Circulating Cell-Free DNA and RNA Analysis as Liquid Biopsy: Optimal Centrifugation Protocol
Abstract
:1. Introduction
2. Results
2.1. Total cfDNA Concentration and Characteristics
2.1.1. cfDNA Yield
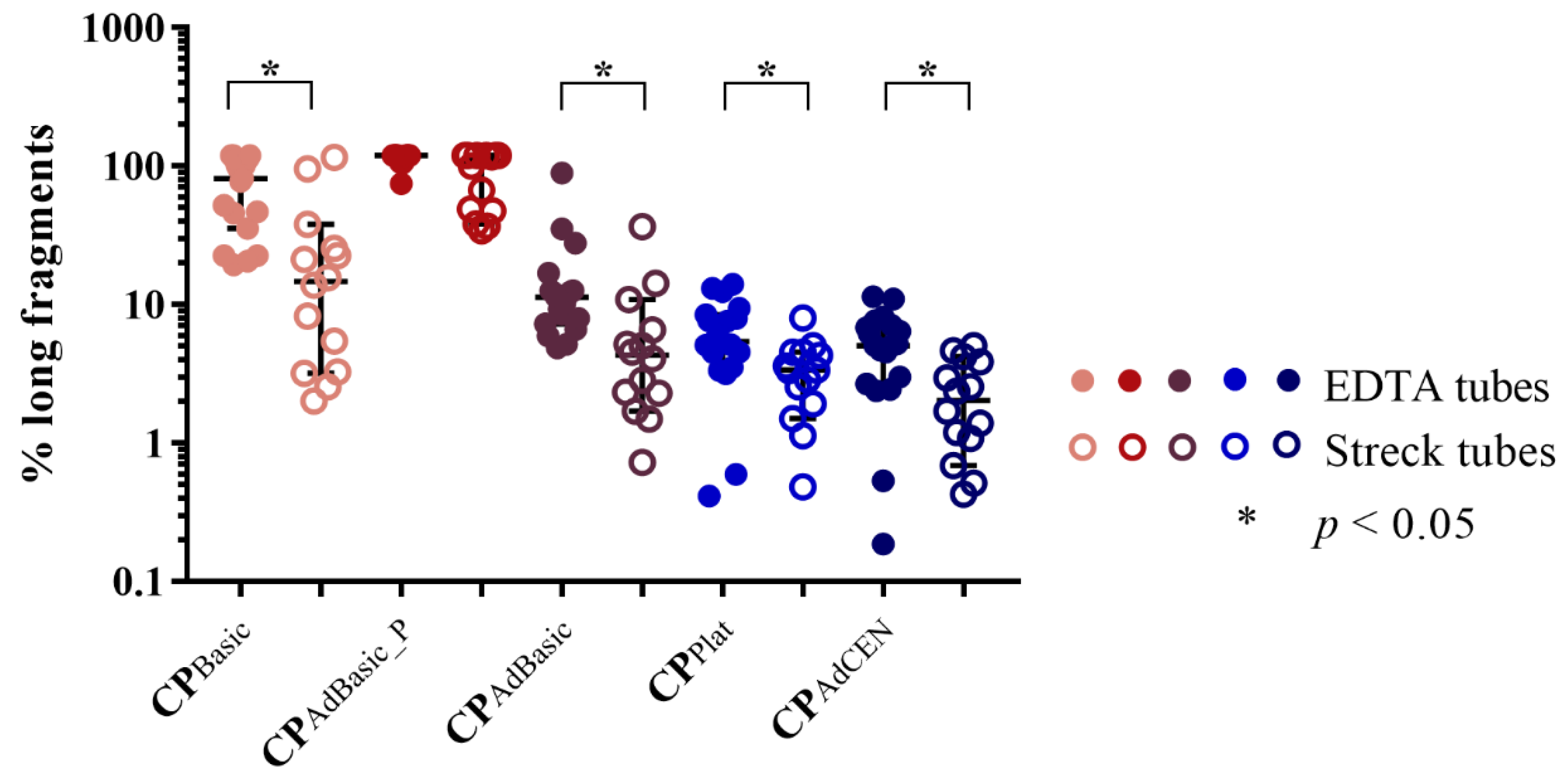
2.1.2. cfDNA Integrity
2.2. KRAS-Mutated ctDNA
2.3. Total cfRNA Concentration
2.4. KRAS-Mutated ctRNA
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. cfDNA Isolation and Analysis
4.3. cfRNA Isolation and Analysis
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Norton, S.E.; Lechner, J.M.; Williams, T.; Fernando, M.R. A stabilizing reagent prevents cell-free DNA contamination by cellular DNA in plasma during blood sample storage and shipping as determined by digital pcr. Clin. Biochem. 2013, 46, 1561–1565. [Google Scholar] [CrossRef]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Warton, K.; Yuwono, N.L.; Cowley, M.J.; McCabe, M.J.; So, A.; Ford, C.E. Evaluation of streck bct and paxgene stabilised blood collection tubes for cell-free circulating DNA studies in plasma. Mol. Diagn Ther. 2017, 21, 563–570. [Google Scholar] [CrossRef]
- Russo, M.; Misale, S.; Wei, G.; Siravegna, G.; Crisafulli, G.; Lazzari, L.; Corti, G.; Rospo, G.; Novara, L.; Mussolin, B.; et al. Acquired resistance to the trk inhibitor entrectinib in colorectal cancer. Cancer Discov. 2016, 6, 36–44. [Google Scholar] [PubMed]
- Nilsson, R.J.; Karachaliou, N.; Berenguer, J.; Gimenez-Capitan, A.; Schellen, P.; Teixido, C.; Tannous, J.; Kuiper, J.L.; Drees, E.; Grabowska, M.; et al. Rearranged eml4-alk fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget 2016, 7, 1066–1075. [Google Scholar] [PubMed]
- Best, M.G.; Wesseling, P.; Wurdinger, T. Tumor-educated platelets as a noninvasive biomarker source for cancer detection and progression monitoring. Cancer Res. 2018, 78, 3407–3412. [Google Scholar] [CrossRef] [PubMed]
- Normanno, N.; Cervantes, A.; Ciardiello, F.; De Luca, A.; Pinto, C. The liquid biopsy in the management of colorectal cancer patients: Current applications and future scenarios. Cancer Treat. Rev. 2018, 70, 1–8. [Google Scholar] [PubMed]
- Stewart, C.M.; Kothari, P.D.; Mouliere, F.; Mair, R.; Somnay, S.; Benayed, R.; Zehir, A.; Weigelt, B.; Dawson, S.J.; Arcila, M.E.; et al. The value of cell-free DNA for molecular pathology. J. Pathol. 2018, 244, 616–627. [Google Scholar] [Green Version]
- Van Dessel, L.F.; Beije, N.; Helmijr, J.C.; Vitale, S.R.; Kraan, J.; Look, M.P.; de Wit, R.; Sleijfer, S.; Jansen, M.P.; Martens, J.W.; et al. Application of circulating tumor DNA in prospective clinical oncology trials—Standardization of preanalytical conditions. Mol. Oncol. 2017, 11, 295–304. [Google Scholar] [CrossRef]
- Sherwood, J.L.; Corcoran, C.; Brown, H.; Sharpe, A.D.; Musilova, M.; Kohlmann, A. Optimised pre-analytical methods improve kras mutation detection in circulating tumour DNA (ctdna) from patients with non-small cell lung cancer (nsclc). PLoS ONE 2016, 11, e0150197. [Google Scholar] [CrossRef] [PubMed]
