Prediction of Colon Cancer Stages and Survival Period with Machine Learning Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Data Preprocessing
2.3. Derivation of Tumor Aggression Score (TAS)
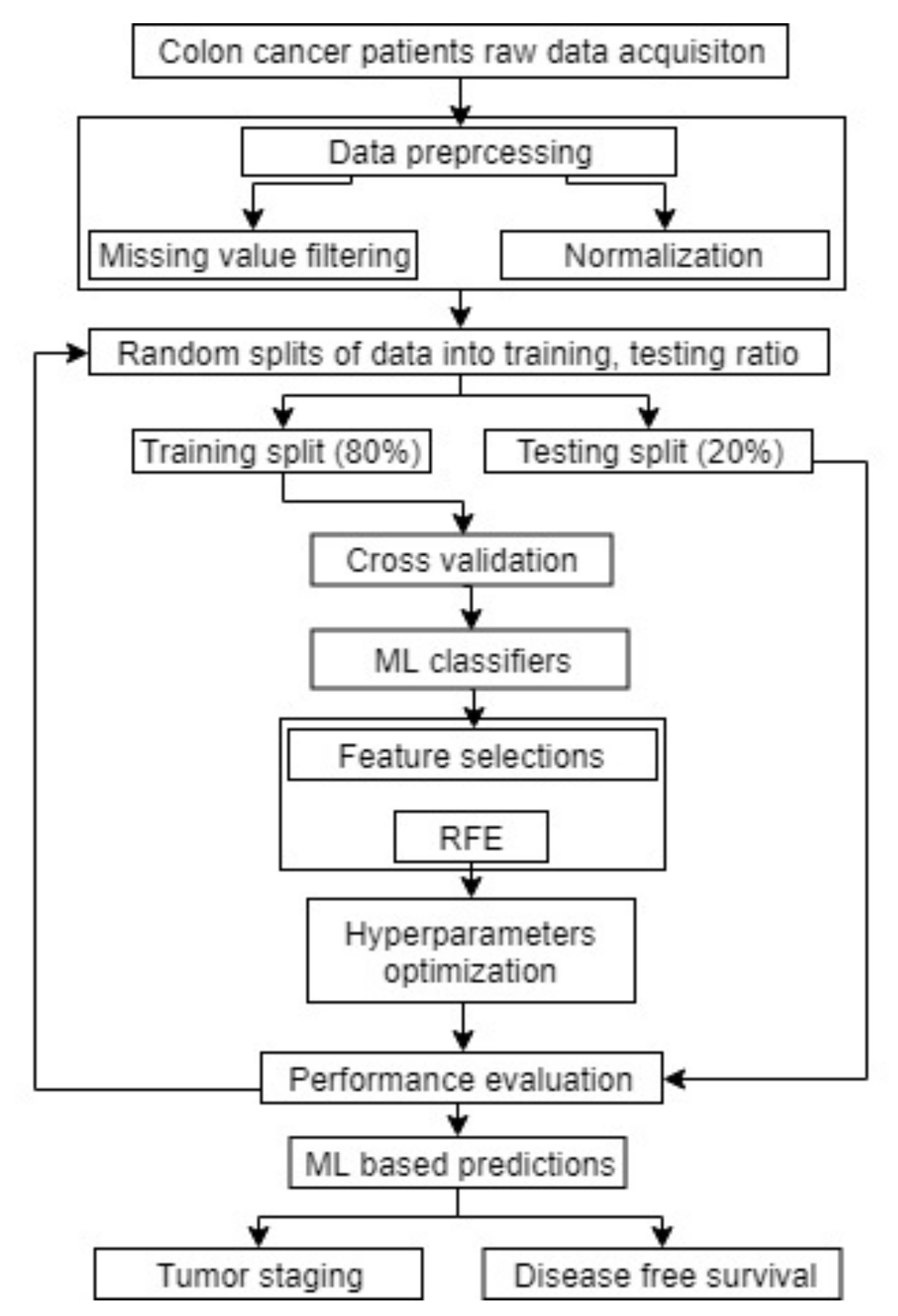
2.4. Machine Learning Analysis
3. Results
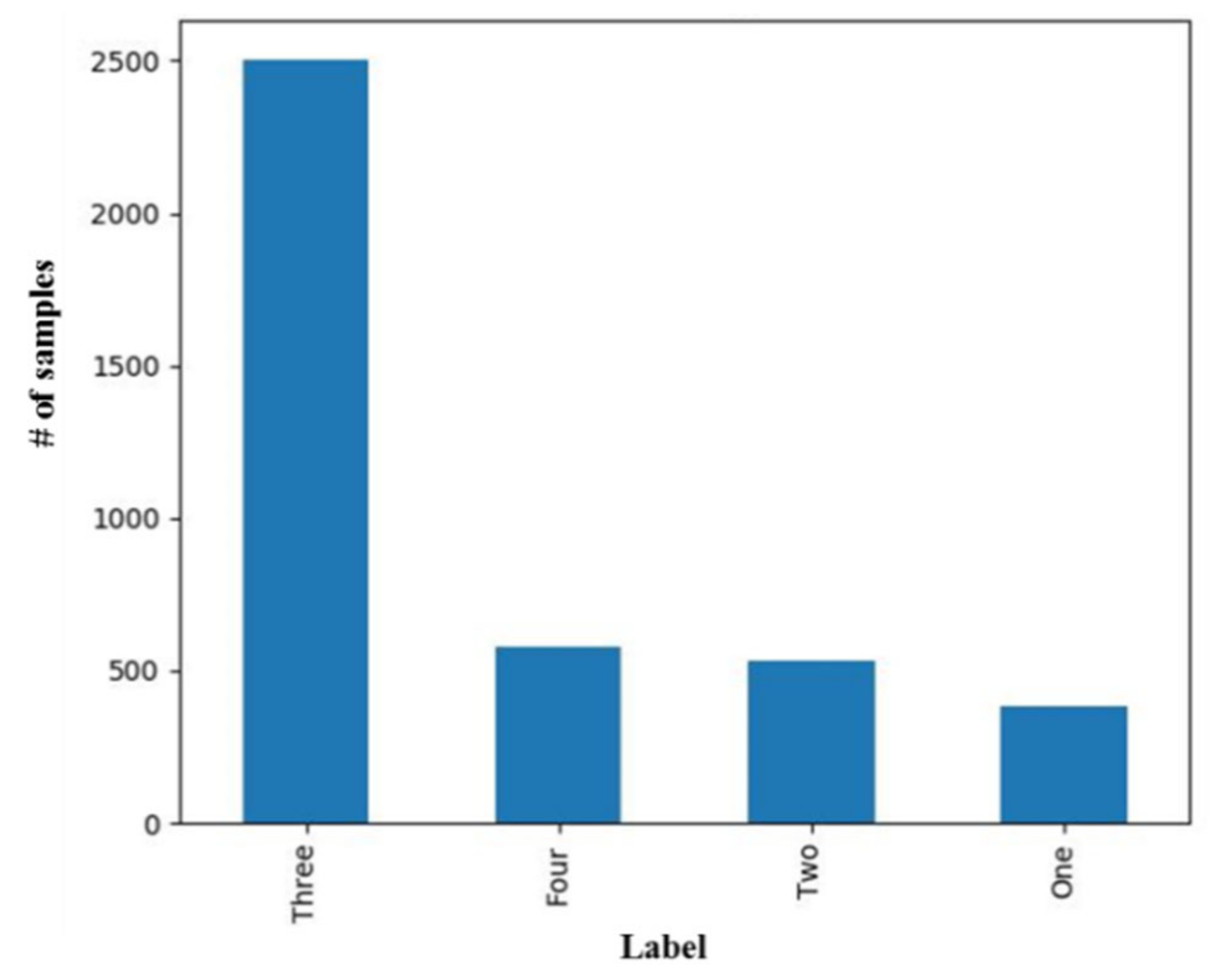
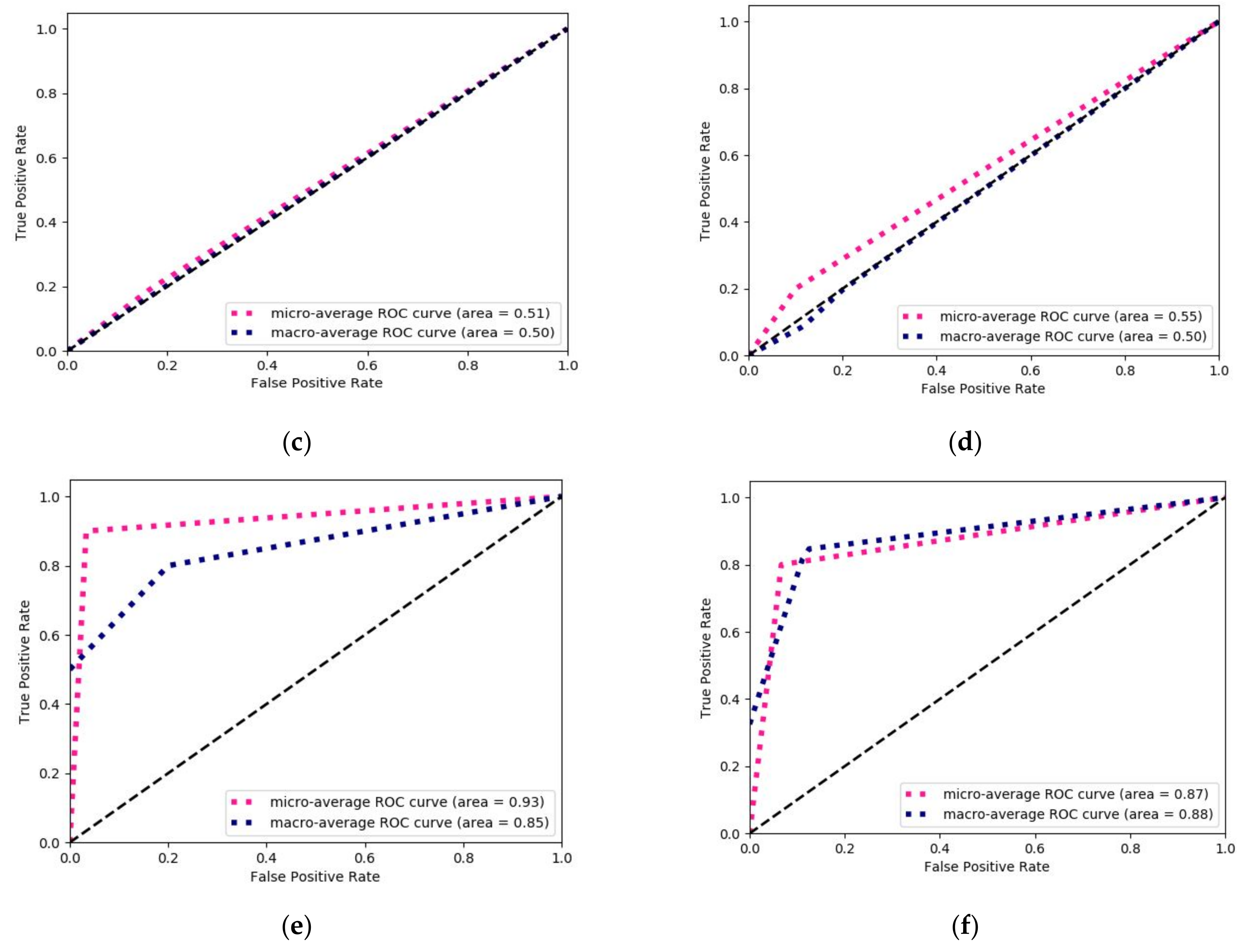
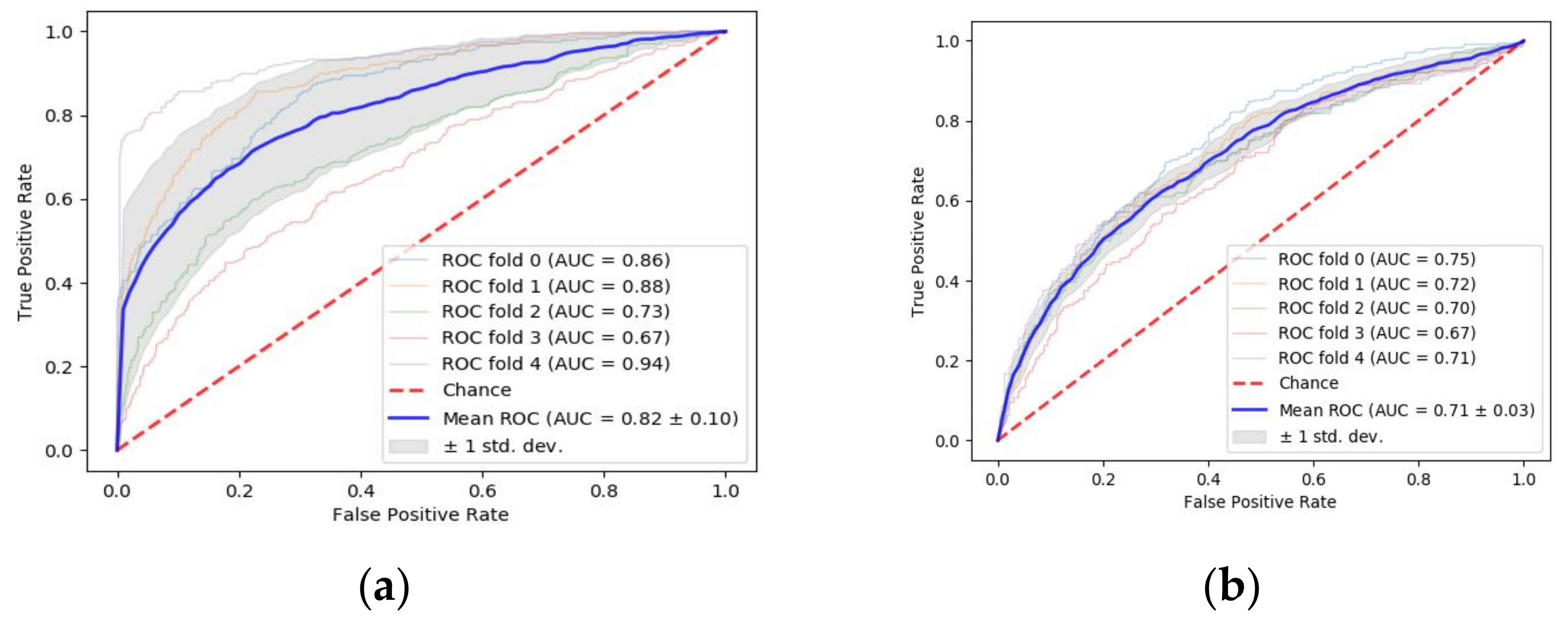
3.1. Prediction of Tumor Stage with Tumor Size as a Prognostic Factor
3.2. Prediction of Tumor Stage with TAS as a Prognostic Factor
3.3. Machine Learning-Based Prediction of DFS Period
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Cancer Research Fund [GB]. Available online: https://www.wcrf.org/dietandcancer/cancer-trends/colorectal-cancer-statistics (accessed on 5 August 2019).
- Health Promotion Administration. Available online: https://www.hpa.gov.tw/Pages/Detail.aspx?nodeid=1405&pid=10232 (accessed on 5 August 2019).
- National Cancer Institute. Surveillance, Epidemiology, and End Results (SEER) Program. Available online: https://seer.cancer.gov/statfacts/html/colorect.html (accessed on 5 August 2019).
- Ministry of Health and Welfare. Available online: https://www.mohw.gov.tw/cp-16-48057-1.html (accessed on 5 August 2019).
- American Cancer Society. Available online: https://www.cancer.org/latest-news/colon-cancer-cases-rising-among-young-adults.html (accessed on 9 July 2019).
- Doi, H.; Uemoto, K.; Suzuki, O.; Yamada, K.; Masai, N.; Tatsumi, D.; Shiomi, H.; Oh, R.J. Effect of primary tumor location and tumor size on the response to radiotherapy for liver metastases from colorectal cancer. Oncol. Lett. 2017, 14, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.Y.; Zhang, K.; Guo, K.B.; Liu, S.; Wasan, H.S.; Jin, H.M.; Yuan, L.; Feng, G.; Shen, F.F.; Shen, M.H.; et al. Value of tumor size as a prognostic factor in metastatic colorectal cancer patients after chemotherapy: A population-based study. Future Oncol. 