RASSF10 Is a TGFβ-Target That Regulates ASPP2 and E-Cadherin Expression and Acts as Tumor Suppressor That Is Epigenetically Downregulated in Advanced Cancer
Abstract
1. Introduction
2. Results
2.1. RASSF10 Inhibts Cell Proliferation and Plays a Role TGFβ Induced Signal Transmission
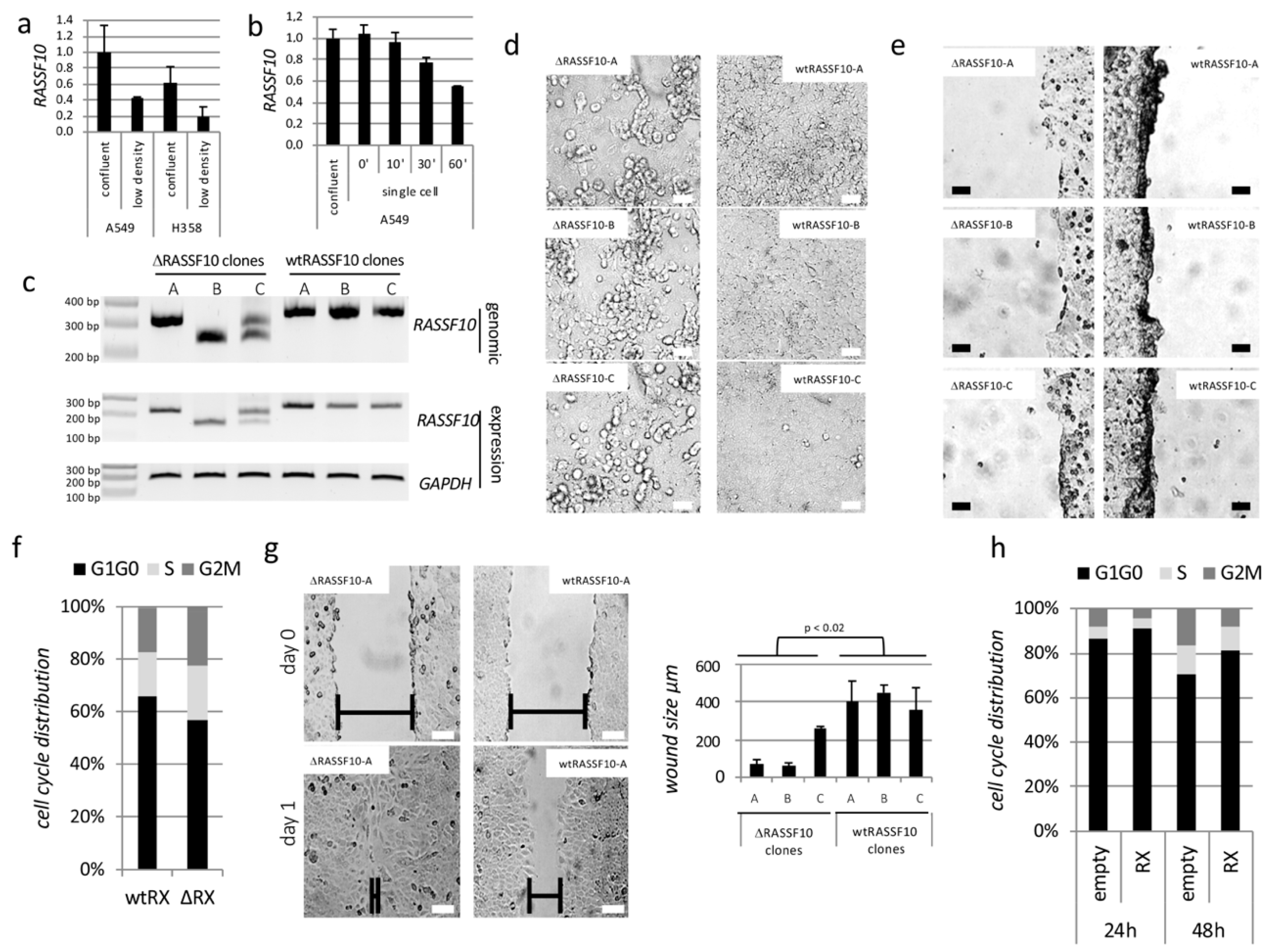
2.1.1. RASSF10 Loss Induces Cell Growth
2.1.2. RASSF10 Is Induced by the EMT Driver TGFβ and RASSF10 Depletion Promotes TGFβ Induced Invasion
2.1.3. RASSF10 Is a Positive Regulator of TGFβ Repressed CDH1 Expression
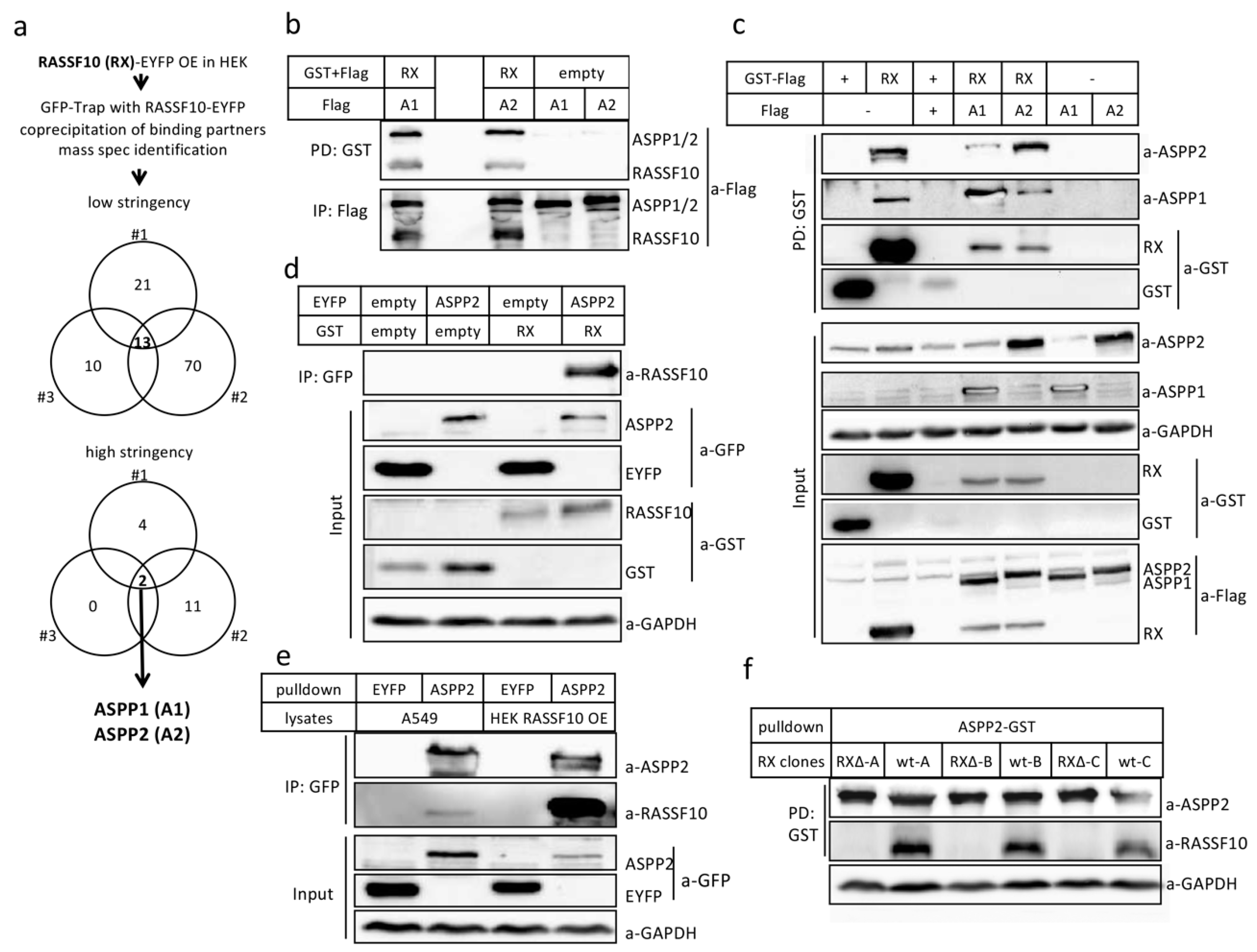
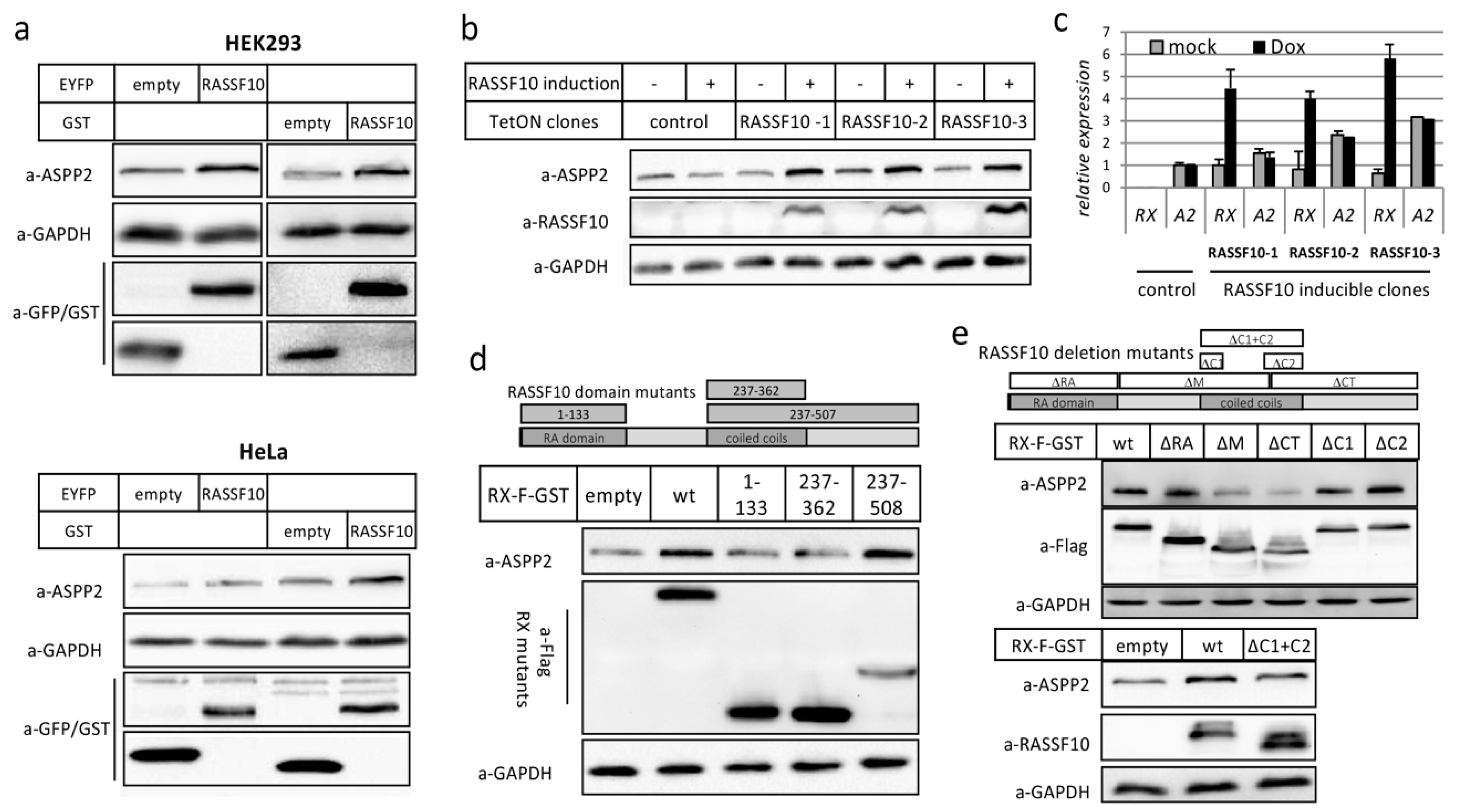
2.2. RASSF10 Interacts and Stabilzes Apoptosis-Stimmlating of p53 Protein 2 (ASPP2)
2.2.1. Verification of RASSF10 Interaction with ASPP1 and ASPP2
2.2.2. RASSF10 Stabilizes ASPP2 Protein through Its Coiled-Coils Domain
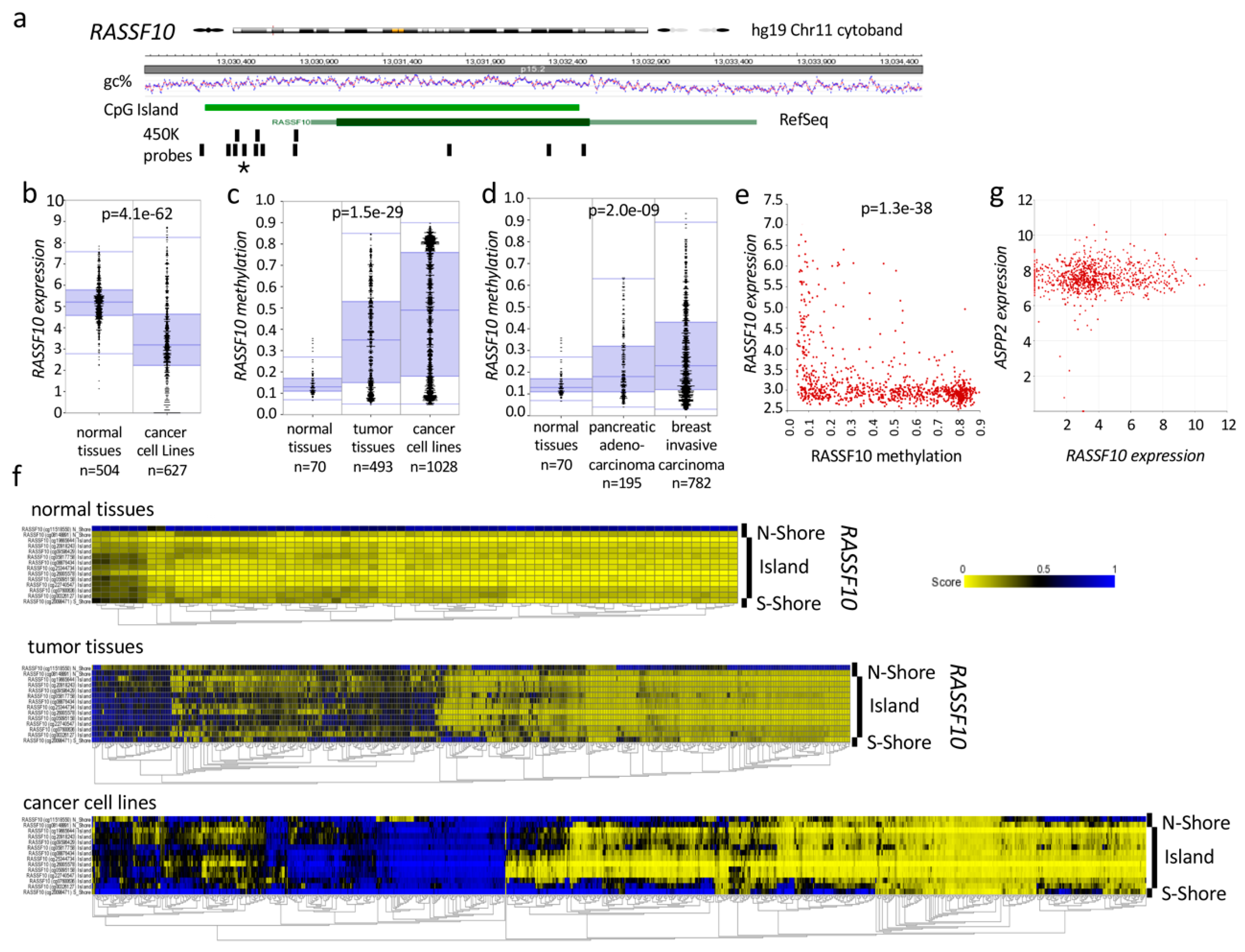
2.3. RASSF10 Is Inactivated across Human Cancers and a Valuable Cancer Biomarker
2.3.1. RASSF10 Is Frequently Hypermethylated in Human Cancers
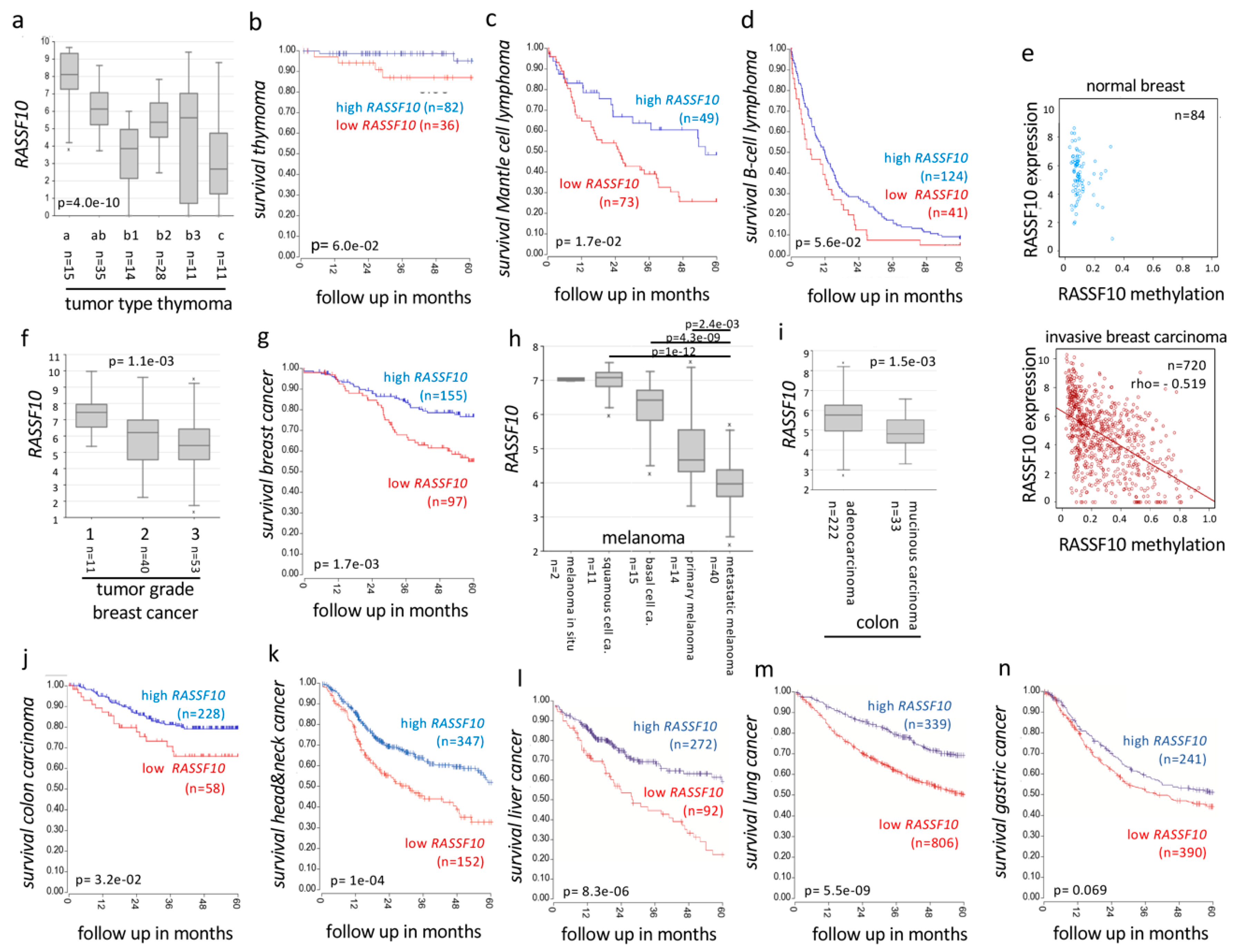
2.3.2. RASSF10 Inactivation Correlates with Clinical Diagnosis and Prognosis of Cancer
2.4. RASSF10 or ASPP2 Depletion Induces Activation of the TGFβ Signaling Pathway
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Treatment of Cell Lines
4.2. Generation of Stable RASSF10 Inducible HEK-Cells
4.3. Mass Spectrometry and Co-immunoprecipitations/Pulldown assays and Western Blotting
4.4. DNA Isolation and Methylation Analysis
4.5. RNA Expression Analysis
4.6. Plasmids
4.7. CRISPR/Cas9
4.8. Invasion Assay
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A. Additional Information on Datasets and Software Used in this Study
References
- Kotiyal, S.; Bhattacharya, S. Events of Molecular Changes in Epithelial-Mesenchymal Transition. Crit. Rev. Eukaryot. Gene Expr. 2016, 26, 163–171. [Google Scholar] [CrossRef]
- Mendonsa, A.M.; Na, T.Y.; Gumbiner, B.M. E-cadherin in contact inhibition and cancer. Oncogene 2018, 37, 4769–4780. [Google Scholar] [CrossRef] [PubMed]
- Cano, A.; Perez-Moreno, M.A.; Rodrigo, I.; Locascio, A.; Blanco, M.J.; del Barrio, M.G.; Portillo, F.; Nieto, M.A. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000, 2, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Peinado, H.; Quintanilla, M.; Cano, A. Transforming growth factor beta-1 induces snail transcription factor in epithelial cell lines: Mechanisms for epithelial mesenchymal transitions. J. Biol. Chem. 2003, 278, 21113–21123. [Google Scholar] [CrossRef] [PubMed]
- Richter, A.M.; Pfeifer, G.P.; Dammann, R.H. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim. Biophys. Acta 2009, 1796, 114–128. [Google Scholar] [CrossRef]
- Sherwood, V.; Recino, A.; Jeffries, A.; Ward, A.; Chalmers, A.D. The N-terminal RASSF family: A new group of Ras-association-domain-containing proteins, with emerging links to cancer formation. Biochem. J. 2010, 425, 303–311. [Google Scholar] [CrossRef]
