Prognostic and Predictive Value of PBRM1 in Clear Cell Renal Cell Carcinoma
Abstract
:1. Introduction
2. Bromodomain-Containing Protein BAF180 Function
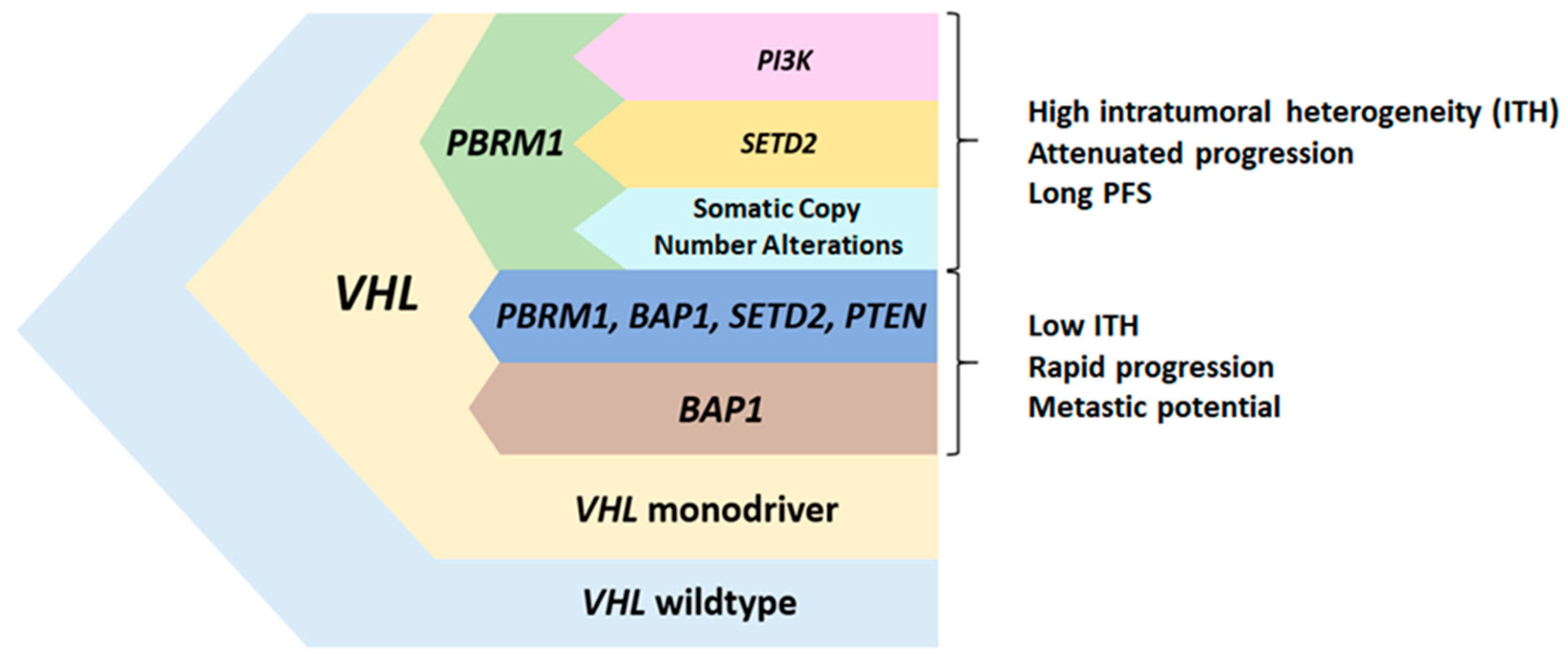
3. PBRM1 Mutations in Clear Cell Renal Cell Carcinoma
4. Prognostic Value of PBRM1 Mutations in Localized and Advanced Disease
4.1. Localized Disease
4.2. Advanced Disease
5. Predictive Value of PBRM1 Mutations
5.1. Targeted Therapy
5.2. Immunotherapy
6. Future Perspectives and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [Green Version]
- Moch, H.; Cubilla, A.L.; Humphrey, P.A.; Reuter, V.E.; Ulbright, T.M. The 2016 WHO classification of tumors of the urinary system and male genital organs-part A: Renal, penile, and testicular tumors. Eur. Urol. 2016, 70, 93–105. [Google Scholar] [CrossRef]
- TNM. Classification of Malignant Tumors, 8th Edition | Wiley [Internet]. Wiley.com. Available online: https://www.wiley.com/en-es/TNM+Classification+of+Malignant+Tumors%2C+8th+Edition-p-9781119263579 (accessed on 1 November 2019).