2019, 15, 1745–1758. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Alonso, S.; Muto, Y.; Perucho, M.; Rikiyama, T. Tumor size is an independent risk predictor for metachronous colorectal cancer. Oncotarget 2016, 7, 17896–17904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossler, O.; Betge, J.; Harbaum, L.; Mrak, K.; Tschmelitsch, J.; Langner, C. Tumor size, tumor location, and antitumor inflammatory response are associated with lymph node size in colorectal cancer patients. Mod. Pathol. 2017, 30, 897–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garapati, S.S.; Hadjiiski, L.; Cha, K.H.; Chan, H.P.; Caoili, E.M.; Cohan, R.H.; Weizer, A.; Alva, A.; Paramagul, C.; Wei, J.; et al. Urinary bladder cancer staging in CT urography using machine learning. Med. Phys. 2017, 44, 5814–5823. [Google Scholar] [CrossRef] [PubMed]
- De Bari, B.; Vallati, M.; Gatta, R.; Lestrade, L.; Manfrida, S.; Carrie, C.; Valentini, V. Development and validation of a machine learning-based predictive model to improve the prediction of inguinal status of anal cancer patients: A preliminary report. Oncotarget 2017, 8, 108509–108521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmieri, L.J.; Fihri, A.; Doat, S.; Dubreuil, O.; Manceau, G.; Karoui, M.; Wagner, M.; Lucidarme, O.; Bachet, J.B. Tumor-size responses to first-line is a predictor of overall survival in metastatic colorectal cancer. Eur. Radiol. 2019, 29, 3871–3880. [Google Scholar] [CrossRef] [Green Version]
- Claret, L.; Pentafragka, C.; Karovic, S.; Zhao, B.; Schwartz, L.H.; Maitland, M.L.; Bruno, R. Comparison of tumor size assessments in tumor growth inhibition-overall survival models with second-line colorectal cancer data from the VELOUR study. Cancer Chemother. Pharmacol. 2018, 82, 49–54. [Google Scholar] [CrossRef]