- Volodko, N.; Gordon, M.; Salla, M.; Ghazaleh, H.A.; Baksh, S. RASSF tumor suppressor gene family: Biological functions and regulation. FEBS Lett. 2014, 588, 2671–2684. [Google Scholar] [CrossRef]
- Wohlgemuth, S.; Kiel, C.; Kramer, A.; Serrano, L.; Wittinghofer, F.; Herrmann, C. Recognizing and defining true Ras binding domains I: Biochemical analysis. J. Mol. Biol. 2005, 348, 741–758. [Google Scholar] [CrossRef]
- Aoyama, Y.; Avruch, J.; Zhang, X.F. Nore1 inhibits tumor cell growth independent of Ras or the MST1/2 kinases. Oncogene 2004, 23, 3426–3433. [Google Scholar] [CrossRef]
- Cooper, W.N.; Hesson, L.B.; Matallanas, D.; Dallol, A.; von Kriegsheim, A.; Ward, R.; Kolch, W.; Latif, F. RASSF2 associates with and stabilizes the proapoptotic kinase MST2. Oncogene 2009, 28, 2988–2998. [Google Scholar] [CrossRef]
- Dittfeld, C.; Richter, A.M.; Steinmann, K.; Klagge-Ulonska, A.; Dammann, R.H. The SARAH Domain of RASSF1A and Its Tumor Suppressor Function. Mol. Biol. Int. 2012, 2012, 196715. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Tommasi, S.; Liu, L.; Yee, J.K.; Dammann, R.; Pfeifer, G.P. RASSF1A is part of a complex similar to the Drosophila Hippo/Salvador/Lats tumor-suppressor network. Curr. Biol. 2007, 17, 700–705. [Google Scholar] [CrossRef] [PubMed]
- Matallanas, D.; Romano, D.; Yee, K.; Meissl, K.; Kucerova, L.; Piazzolla, D.; Baccarini, M.; Vass, J.K.; Kolch, W.; O’Neill, E. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol. Cell 2007, 27, 962–975. [Google Scholar] [CrossRef] [PubMed]
- Donninger, H.; Allen, N.; Henson, A.; Pogue, J.; Williams, A.; Gordon, L.; Kassler, S.; Dunwell, T.; Latif, F.; Clark, G.J. Salvador protein is a tumor suppressor effector of RASSF1A with hippo pathway-independent functions. J. Biol. Chem. 2011, 286, 18483–18491. [Google Scholar] [CrossRef]
- Jimenez, A.P.; Traum, A.; Boettger, T.; Hackstein, H.; Richter, A.M.; Dammann, R.H. The tumor suppressor RASSF1A induces the YAP1 target gene ANKRD1 that is epigenetically inactivated in human cancers and inhibits tumor growth. Oncotarget 2017, 8, 88437–88452. [Google Scholar] [CrossRef]
- Dammann, R.; Li, C.; Yoon, J.H.; Chin, P.L.; Bates, S.; Pfeifer, G.P. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat. Genet. 2000, 25, 315–319. [Google Scholar] [CrossRef]
- Schagdarsurengin, U.; Richter, A.M.; Wohler, C.; Dammann, R.H. Frequent epigenetic inactivation of RASSF10 in thyroid cancer. Epigenetics 2009, 4, 571–576. [Google Scholar] [CrossRef]
- Underhill-Day, N.; Hill, V.; Latif, F. N-terminal RASSF family: RASSF7-RASSF10. Epigenetics 2011, 6, 284–292. [Google Scholar] [CrossRef]
- Richter, A.M.; Walesch, S.K.; Wurl, P.; Taubert, H.; Dammann, R.H. The tumor suppressor RASSF10 is upregulated upon contact inhibition and frequently epigenetically silenced in cancer. Oncogenesis 2012, 1, e18. [Google Scholar] [CrossRef]