- Van Ginkel, J.H.; van den Broek, D.A.; van Kuik, J.; Linders, D.; de Weger, R.; Willems, S.M.; Huibers, M.M.H. Preanalytical blood sample workup for cell-free DNA analysis using droplet digital pcr for future molecular cancer diagnostics. Cancer Med. 2017, 6, 2297–2307. [Google Scholar]
- Fernando, M.R.; Norton, S.E.; Luna, K.K.; Lechner, J.M.; Qin, J. Stabilization of cell-free rna in blood samples using a new collection device. Clin. Biochem. 2012, 45, 1497–1502. [Google Scholar] [CrossRef] [PubMed]
- Chiu, R.W.; Poon, L.L.; Lau, T.K.; Leung, T.N.; Wong, E.M.; Lo, Y.M. Effects of blood-processing protocols on fetal and total DNA quantification in maternal plasma. Clin. Chem. 2001, 47, 1607–1613. [Google Scholar]
- El Messaoudi, S.; Rolet, F.; Mouliere, F.; Thierry, A.R. Circulating cell free DNA: Preanalytical considerations. Clin. Chim. Acta 2013, 424, 222–230. [Google Scholar] [CrossRef]
- Risberg, B.; Tsui, D.W.Y.; Biggs, H.; Ruiz-Valdepenas Martin de Almagro, A.; Dawson, S.J.; Hodgkin, C.; Jones, L.; Parkinson, C.; Piskorz, A.; Marass, F.; et al. Effects of collection and processing procedures on plasma circulating cell-free DNA from cancer patients. J. Mol. Diagn. 2018, 20, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Demuth, C.; Spindler, K.G.; Johansen, J.S.; Pallisgaard, N.; Nielsen, D.; Hogdall, E.; Vittrup, B.; Sorensen, B.S. Measuring kras mutations in circulating tumor DNA by droplet digital pcr and next-generation sequencing. Transl. Oncol. 2018, 11, 1220–1224. [Google Scholar] [CrossRef]
- Volckmar, A.L.; Sultmann, H.; Riediger, A.; Fioretos, T.; Schirmacher, P.; Endris, V.; Stenzinger, A.; Dietz, S. A field guide for cancer diagnostics using cell-free DNA: From principles to practice and clinical applications. Genes Chromosomes Cancer 2018, 57, 123–139. [Google Scholar]
- Sorber, L.; Zwaenepoel, K.; De Winne, K.; Van Casteren, K.; Augustus, E.; Jacobs, J.; Zhang, X.H.; Galdermans, D.; De Droogh, E.; Lefebure, A.; et al. A multicenter study to assess egfr mutational status in plasma: Focus on an optimized workflow for liquid biopsy in a clinical setting. Cancers 2018, 10, 290. [Google Scholar]
- Streck. Cell-Free DNA bct: Instructions for Use; Streck: Omaha, NE, USA, 2014. [Google Scholar]
- Devonshire, A.S.; Whale, A.S.; Gutteridge, A.; Jones, G.; Cowen, S.; Foy, C.A.; Huggett, J.F. Towards standardisation of cell-free DNA measurement in plasma: Controls for extraction efficiency, fragment size bias and quantification. Anal. Bioanal. Chem. 2014, 406, 6499–6512. [Google Scholar] [PubMed]
- Cheng, H.H.; Yi, H.S.; Kim, Y.; Kroh, E.M.; Chien, J.W.; Eaton, K.D.; Goodman, M.T.; Tait, J.F.; Tewari, M.; Pritchard, C.C. Plasma processing conditions substantially influence circulating microrna biomarker levels. PLoS ONE 2013, 8, e64795. [Google Scholar]