- Vidyasagar, M. Machine learning methods in computational cancer biology. Annu. Rev. Control. 2017, 43, 107–127. [Google Scholar] [CrossRef]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef] [Green Version]
- Hornbrook, M.C.; Goshen, R.; Choman, E.; O’Keeffe-Rosetti, M.; Kinar, Y.; Liles, E.G.; Rust, K.C. Early Colorectal Cancer Detected by Machine Learning Model Using Gender, Age, and Complete Blood Count Data. Digestive Dis. Sci. 2017, 62, 2719–2727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, M.; Hon, S.; Gu, Y.S.; Bounsanga, J.; Hon, E.; Hansen, A.R.; Nielson, D.; Voss, M. Development of predictive models for personalized, precision medicine in colorectal cancer using machine learning. Qual. Life Res. 2017, 26, 65. [Google Scholar]
- Kinar, Y.; Akiva, P.; Choman, E.; Kariv, R.; Shalev, V.; Levin, B.; Narod, S.A.; Goshen, R. Performance analysis of a machine learning flagging system used to identify a group of individuals at a high risk for colorectal cancer. PLoS ONE 2017, 12, e0171759. [Google Scholar] [CrossRef] [PubMed]
- Jovic, S.; Miljkovic, M.; Ivanovic, M.; Saranovic, M.; Arsic, M. Prostate Cancer Probability Prediction By Machine Learning Technique. Cancer Investig. 2017, 35, 647–651. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.H.; Chang, P.M.H.; Hsu, C.W.; Huang, C.Y.F.; Ng, K.L. Drug repositioning for non-small cell lung cancer by using machine learning algorithms and topological graph theory. BMC Bioinform. 2016, 17, S2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papp, L.; Nakuz, T.; Magometschnigg, H.; Grahovac, M.; Helbich, T.; Karanikas, G.; Haug, A.; Pinker, K.; Beyer, T.; Hacker, M. In-vivo tumour characterization of breast cancer using [F-18]FDG-PET/CT and supervised machine-learning. Eur. J. Nucl. Med. Mol. Imaging 2017, 44, S414. [Google Scholar]