- Richter, A.M.; Haag, T.; Walesch, S.; Herrmann-Trost, P.; Marsch, W.C.; Kutzner, H.; Helmbold, P.; Dammann, R.H. Aberrant Promoter Hypermethylation of RASSF Family Members in Merkel Cell Carcinoma. Cancers 2013, 5, 1566–1576. [Google Scholar] [CrossRef]
- Richter, A.M.; Zimmermann, T.; Haag, T.; Walesch, S.K.; Dammann, R.H. Promoter methylation status of Ras-association domain family members in pheochromocytoma. Front. Endocrinol. 2015, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Richter, A.M.; Walesch, S.K.; Dammann, R.H. Aberrant Promoter Methylation of the Tumour Suppressor RASSF10 and Its Growth Inhibitory Function in Breast Cancer. Cancers 2016, 8, 26. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, J.; Wang, L.; Qian, C.; Qian, Y.; Xuan, H.; Zhuo, W.; Li, X.; Yu, J.; Si, J. Ras-association domain family 10 acts as a novel tumor suppressor through modulating MMP2 in hepatocarcinoma. Oncogenesis 2016, 5, e237. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Wang, W.; Jin, J.; Yu, Z.; Xin, X. RASSF10 is Epigenetically Inactivated and Suppresses Cell Proliferation and Induces Cell Apoptosis by Activating the p53 Signalling Pathway in Papillary Thyroid Carcinoma Cancer. Cell Physiol. Biochem. 2017, 41, 1229–1239. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liang, Q.; Liu, W.; Zhang, N.; Xu, L.; Zhang, X.; Zhang, J.; Sung, J.J.; Yu, J. Ras association domain family member 10 suppresses gastric cancer growth by cooperating with GSTP1 to regulate JNK/c-Jun/AP-1 pathway. Oncogene 2016, 35, 2453–2464. [Google Scholar] [CrossRef]
- Wang, Y.; Bu, F.; Royer, C.; Serres, S.; Larkin, J.R.; Soto, M.S.; Sibson, N.R.; Salter, V.; Fritzsche, F.; Turnquist, C.; et al. ASPP2 controls epithelial plasticity and inhibits metastasis through beta-catenin-dependent regulation of ZEB1. Nat. Cell Biol. 2014, 16, 1092–1104. [Google Scholar] [CrossRef]
- Gen, Y.; Yasui, K.; Kitaichi, T.; Iwai, N.; Terasaki, K.; Dohi, O.; Hashimoto, H.; Fukui, H.; Inada, Y.; Fukui, A.; et al. ASPP2 suppresses invasion and TGF-beta1-induced epithelial-mesenchymal transition by inhibiting Smad7 degradation mediated by E3 ubiquitin ligase ITCH in gastric cancer. Cancer Lett. 2017, 398, 52–61. [Google Scholar] [CrossRef]
- Molenaar, J.J.; Koster, J.; Zwijnenburg, D.A.; van Sluis, P.; Valentijn, L.J.; van der Ploeg, I.; Hamdi, M.; van Nes, J.; Westerman, B.A.; van Arkel, J.; et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 2012, 483, 589–593. [Google Scholar] [CrossRef]
- Strzyz, P. TGF beta and EMT as double agents. Nat. Rev. Mol. Cell Biol. 2016, 17, 202–203. [Google Scholar] [CrossRef]
- Aomatsu, K.; Arao, T.; Sugioka, K.; Matsumoto, K.; Tamura, D.; Kudo, K.; Kaneda, H.; Tanaka, K.; Fujita, Y.; Shimomura, Y.; et al. TGF-beta Induces Sustained Upregulation of SNAI1 and SNAI2 through Smad and Non-Smad Pathways in a Human Corneal Epithelial Cell Line. Investig. Ophthalmol. Vis. Sci. 2011, 52, 2437–2443. [Google Scholar] [CrossRef]