- Janzen, N.K.; Kim, H.L.; Figlin, R.A.; Belldegrun, A.S. Surveillance after radical or partial nephrectomy for localized renal cell carcinoma and management of recurrent disease. Urol. Clin. N. Am. 2003, 30, 843–852. [Google Scholar] [CrossRef]
- Meskawi, M.; Sun, M.; Trinh, Q.-D.; Bianchi, M.; Hansen, J.; Tian, Z.; Rink, M.; Ismail, S.; Shariat, S.F.; Montorsi, F.; et al. A review of integrated staging systems for renal cell carcinoma. Eur. Urol. 2012, 62, 303–314. [Google Scholar] [CrossRef]
- Patard, J.-J.; Kim, H.L.; Lam, J.S.; Dorey, F.J.; Pantuck, A.J.; Zisman, A.; Ficarra, V.; Han, K.R.; Cindolo, L.; De La Taille, A.; et al. Use of the University of California Los Angeles integrated staging system to predict survival in renal cell carcinoma: An international multicenter study. J. Clin. Oncol. 2004, 22, 3316–3322. [Google Scholar] [CrossRef] [PubMed]
- Leibovich, B.C.; Blute, M.L.; Cheville, J.C.; Lohse, C.M.; Frank, I.; Kwon, E.D.; Weaver, A.L.; Parker, A.S.; Zincke, H. Prediction of progression after radical nephrectomy for patients with clear cell renal cell carcinoma: A stratification tool for prospective clinical trials. Cancer 2003, 97, 1663–1671. [Google Scholar] [CrossRef] [PubMed]
- Gnarra, J.R.; Tory, K.; Weng, Y.; Schmidt, L.; Wei, M.H.; Li, H.; Latif, F.; Liu, S.; Chen, F.; Duh, F.M.; et al. Mutations of the VHL tumor suppressor gene in renal carcinoma. Nat. Genet. 1994, 7, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Brugarolas, J. Molecular genetics of clear-cell renal cell carcinoma. J. Clin. Oncol. 2014, 32, 1968–1976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerlinger, M.; Horswell, S.; Larkin, J.; Rowan, A.J.; Salm, M.P.; Varela, I.; Fisher, R.; McGranahan, N.; Matthews, N.; Santos, C.R.; et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 2014, 46, 225–233. [Google Scholar] [CrossRef]
- Linehan, W.M.; Ricketts, C.J. The Cancer Genome Atlas of renal cell carcinoma: Findings and clinical implications. Nat. Rev. Urol. 2019, 16, 539–552. [Google Scholar] [CrossRef]
- Ljungberg, B.; Albiges, L.; Abu-Ghanem, Y.; Bensalah, K.; Dabestani, S.; Fernández-Pello, S.; Rachel, H.G.; Fabian, H.; Milan, H.; Markus, A.K.; et al. European association of urology guidelines on renal cell carcinoma: The 2019 update. Eur. Urol. 2019, 75, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Choueiri, T.K.; Motzer, R.J. Systemic therapy for metastatic renal-cell carcinoma. N. Engl. J. Med. 2017, 376, 354–366. [Google Scholar] [CrossRef] [PubMed]
- de Velasco, G.; Bex, A.; Albiges, L.; Powles, T.; Rini, B.I.; Motzer, R.J.; Heng, D.Y.C.; Escudier, B. Sequencing and combination of systemic therapy in metastatic renal cell carcinoma. Eur. Urol. Oncol. 2019, 2, 505–514. [Google Scholar] [CrossRef]
- Heng, D.Y.C.; Xie, W.; Regan, M.M.; Warren, M.A.; Golshayan, A.R.; Sahi, C.; Eigl, B.J.; Ruether, J.D.; Cheng, T.; North, S.; et al. Prognostic factors for overall survival in patients with metastatic renal cell carcinoma treated with vascular endothelial growth factor-targeted agents: Results from a large, multicenter study. J. Clin. Oncol. 2009, 27, 5794–5799. [Google Scholar] [CrossRef]