- Ferroni, P.; Zanzotto, F.M.; Riondino, S.; Scarpato, N.; Guadagni, F.; Roselli, M. Breast Cancer Prognosis Using a Machine Learning Approach. Cancers (Basel) 2019, 11, 328. [Google Scholar] [CrossRef] [Green Version]
- Rehman, O.; Zhuang, H.; Muhamed Ali, A.; Ibrahim, A.; Li, Z. Validation of miRNAs as Breast Cancer Biomarkers with a Machine Learning Approach. Cancers (Basel) 2019, 11, 431. [Google Scholar] [CrossRef]
- Lu, T.P.; Kuo, K.T.; Chen, C.H.; Chang, M.C.; Lin, H.P.; Hu, Y.H.; Chiang, Y.C.; Cheng, W.F.; Chen, C.A. Developing a Prognostic Gene Panel of Epithelial Ovarian Cancer Patients by a Machine Learning Model. Cancers (Basel) 2019, 11, 270. [Google Scholar] [CrossRef] [Green Version]
- Barlow, H.M.S.; Khushi, M. Predicting High-Risk Prostate Cancer Using Machine Learning Methods. Data 2019. [Google Scholar] [CrossRef] [Green Version]
- Hueman, M.; Wang, H.; Henson, D.; Chen, D. Expanding the TNM for cancers of the colon and rectum using machine learning: A demonstration. ESMO Open 2019, 4, e000518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Huang, S.; Yang, J.; Fong, S.; Zhao, Q. Mining Prognosis Index of Brain Metastases Using Artificial Intelligence. Cancers (Basel) 2019, 11, 1140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, B.; Feng, Y.; Mo, S.B.; Cai, S.J.; Huang, L.Y. Smaller tumor size is associated with poor survival in T4b colon cancer. World J. Gastroenterol. 2016, 22, 6726–6735. [Google Scholar] [CrossRef]




| Sources | Parameters Collected |
|---|---|
| Chart records | Age, gender, adjuvant therapy, status of follow-up, medical illness, pre-operation lab data |
| History taking | Smoking history, coffee consumption, alcohol consumption, physical activity |
| Intra-operative finding | Operation date, intent of resection, operation timing, operation finding, operation type, early morbidity, late morbidity, mortality |