- Johansson, J.; Tabor, V.; Wikell, A.; Jalkanen, S.; Fuxe, J. TGF-beta1-Induced Epithelial-Mesenchymal Transition Promotes Monocyte/Macrophage Properties in Breast Cancer Cells. Front. Oncol. 2015, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Hocevar, B.A.; Howe, P.H. Mechanisms of TGF-beta-induced cell cycle arrest. Min. electrolyte Metab. 1998, 24, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Rath, A.; Glibowicka, M.; Nadeau, V.G.; Chen, G.; Deber, C.M. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. USA 2009, 106, 1760–1765. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Zhu, Q.; Huang, D.; Zhao, S.; Jan Lo, L.; Peng, J. An equation to estimate the difference between theoretically predicted and SDS PAGE-displayed molecular weights for an acidic peptide. Sci. Rep. 2015, 5, 13370. [Google Scholar] [CrossRef]
- Trigiante, G.; Lu, X. ASPP [corrected] and cancer. Nat. Rev. Cancer 2006, 6, 217–226. [Google Scholar] [CrossRef]
- Herman, J.G. Hypermethylation of tumor suppressor genes in cancer. Semin. Cancer Biol. 1999, 9, 359–367. [Google Scholar] [CrossRef]
- Chen, Y.J.; Tang, Q.B.; Zou, S.Q. Inactivation of RASSF1A, the tumor suppressor gene at 3p21.3 in extrahepatic cholangiocarcinoma. World J. Gastroenterol. 2005, 11, 1333–1338. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef]
- Muller, P.A.J.; Vousden, K.H. p53 mutations in cancer. Nat. Cell Biol. 2013, 15, 2–8. [Google Scholar] [CrossRef]
- Kasai, H.; Allen, J.T.; Mason, R.M.; Kamimura, T.; Zhang, Z. TGF-beta1 induces human alveolar epithelial to mesenchymal cell transition (EMT). Respir. Res. 2005, 6, 56. [Google Scholar] [CrossRef] [PubMed]
- Jahn, S.C.; Law, M.E.; Corsino, P.E.; Parker, N.N.; Pham, K.; Davis, B.J.; Lu, J.R.; Law, B.K. An in vivo model of epithelial to mesenchymal transition reveals a mitogenic switch. Cancer Lett. 2012, 326, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Seoane, J. Escaping from the TGF beta anti-proliferative control. Carcinogenesis 2006, 27, 2148–2156. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Zhang, Y.E. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 2003, 425, 577–584. [Google Scholar] [CrossRef]
- Shi, Y.G.; Massague, J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 2003, 113, 685–700. [Google Scholar] [CrossRef]
- Bachman, K.E.; Park, B.H. Duel nature of TGF-beta signaling: Tumor suppressor vs. tumor promoter. Curr. Opin. Oncol. 2005, 17, 49–54. [Google Scholar] [CrossRef]
- Miao, L.Y.; Wang, Y.S.; Xia, H.P.; Yao, C.Y.; Cai, H.R.; Song, Y. SPOCK1 is a novel transforming growth factor-beta target gene that regulates lung cancer cell epithelial-mesenchymal transition. Biochem. Biophys. Res. Commun. 2013, 440, 792–797. [Google Scholar] [CrossRef]