- Graham, J.; Dudani, S.; Heng, D.Y.C. Prognostication in kidney cancer: Recent advances and future directions. J. Clin. Oncol. 2018, 36, 3567–3573. [Google Scholar] [CrossRef]
- Hargreaves, D.C.; Crabtree, G.R. ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res. 2011, 21, 396–420. [Google Scholar] [CrossRef]
- Kadoch, C.; Hargreaves, D.C.; Hodges, C.; Elias, L.; Ho, L.; Ranish, J.; Crabtree, G.R. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 2013, 45, 592–601. [Google Scholar] [CrossRef]
- Masliah-Planchon, J.; Bièche, I.; Guinebretière, J.-M.; Bourdeaut, F.; Delattre, O. SWI/SNF chromatin remodeling and human malignancies. Annu. Rev. Pathol. Mech. Dis. 2015, 10, 145–171. [Google Scholar] [CrossRef]
- Varela, I.; Tarpey, P.; Raine, K.; Huang, D.; Ong, C.K.; Stephens, P.; Davies, H.; Jones, D.; Lin, M.L.; Teague, J.; et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 2011, 469, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, B.; Porter, E.G.; Stewart, J.C.; Ferreira, C.R.; Schipma, M.J.; Dykhuizen, E.C. PBRM1 regulates the expression of genes involved in metabolism and cell adhesion in renal clear cell carcinoma. PLoS ONE 2016, 11, e0153718. [Google Scholar] [CrossRef] [Green Version]
- Kakarougkas, A.; Ismail, A.; Chambers, A.L.; Riballo, E.; Herbert, A.D.; Künzel, J.; Löbrich, M.; Jeggo, P.A.; Downs, J.A. Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol. Cell 2014, 55, 723–732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brownlee, P.M.; Chambers, A.L.; Cloney, R.; Bianchi, A.; Downs, J.A. BAF180 promotes cohesion and prevents genome instability and aneuploidy. Cell Rep. 2014, 6, 973–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macher-Goeppinger, S.; Keith, M.; Tagscherer, K.E.; Singer, S.; Winkler, J.; Hofmann, T.G.; Pahernik, S.; Duensing, S.; Hohenfellner, M.; Kopitz, J.; et al. PBRM1 (BAF180) protein is functionally regulated by p53-induced protein degradation in renal cell carcinomas. J Pathol. 2015, 237, 460–471. [Google Scholar] [CrossRef]
- Burrows, A.E.; Smogorzewska, A.; Elledge, S.J. Polybromo-associated BRG1-associated factor components BRD7 and BAF180 are critical regulators of p53 required for induction of replicative senescence. Proc. Natl. Acad. Sci. USA 2010, 107, 14280–14285. [Google Scholar] [CrossRef] [Green Version]
- Murakami, A.; Wang, L.; Kalhorn, S.; Schraml, P.; Rathmell, W.K.; Tan, A.C.; Nemenoff, R.; Stenmark, K.; Jiang, B.H.; Reyland, M.E.; et al. Context-dependent role for chromatin remodeling component PBRM1/BAF180 in clear cell renal cell carcinoma. Oncogenesis 2017, 6, e287. [Google Scholar] [CrossRef] [Green Version]
- Gordan, J.D.; Bertout, J.A.; Hu, C.-J.; Diehl, J.A.; Simon, M.C. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell 2007, 11, 335–347. [Google Scholar] [CrossRef] [Green Version]
- Linehan, W.M.; Srinivasan, R.; Schmidt, L.S. The genetic basis of kidney cancer: A metabolic disease. Nat. Rev. Urol. 2010, 7, 277–285. [Google Scholar] [CrossRef]