| Histo-pathologyReports | Tumor location, gross appearance, circumferential involvement, tumor size, histologic type, histologic grade, tumor extension, examined lymph node number, total positive lymph node number, TNM staging |
| Parameters | Tumor Aggression Score | p- | |
|---|---|---|---|
| <9.8 (3709) | ≥9.8 (294) | ||
| BMI | 0.004 | ||
| <18.5 | 215 (5.8) | 35 (11.90) | |
| 18.5–23.9 | 1665 (44.89) | 151 (51.36) | |
| 24.0–26.9 | 1070 (28.85) | 66 (22.45) | |
| ≥27 | 759 (20.46) | 42 (14.29) | |
| Family History (FH) | <0.001 | ||
| No | 2145 (57.83) | 180 (61.23) | |
| Yes | 1429 (38.53) | 104 (35.37) | |
| Unknown | 135 (3.64) | 10 (3.4) | |
| Age | 0.007 | ||
| <50 | 527 (14.20) | 50 (17) | |
| ≥50 | 3182 (85.80) | 244 (83) | |
| Gender | <0.001 | ||
| Male | 2114 (57) | 165 (56.12) | |
| Female | 1595 (43) | 129 (43.88) | |
| Hypertension | <0.001 | ||
| Yes | 2447 (65.97) | 191 (64.96) | |
| No | 1262 (34.03) | 103 (35.04) | |
| Diabetes | <0.001 | ||
| Yes | 3136 (84.55) | 231 (78.57) | |
| No | 573 (15.45) | 63 (21.43) | |
| Smoking | 0.001 | ||
| Never | 2324 (62.66) | 174 (59.18) | |
| Ex-Smoker | 546 (14.72) | 42 (14.29) | |
| Current | 839 (22.62) | 78 (26.53) | |
| Alcohol | <0.001 | ||
| Never | 2622 (70.69) | 213 (72.45) | |
| Ex-Drinker | 218 (5.88) | 18 (6.12) | |
| Current | 869 (23.43) | 63 (21.43) | |
| CEA Level | <0.001 | ||
| <5 | 2424 (65.35) | 145 (49.32) | |
| ≥5 | 1285 (34.65) | 149 (50.68) | |
| Hemoglobin | 0.9 | ||
| Low (<11) | 853 (23) | 182 (61.90) | |
| Normal | 2856 (77) | 112 (38.10) | |
| LAB_ALB | <0.001 | ||
| ≤3.5 | 424 (11.43) | 128 (43.54) | |
| ˃3.5 | 3285 (88.57) | 166 (56.46) | |
| LAB_CR | <0.001 | ||
| ≤1.1 | 2954 (79.64) | 233 (79.25) | |
| ˃1.1 | 755 (20.36) | 61 (20.75) | |
| WBC | <0.001 | ||
| ≤5500 | 202 (5.5) | 14(4.8) | |