- Mitchell, A.J.; Gray, W.D.; Hayek, S.S.; Ko, Y.A.; Thomas, S.; Rooney, K.; Awad, M.; Roback, J.D.; Quyyumi, A.; Searles, C.D. Platelets confound the measurement of extracellular mirna in archived plasma. Sci. Rep. 2016, 6, 32651. [Google Scholar] [CrossRef] [PubMed]
- Cerkovnik, P.; Perhavec, A.; Zgajnar, J.; Novakovic, S. Optimization of an rna isolation procedure from plasma samples. Int. J. Mol. Med. 2007, 20, 293–300. [Google Scholar] [CrossRef]
- Decock, A.; De Wever, O.; Dhondt, B.; D’huyvetter, T.; Helsmoortel, H.; Hendrix, A.; Philippron, A.; Verniers, K.; Mestdagh, P.; Vandesompele, J. The extracellular rna quality control (exrnaqc) study: Testing and controlling pre-analytical variables of rna sequencing on liquid biopsies. In LKI Symposium, Leuven, Belgium—Abstract; http://hdl.handle.net/1854/LU-8552649; Ghent University Library: Ghent, Belgium, 2018. [Google Scholar]
- Wrzyszcz, A.; Urbaniak, J.; Sapa, A.; Wozniak, M. An efficient method for isolation of representative and contamination-free population of blood platelets for proteomic studies. Platelets 2017, 28, 43–53. [Google Scholar] [PubMed]
- Doss, J.F.; Corcoran, D.L.; Jima, D.D.; Telen, M.J.; Dave, S.S.; Chi, J.T. A comprehensive joint analysis of the long and short rna transcriptomes of human erythrocytes. BMC Genom. 2015, 16, 952. [Google Scholar] [CrossRef]
- Rikkert, L.G.; van der Pol, E.; van Leeuwen, T.G.; Nieuwland, R.; Coumans, F.A.W. Centrifugation affects the purity of liquid biopsy-based tumor biomarkers. Cytom. A 2018, 93, 1207–1212. [Google Scholar]
- Lin, J.J.; Riely, G.J.; Shaw, A.T. Targeting alk: Precision medicine takes on drug resistance. Cancer Discov. 2017, 7, 137–155. [Google Scholar]
- Gainor, J.F.; Dardaei, L.; Yoda, S.; Friboulet, L.; Leshchiner, I.; Katayama, R.; Dagogo-Jack, I.; Gadgeel, S.; Schultz, K.; Singh, M.; et al. Molecular mechanisms of resistance to first- and second-generation alk inhibitors in alk-rearranged lung cancer. Cancer Discov. 2016, 6, 1118–1133. [Google Scholar] [CrossRef]
- Best, M.G.; Sol, N.; Kooi, I.; Tannous, J.; Westerman, B.A.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Koster, J.; et al. Rna-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell 2015, 28, 666–676. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Williams, T.L.; Fernando, M.R. A novel blood collection device stabilizes cell-free rna in blood during sample shipping and storage. BMC Res. Notes 2013, 6, 380. [Google Scholar] [CrossRef] [PubMed]
- George, N. Evaluation of available blood collection tubes for use in stabilizing concentrations of extracellular vesicles/exosomes and associated cell-free rna. In Proceedings of the American Association for Cancer Research Annual Meeting 2018, Chicago, IL, USA, 14–18 April 2018. [Google Scholar]
- Goethals, S.; De Wilde, A.; Lesage, K.; Smits, E.; Pauwels, P.; Peeters, M. Tumorbank@uza: A collection of tissue, fluid samples and associated data of oncology patients for the use in translational research. Open J. Bioresour. 2018, 5, 4. [Google Scholar] [CrossRef]