- Kourtidis, A.; Lu, R.; Pence, L.J.; Anastasiadis, P.Z. A central role for cadherin signaling in cancer. Exp. Cell Res. 2017, 358, 78–85. [Google Scholar] [CrossRef]
- Vigneron, A.M.; Ludwig, R.L.; Vousden, K.H. Cytoplasmic ASPP1 inhibits apoptosis through the control of YAP. Genes Dev. 2010, 24, 2430–2439. [Google Scholar] [CrossRef]
- Bergamaschi, D.; Samuels, Y.; Jin, B.; Duraisingham, S.; Crook, T.; Lu, X. ASPP1 and ASPP2: Common activators of p53 family members. Mol. Cell Biol. 2004, 24, 1341–1350. [Google Scholar] [CrossRef]
- Cong, W.; Hirose, T.; Harita, Y.; Yamashita, A.; Mizuno, K.; Hirano, H.; Ohno, S. ASPP2 regulates epithelial cell polarity through the PAR complex. Curr. Biol. 2010, 20, 1408–1414. [Google Scholar] [CrossRef] [PubMed]
- Sottocornola, R.; Royer, C.; Vives, V.; Tordella, L.; Zhong, S.; Wang, Y.; Ratnayaka, I.; Shipman, M.; Cheung, A.; Gaston-Massuet, C.; et al. ASPP2 binds Par-3 and controls the polarity and proliferation of neural progenitors during CNS development. Dev. Cell 2010, 19, 126–137. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, N. Cell polarity: ASPP2 gets a polarity complex. Nat. Rev. Cancer 2010, 10, 528. [Google Scholar] [CrossRef] [PubMed]
- Hauri, S.; Wepf, A.; van Drogen, A.; Varjosalo, M.; Tapon, N.; Aebersold, R.; Gstaiger, M. Interaction proteome of human Hippo signaling: Modular control of the co-activator YAP1. Mol. Syst. Biol. 2013, 9, 713. [Google Scholar] [CrossRef] [PubMed]
- Royer, C.; Koch, S.; Qin, X.; Zak, J.; Buti, L.; Dudziec, E.; Zhong, S.; Ratnayaka, I.; Srinivas, S.; Lu, X. ASPP2 links the apical lateral polarity complex to the regulation of YAP activity in epithelial cells. PLoS ONE 2014, 9, e111384. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Abdollah, S.; Qiu, Y.; Cai, J.; Xu, Y.Y.; Grinnell, B.W.; Richardson, M.A.; Topper, J.N.; Gimbrone, M.A., Jr.; Wrana, J.L.; et al. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell 1997, 89, 1165–1173. [Google Scholar] [CrossRef]
- Wells, J.E.; Howlett, M.; Cole, C.H.; Kees, U.R. Deregulated expression of connective tissue growth factor (CTGF/CCN2) is linked to poor outcome in human cancer. Int. J. Cancer 2015, 137, 504–511. [Google Scholar] [CrossRef]
- Hill, V.K.; Underhill-Day, N.; Krex, D.; Robel, K.; Sangan, C.B.; Summersgill, H.R.; Morris, M.; Gentle, D.; Chalmers, A.D.; Maher, E.R.; et al. Epigenetic inactivation of the RASSF10 candidate tumor suppressor gene is a frequent and an early event in gliomagenesis. Oncogene 2010, 30, 978–989. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Wei, Y.; Ma, J.; Peng, J.; Wumaier, R.; Shen, S.; Zhang, P.; Yu, L. The tumor suppressor proteins ASPP1 and ASPP2 interact with C-Nap1 and regulate centrosome linker reassembly. Biochem. Biophys. Res. Commun. 2015, 458, 494–500. [Google Scholar] [CrossRef]