| ˃5500 | 3507 (94.5) | 280(95.2) | |
| OP Time | 0.001 | ||
| Elective | 3635 (98) | 284 (96.6) | |
| Emergency | 74 (2) | 10 (3.4) | |
| OP Find | <0.001 | ||
| None | 3199 (86.25) | 205 (69.73) | |
| Combined | 470 (12.67) | 84 (28.57) | |
| Any one | 40 (1.08) | 5 (1.7) | |
| CirInvo | <0.001 | ||
| No | 1972 (53.17) | 26 (8.84) | |
| Yes | 1737 (46.83) | 268 (91.16) | |
| Tumor Differentiation | <0.001 | ||
| Grade I | 477 (12.86) | 7 (2.38) | |
| Grade II | 3001 (80.91) | 183 (62.24) | |
| Grade III | 231 (6.22) | 104 (35.37) | |
| Tumor Width | <0.001 | ||
| ≤4.4 | 2582 (69.61) | 8 (2.73) | |
| ˃4.4 | 1127 (30.39) | 286 (97.27) | |
| Tumor Length | <0.001 | ||
| ≤4.4 | 2679 (72.22) | 10 (3.4) | |
| ˃4.4 | 1030 (27.78) | 284 (96.6) | |
| T stage | <0.001 | ||
| T1 | 377 (10.16) | 5 (1.70) | |
| T2 | 531 (14.32) | 4 (1.36) | |
| T3 | 2322 (62.61) | 184 (62.59) | |
| T4 | 479 (12.91) | 101 (34.35) | |
| N stage | <0.001 | ||
| N0 | 2062 (55.6) | 179 (60.89) | |
| N1 | 1010 (27.23) | 57 (19.39) | |
| N2 | 522 (14.07) | 46 (1.24) | |
| N3 | 115 (3.10) | 12 (4.08) | |
| Algorithms | Evaluation Metrics (Average(± sd)) | |||
|---|---|---|---|---|
| Accuracy | Precision | Recall | F-Measure | |
| Random Forest | 0.73 (± 0.01) | 0.70 (± 0.03) | 0.74 (± 0.01) | 0.67 (± 0.01) |
| Support Vector Machines | 0.63 (± 0.00) | 0.39 (± 0.00) | 0.63 (± 0.00) | 0.48 (± 0.00) |
| Logistic Regression | 0.63 (± 0.00) | 0.39 (± 0.00) | 0.63 (± 0.00) | 0.48 (± 0.00) |
| Multilayer Perceptron | 0.63 (± 0.00) | 0.44 (± 0.12) | 0.63 (± 0.02) | 0.48 (± 0.00) |
| K-Nearest Neighbor | 0.64 (± 0.01) | 0.57 (± 0.01) | 0.64 (± 0.01) | 0.53 (± 0.02) |
| Adaptive Boosting | 0.73 (± 0.01) | 0.72 (± 0.08) | 0.73 (± 0.01) | 0.66 (± 0.01) |
| Algorithms | Evaluation Metrics | |||
|---|---|---|---|---|
| Accuracy | Precision | Recall | F-Measure | |