- Xia, W.; Nagase, S.; Montia, A.G.; Kalachikov, S.M.; Keniry, M.; Su, T.; Memeo, L.; Hibshoosh, H.; Parsons, R. BAF180 is a critical regulator of p21 induction and a tumor suppressor mutated in breast cancer. Cancer Res. 2008, 68, 1667–1674. [Google Scholar] [CrossRef] [Green Version]
- Benusiglio, P.R.; Couvé, S.; Gilbert-Dussardier, B.; Deveaux, S.; Le Jeune, H.; Da Costa, M.; Fromont, G.; Memeteau, F.; Yacoub, M.; Coupier, I.; et al. A germline mutation in PBRM1 predisposes to renal cell carcinoma. J. Med. Genet. 2015, 52, 426–430. [Google Scholar] [CrossRef]
- Ricketts, C.J.; De Cubas, A.A.; Fan, H.; Smith, C.C.; Lang, M.; Reznik, E.; Bowlby, R.; Gibb, E.A.; Akbani, R.; Beroukhim, R.; et al. The cancer genome atlas comprehensive molecular characterization of renal cell carcinoma. Cell Rep. 2018, 23, 313.e5–326.e5. [Google Scholar] [CrossRef] [Green Version]
- Bailey, M.H.; Tokheim, C.; Porta-Pardo, E.; Sengupta, S.; Bertrand, D.; Weerasinghe, A.; Colaprico, A.; Wendl, M.C.; Kim, J.; Reardon, B.; et al. Comprehensive characterization of cancer driver genes and mutations. Cell 2018, 173, 371.e18–385.e18. [Google Scholar] [CrossRef] [Green Version]
- cBioPortal for Cancer Genomics [Internet]. Available online: http://www.cbioportal.org/ (accessed on 27 November 2019).
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [Green Version]
- Turajlic, S.; Xu, H.; Litchfield, K.; Rowan, A.; Chambers, T.; Lopez, J.I.; Nicol, D.; O’Brien, T.; Larkin, J.; Horswell, S.; et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx renal. Cell 2018, 173, 581.e12–594.e12. [Google Scholar] [CrossRef]
- Joseph, R.W.; Kapur, P.; Serie, D.J.; Parasramka, M.; Ho, T.H.; Cheville, J.C.; Frenkel, E.; Parker, A.S.; Brugarolas, J. Clear cell renal cell carcinoma subtypes identified by BAP1 and PBRM1 expression. J. Urol. 2016, 195, 180–187. [Google Scholar] [CrossRef] [Green Version]
- Peña-Llopis, S.; Vega-Rubín-de-Celis, S.; Liao, A.; Leng, N.; Pavía-Jiménez, A.; Wang, S.; Yamasaki, T.; Zhrebker, L.; Sivanand, S.; Spence, P.; et al. BAP1 loss defines a new class of renal cell carcinoma. Nat. Genet. 2012, 44, 751–759. [Google Scholar] [CrossRef]
- Peña-Llopis, S.; Christie, A.; Xie, X.-J.; Brugarolas, J. Cooperation and antagonism among cancer genes: The renal cancer paradigm. Cancer Res. 2013, 73, 4173–4179. [Google Scholar] [CrossRef] [Green Version]
- Gu, Y.-F.; Cohn, S.; Christie, A.; McKenzie, T.; Wolff, N.; Do, Q.N.; Madhuranthakam, A.J.; Pedrosa, I.; Wang, T.; Dey, A.; et al. Modeling renal cell carcinoma in mice: Bap1 and Pbrm1 inactivation drive tumor grade. Cancer Discov. 2017, 7, 900–917. [Google Scholar] [CrossRef] [Green Version]
- Kapur, P.; Peña-Llopis, S.; Christie, A.; Zhrebker, L.; Pavía-Jiménez, A.; Rathmell, W.K.; Xie, X.J.; Brugarolas, J. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: A retrospective analysis with independent validation. Lancet Oncol. 2013, 14, 159–167. [Google Scholar] [CrossRef] [Green Version]
- Gossage, L.; Murtaza, M.; Slatter, A.F.; Lichtenstein, C.P.; Warren, A.; Haynes, B.; Marass, F.; Roberts, I.; Shanahan, S.J.; Claas, A.; et al. Clinical and pathological impact of VHL, PBRM1, BAP1, SETD2, KDM6A, and JARID1c in clear cell renal cell carcinoma. Genes Chromosomes Cancer 2014, 53, 38–51. [Google Scholar] [CrossRef]