- Moore, J.K.; Cooper, J.A. Coordinating mitosis with cell polarity: Molecular motors at the cell cortex. Semin. Cell Dev. Biol. 2010, 21, 283–289. [Google Scholar] [CrossRef]
- Lee, M.; Vasioukhin, V. Cell polarity and cancer—Cell and tissue polarity as a non-canonical tumor suppressor. J. Cell Sci. 2008, 121, 1141–1150. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.; Bestor, T.H. Altered methylation patterns in cancer cell genomes: Cause or consequence? Cancer Cell 2002, 1, 299–305. [Google Scholar] [CrossRef]
- Herman, J.G.; Baylin, S.B. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003, 349, 2042–2054. [Google Scholar] [CrossRef] [PubMed]
- Strunnikova, M.; Schagdarsurengin, U.; Kehlen, A.; Garbe, J.C.; Stampfer, M.R.; Dammann, R. Chromatin inactivation precedes de novo DNA methylation during the progressive epigenetic silencing of the RASSF1A promoter. Mol. Cell Biol. 2005, 25, 3923–3933. [Google Scholar] [CrossRef]
- Helmbold, P.; Richter, A.M.; Walesch, S.; Skorokhod, A.; Marsch, W.; Enk, A.; Dammann, R.H. RASSF10 promoter hypermethylation is frequent in malignant melanoma of the skin but uncommon in nevus cell nevi. J. Investig. Derm. 2012, 132, 687–694. [Google Scholar] [CrossRef]
- Palmirotta, R.; Lovero, D.; Cafforio, P.; Felici, C.; Mannavola, F.; Pelle, E.; Quaresmini, D.; Tucci, M.; Silvestris, F. Liquid biopsy of cancer: A multimodal diagnostic tool in clinical oncology. Ther. Adv. Med. Oncol. 2018, 10, 1–24. [Google Scholar] [CrossRef]
- Konstantinidi, E.M.; Lappas, A.S.; Tzortzi, A.S.; Behrakis, P.K. Exhaled Breath Condensate: Technical and Diagnostic Aspects. Sci. World J. 2015, 2015, 435160. [Google Scholar] [CrossRef]
- Han, W.G.; Wang, T.; Reilly, A.A.; Keller, S.M.; Spivack, S.D. Gene promoter methylation assayed in exhaled breath, with differences in smokers and lung cancer patients. Respir. Res. 2009, 10. [Google Scholar] [CrossRef]
- Rindlisbacher, B.; Strebel, C.; Guler, S.; Kollar, A.; Geiser, T.; Martin Fiedler, G.; Benedikt Leichtle, A.; Bovet, C.; Funke-Chambour, M. Exhaled breath condensate as a potential biomarker tool for idiopathic pulmonary fibrosis-a pilot study. J. Breath Res. 2017, 12, 016003. [Google Scholar] [CrossRef]
- Kwapisz, D. The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Ann. Transl. Med. 2017, 5, 46. [Google Scholar] [CrossRef]
- Kint, S.; De Spiegelaere, W.; De Kesel, J.; Vandekerckhove, L.; Van Criekinge, W. Evaluation of bisulfite kits for DNA methylation profiling in terms of DNA fragmentation and DNA recovery using digital PCR. PLoS ONE 2018, 13, e0199091. [Google Scholar] [CrossRef]
- Wreczycka, K.; Gosdschan, A.; Yusuf, D.; Gruning, B.; Assenov, Y.; Akalin, A. Strategies for analyzing bisulfite sequencing data. J. Biotechnol. 2017, 261, 105–115. [Google Scholar] [CrossRef]
- Taylor, S.C.; Laperriere, G.; Germain, H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: From variable nonsense to publication quality data. Sci. Rep. 2017, 7, 2409. [Google Scholar] [CrossRef]