| Random Forest | 0.74 | 0.77 | 0.74 | 0.67 |
| Support Vector Machines | 0.64 | 0.47 | 0.64 | 0.51 |
| Logistic Regression | 0.65 | 0.48 | 0.65 | 0.54 |
| Multilayer Perceptron | 0.67 | 0.55 | 0.67 | 0.58 |
| K-Nearest Neighbor | 0.63 | 0.50 | 0.63 | 0.51 |
| Adaptive Boosting | 0.67 | 0.54 | 0.67 | 0.57 |
| Algorithms | Evaluation Metrics (Average (± sd)) | |||
|---|---|---|---|---|
| Accuracy | Precision | Recall | F-Measure | |
| Random Forest | 0.90 (± 0.01) | 0.90 (± 0.02) | 0.90 (± 0.02) | 0.90 (± 0.02) |
| Support Vector Machines | 0.73 (± 0.02) | 0.58 (± 0.08) | 0.73 (± 0.02) | 0.63 (± 0.02) |
| Logistic Regression | 0.63 (± 0.00) | 0.41 (± 0.00) | 0.63 (± 0.00) | 0.49 (± 0.00) |
| Multilayer Perceptron | 0.63 (± 0.02) | 0.41 (± 0.07) | 0.63 (± 0.02) | 0.50 (± 0.03) |
| K-Nearest Neighbor | 0.86 (± 0.01) | 0.88 (± 0.01) | 0.86 (± 0.01) | 0.85 (± 0.01) |
| Adaptive Boosting | 0.89 (± 0.01) | 0.89 (± 0.01) | 0.89 (± 0.01) | 0.89 (± 0.01) |
| Algorithms | Evaluation Metrics | |||
|---|---|---|---|---|
| Accuracy | Precision | Recall | F-Measure | |
| Random Forest | 0.89 | 0.89 | 0.88 | 0.89 |
| Support Vector Machines | 0.73 | 0.65 | 0.73 | 0.64 |
| Logistic Regression | 0.62 | 0.38 | 0.62 | 0.48 |
| Multilayer Perceptron | 0.62 | 0.52 | 0.64 | 0.48 |
| K-Nearest Neighbor | 0.85 | 0.87 | 0.85 | 0.84 |
| Adaptive Boosting | 0.81 | 0.81 | 0.81 | 0.78 |
| Algorithms | Evaluation Metrics (Average (± sd)) | |||
|---|---|---|---|---|
| Accuracy | Precision | Recall | F-Measure | |
| Random Forest | 0.84 (± 0.12) | 0.82 (± 0.14) | 0.83 (± 0.12) | 0.81 (± 0.14) |
| Support Vector Machines | 0.77 (± 0.03) | 0.74 (± 0.07) | 0.77 (± 0.03) | 0.71 (± 0.05) |
| Logistic Regression | 0.76 (± 0.02) | 0.73 (± 0.04) | 0.76 (± 0.02) | 0.71 (± 0.02) |
| Multilayer Perceptron | 0.78 (± 0.11) | 0.77 (± 0.10) | 0.77 (± 0.11) | 0.77 (± 0.12) |
