillus amyloliquefaciens D-1 Strain Is Responsible for Zearalenone Detoxifying in Coix Semen. Toxins. 2023; 15(12):674. https://doi.org/10.3390/toxins15120674
Chicago/Turabian StyleDeng, Tao, Yefei Chen, Jinqiang Zhang, Yanping Gao, Changgui Yang, Weike Jiang, Xiaohong Ou, Yanhong Wang, Lanping Guo, Tao Zhou, and et al. 2023. "A Probiotic Bacillus amyloliquefaciens D-1 Strain Is Responsible for Zearalenone Detoxifying in Coix Semen" Toxins 15, no. 12: 674. https://doi.org/10.3390/toxins15120674