- Dammann, R.H.; Richter, A.M.; Jimenez, A.P.; Woods, M.; Kuster, M.; Witharana, C. Impact of Natural Compounds on DNA Methylation Levels of the Tumor Suppressor Gene RASSF1A in Cancer. Int. J. Mol. Sci. 2017, 18, 2160. [Google Scholar] [CrossRef]
- Kruger, M.; Moser, M.; Ussar, S.; Thievessen, I.; Luber, C.A.; Forner, F.; Schmidt, S.; Zanivan, S.; Fassler, R.; Mann, M. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 2008, 134, 353–364. [Google Scholar] [CrossRef]
- Konzer, A.; Ruhs, A.; Braun, H.; Jungblut, B.; Braun, T.; Kruger, M. Stable isotope labeling in zebrafish allows in vivo monitoring of cardiac morphogenesis. Mol. Cell. Proteom. 2013, 12, 1502–1512. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Diez-Villanueva, A.; Mallona, I.; Peinado, M.A. Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenet. Chromatin 2015, 8, 22. [Google Scholar] [CrossRef]
- Szasz, A.M.; Lanczky, A.; Nagy, A.; Forster, S.; Hark, K.; Green, J.E.; Boussioutas, A.; Busuttil, R.; Szabo, A.; Gyorffy, B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 2016, 7, 49322–49333. [Google Scholar] [CrossRef]
- Gyorffy, B.; Surowiak, P.; Budczies, J.; Lanczky, A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE 2013, 8, e82241. [Google Scholar] [CrossRef]
- Gyorffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef]
- Nagy, A.; Lanczky, A.; Menyhart, O.; Gyorffy, B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 2018, 8, 9227. [Google Scholar] [CrossRef] [PubMed]
- Modhukur, V.; Iljasenko, T.; Metsalu, T.; Lokk, K.; Laisk-Podar, T.; Vilo, J. MethSurv: A web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 2018, 10, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Sartor, M.A.; Mahavisno, V.; Keshamouni, V.G.; Cavalcoli, J.; Wright, Z.; Karnovsky, A.; Kuick, R.; Jagadish, H.; Mirel, B.; Weymouth, T.; et al. ConceptGen: A gene set enrichment and gene set relation mapping tool. Bioinformatics 2010, 26, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Hesling, C.; Fattet, L.; Teyre, G.; Jury, D.; Gonzalo, P.; Lopez, J.; Vanbelle, C.; Morel, A.P.; Gillet, G.; Mikaelian, I.; et al. Antagonistic regulation of EMT by TIF1 gamma and Smad4 in mammary epithelial cells. EMBO Rep. 2011, 12, 665–672. [Google Scholar] [CrossRef]




| GO Term | GOPath | p-Value |
|---|---|---|
| cell periphery | GO:0071944 | 1.2 × 10−32 |
| plasma membrane | GO:0005886 | 7.1 × 10−31 |
| epidermis development | GO:0008544 | 1.5 × 10−30 |
| skin development | GO:0043588 | 4.5 × 10−30 |
| membrane part | GO:0044425 | 8.4 × 10−28 |
| epidermal cell differentiation | GO:0009913 | 1.3 × 10−25 |
| epithelial cell differentiation | GO:0030855 | 1.4 × 10−24 |
| apical plasma membrane | GO:0016324 | 8.7 × 10−18 |
| cell–cell junction | GO:0005911 | 5.0 × 10−16 |