| K-Nearest Neighbor | 0.75 (± 0.06) | 0.72 (± 0.08) | 0.75 (± 0.06) | 0.71 (± 0.02) |
| Adaptive Boosting | 0.77 (± 0.03) | 0.75 (± 0.04) | 0.77 (± 0.03) | 0.74 (± 0.03) |
| Algorithms | Evaluation Metrics | |||
|---|---|---|---|---|
| Accuracy | Precision | Recall | F-Measure | |
| Random Forest | 0.76 | 0.74 | 0.76 | 0.71 |
| Support Vector Machines | 0.74 | 0.71 | 0.74 | 0.64 |
| Logistic Regression | 0.73 | 0.70 | 0.73 | 0.71 |
| Multilayer Perceptron | 0.64 | 0.66 | 0.64 | 0.65 |
| K-Nearest Neighbor | 0.73 | 0.70 | 0.73 | 0.70 |
| Adaptive Boosting | 0.66 | 0.70 | 0.66 | 0.67 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, P.; Chiang, S.-F.; Sahoo, P.K.; Mohapatra, S.K.; You, J.-F.; Onthoni, D.D.; Hung, H.-Y.; Chiang, J.-M.; Huang, Y.; Tsai, W.-S. Prediction of Colon Cancer Stages and Survival Period with Machine Learning Approach. Cancers 2019, 11, 2007. https://doi.org/10.3390/cancers11122007
Gupta P, Chiang S-F, Sahoo PK, Mohapatra SK, You J-F, Onthoni DD, Hung H-Y, Chiang J-M, Huang Y, Tsai W-S. Prediction of Colon Cancer Stages and Survival Period with Machine Learning Approach. Cancers. 2019; 11(12):2007. https://doi.org/10.3390/cancers11122007
Chicago/Turabian StyleGupta, Pushpanjali, Sum-Fu Chiang, Prasan Kumar Sahoo, Suvendu Kumar Mohapatra, Jeng-Fu You, Djeane Debora Onthoni, Hsin-Yuan Hung, Jy-Ming Chiang, Yenlin Huang, and Wen-Sy Tsai. 2019. "Prediction of Colon Cancer Stages and Survival Period with Machine Learning Approach" Cancers 11, no. 12: 2007. https://doi.org/10.3390/cancers11122007
APA StyleGupta, P., Chiang, S.-F., Sahoo, P. K., Mohapatra, S. K., You, J.-F., Onthoni, D. D., Hung, H.-Y., Chiang, J.-M., Huang, Y., & Tsai, W.-S. (2019). Prediction of Colon Cancer Stages and Survival Period with Machine Learning Approach. Cancers, 11(12), 2007. https://doi.org/10.3390/cancers11122007