- Sorber, L.; Zwaenepoel, K.; Deschoolmeester, V.; Roeyen, G.; Lardon, F.; Rolfo, C.; Pauwels, P. A comparison of cell-free DNA isolation kits: Isolation and quantification of cell-free DNA in plasma. J. Mol. Diagn. 2017, 19, 162–168. [Google Scholar] [PubMed]
- Rago, C.; Huso, D.L.; Diehl, F.; Karim, B.; Liu, G.; Papadopoulos, N.; Samuels, Y.; Velculescu, V.E.; Vogelstein, B.; Kinzler, K.W.; et al. Serial assessment of human tumor burdens in mice by the analysis of circulating DNA. Cancer Res. 2007, 67, 9364–9370. [Google Scholar] [CrossRef] [PubMed]
- Diaz, I.M.; Nocon, A.; Mehnert, D.H.; Fredebohm, J.; Diehl, F.; Holtrup, F. Performance of streck cfdna blood collection tubes for liquid biopsy testing. PLoS ONE 2016, 11, e0166354. [Google Scholar]




| ID | Protocol | Specifications | Details | Temperature | Matrix |
|---|---|---|---|---|---|
| CPBasic | Basic (Biobank UZA/UAntwerpen) | 1. 10’ – 400 g | RT | Plasma | |
| CPAdBasic_P | Basic (adapted) | 1. 10’ – 400 g 2. 1’ – max speed | 2nd centrifugation step after storage at −80 °C | RT | Pellet |
| CPAdBasic | Plasma | ||||
| CPPlat | Platelet | 1. 20’ – 120 g 2. 20’ – 360 g 3. 5’ – 360 g | Platelets are washed with PBS in 3rd centrifugation step | RT | Plasma |
| CPPlat_P | Platelets | ||||
| CPStreck | Streck | 1. 10’ – 1600 g 2. 10’ – 6000 g | RT | Plasma | |
| CPCEN | CEN | 1. 10’ – 1900 g 2. 10’ – 16,000 g | Blood samples on ice immediately after blood collection | 4 °C | Plasma |
| CPAdCEN | CEN (adapted) | 1. 10’ – 1900 g 2. 10’ – 16,000 g | RT | Plasma |
| Phase | Participants | EDTA (N = 19) | Streck (N = 14) | ||
|---|---|---|---|---|---|
| cfDNA Samples | cfRNA Samples | cfDNA Samples | cfRNA Samples | ||
| 1 | Healthy volunteers | n° = 48 | n° = 55 | n° = 12 | n° = 7 |
| |||||
| 2 | Healthy volunteers—spiked with KRAS-mutated DNA | n° = 24 | n° = 28 | n° = 24 | n° = 28 |
| |||||
| 3 | KRAS-mutated metastatic cancer patients | n° = 42 | n° = 47 | n° = 48 | n° = 53 |
| |||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sorber, L.; Zwaenepoel, K.; Jacobs, J.; De Winne, K.; Goethals, S.; Reclusa, P.; Van Casteren, K.; Augustus, E.; Lardon, F.; Roeyen, G.; et al. Circulating Cell-Free DNA and RNA Analysis as Liquid Biopsy: Optimal Centrifugation Protocol. Cancers 2019, 11, 458. https://doi.org/10.3390/cancers11040458
Sorber L, Zwaenepoel K, Jacobs J, De Winne K, Goethals S, Reclusa P, Van Casteren K, Augustus E, Lardon F, Roeyen G, et al. Circulating Cell-Free DNA and RNA Analysis as Liquid Biopsy: Optimal Centrifugation Protocol. Cancers. 2019; 11(4):458. https://doi.org/10.3390/cancers11040458
Chicago/Turabian StyleSorber, Laure, Karen Zwaenepoel, Julie Jacobs, Koen De Winne, Sofie Goethals, Pablo Reclusa, Kaat Van Casteren, Elien Augustus, Filip Lardon, Geert Roeyen, and et al. 2019. "Circulating Cell-Free DNA and RNA Analysis as Liquid Biopsy: Optimal Centrifugation Protocol" Cancers 11, no. 4: 458. https://doi.org/10.3390/cancers11040458