| A549 Clone | Treatment | Mean Number of Invasive Cells (± SD, n = 3) | Fold Induction (p-Value) a |
|---|---|---|---|
| A ΔRASSF10 | 5 ng/mL TGFβ | 523 (±237) | 870 (0.02) |
| A ΔRASSF10 | mock | 0.3 (±0.6) | |
| B ΔRASSF10 | 5 ng/mL TGFβ | 727 (±363) | 15 (0.03) |
| B ΔRASSF10 | mock | 48 (±73) | |
| C ΔRASSF10 | 5 ng/mL TGFβ | 59 (±27) | 197 (0.02) |
| C ΔRASSF10 | mock | 0.3 (±0.6) | |
| A wtRASSF10 | 5 ng/mL TGFβ | 14 (±10) | 1.2 (0.87) |
| A wtRASSF10 | mock | 12 (±15) | |
| B wtRASSF10 | 5 ng/mL TGFβ | 15 (±13) | 21 (0.14) |
| B wtRASSF10 | mock | 0.7 (±1.2) |
| No | Protein ID | Enrichment Above Control | Full Protein Name |
|---|---|---|---|
| IP | RASSF10 | 286 | Ras-association domain family 10 |
| 1 | ASPP1 | 185 | Apoptosis-stimulating of p53 protein 1 |
| 2 | ASPP2 | 184 | Apoptosis-Stimulating of p53 protein 2 |
| 3 | SMU1 | 70 | WD40 repeat-containing protein SMU1 |
| 4 | EHD4 | 50 | EH-domain containing 4 |
| 5 | SDC2 | 42 | Syndecan 2 |
| 6 | PLEKHA5 | 42 | Pleckstrin Homology Domain Containing, Family A Member 5 |
| 7 | PTPN13 | 32 | Protein tyrosine phosphatase, non-receptor type 13 |
| 8 | HRC1 | 31 | HRAS1-related cluster protein 1 |
| 9 | CASK | 25 | Calcium/calmodulin-dependent serine protein kinase |
| 10 | NID1 | 23 | Nidogen 1 |
| 11 | DLG1 | 18 | Discs, large homolog 1 (Drosophila) |
| 12 | PPP1CA | 17 | Protein phosphatase 1, catalytic subunit, alpha isoform |
| 13 | FBXW11 | 16 | F-box and WD-40 domain protein 11 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Richter, A.M.; Küster, M.M.; Woods, M.L.; Walesch, S.K.; Gökyildirim, M.Y.; Krueger, M.; Dammann, R.H. RASSF10 Is a TGFβ-Target That Regulates ASPP2 and E-Cadherin Expression and Acts as Tumor Suppressor That Is Epigenetically Downregulated in Advanced Cancer. Cancers 2019, 11, 1976. https://doi.org/10.3390/cancers11121976
Richter AM, Küster MM, Woods ML, Walesch SK, Gökyildirim MY, Krueger M, Dammann RH. RASSF10 Is a TGFβ-Target That Regulates ASPP2 and E-Cadherin Expression and Acts as Tumor Suppressor That Is Epigenetically Downregulated in Advanced Cancer. Cancers. 2019; 11(12):1976. https://doi.org/10.3390/cancers11121976
Chicago/Turabian StyleRichter, Antje M., Miriam M. Küster, Michelle L. Woods, Sara K. Walesch, Mira Y. Gökyildirim, Marcus Krueger, and Reinhard H. Dammann. 2019. "RASSF10 Is a TGFβ-Target That Regulates ASPP2 and E-Cadherin Expression and Acts as Tumor Suppressor That Is Epigenetically Downregulated in Advanced Cancer" Cancers 11, no. 12: 1976. https://doi.org/10.3390/cancers11121976
APA StyleRichter, A. M., Küster, M. M., Woods, M. L., Walesch, S. K., Gökyildirim, M. Y., Krueger, M., & Dammann, R. H. (2019). RASSF10 Is a TGFβ-Target That Regulates ASPP2 and E-Cadherin Expression and Acts as Tumor Suppressor That Is Epigenetically Downregulated in Advanced Cancer. Cancers, 11(12), 1976. https://doi.org/10.3390/cancers11121976