- Pawłowski, R.; Mühl, S.M.; Sulser, T.; Krek, W.; Moch, H.; Schraml, P. Loss of PBRM1 expression is associated with renal cell carcinoma progression. Int. J. Cancer 2013, 132, E11–E17. [Google Scholar] [CrossRef]
- da Costa, W.H.; Rezende, M.; Carneiro, F.C.; Rocha, R.M.; da Cunha, I.W.; Carraro, D.M.; Guimaraes, G.C.; de Cassio Zequi, S. Polybromo-1 (PBRM1), a SWI/SNF complex subunit is a prognostic marker in clear cell renal cell carcinoma. BJU Int. 2014, 113, E157–E163. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Peng, S.; Guo, L.; Xie, H.; Wang, A.; Shang, Z.; Niu, Y. Prognostic and clinicopathological value of PBRM1 expression in renal cell carcinoma. Clin. Chim. Acta 2018, 486, 9–17. [Google Scholar] [CrossRef]
- Nam, S.J.; Lee, C.; Park, J.H.; Moon, K.C. Decreased PBRM1 expression predicts unfavorable prognosis in patients with clear cell renal cell carcinoma. Urol. Oncol. 2015, 33, 340.e9–340.e16. [Google Scholar] [CrossRef]
- Hakimi, A.A.; Ostrovnaya, I.; Reva, B.; Schultz, N.; Chen, Y.-B.; Gonen, M.; Liu, H.; Takeda, S.; Voss, M.H.; Tickoo, S.K.; et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: A report by MSKCC and the KIRC TCGA research network. Clin. Cancer Res. 2013, 19, 3259–3267. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; Dulaimi, E.; Devarajan, K.; Parsons, T.; Wang, Q.; O’Neill, R.; Solomides, C.; Peiper, S.C.; Testa, J.R.; Uzzo, R.; et al. Intratumoral heterogeneity analysis reveals hidden associations between protein expression losses and patient survival in clear cell renal cell carcinoma. Oncotarget 2017, 8, 37423–37434. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-Y.; Lee, S.-H.; Moon, K.C.; Kwak, C.; Kim, H.H.; Keam, B.; Kim, T.M.; Heo, D.S. The impact of PBRM1 expression as a prognostic and predictive marker in metastatic renal cell carcinoma. J. Urol. 2015, 194, 1112–1119. [Google Scholar] [CrossRef]
- Carlo, M.I.; Manley, B.; Patil, S.; Woo, K.M.; Coskey, D.T.; Redzematovic, A.; Arcila, M.; Ladanyi, M.; Lee, W.; Chen, Y.B.; et al. Genomic alterations and outcomes with VEGF-targeted therapy in patients with clear cell renal cell carcinoma. Kidney Cancer 2017, 1, 49–56. [Google Scholar] [CrossRef] [Green Version]
- Tennenbaum, D.M.; Manley, B.J.; Zabor, E.; Becerra, M.F.; Carlo, M.I.; Casuscelli, J.; Redzematovic, A.; Khan, N.; Arcila, M.E.; Voss, M.H.; et al. Genomic alterations as predictors of survival among patients within a combined cohort with clear cell renal cell carcinoma undergoing cytoreductive nephrectomy. Urol. Oncol. 2017, 35, 532.e7–532.e13. [Google Scholar] [CrossRef]
- Voss, M.H.; Reising, A.; Cheng, Y.; Patel, P.; Marker, M.; Kuo, F.; Chan, T.A.; Choueiri, T.K.; Hsieh, J.J.; Hakimi, A.A.; et al. Genomically annotated risk model for advanced renal-cell carcinoma: A retrospective cohort study. Lancet Oncol. 2018, 19, 1688–1698. [Google Scholar] [CrossRef]
- Kim, S.H.; Park, W.S.; Park, E.Y.; Park, B.; Joo, J.; Joung, J.Y.; Seo, H.K.; Lee, K.H.; Chung, J. The prognostic value of BAP1, PBRM1, pS6, PTEN, TGase2, PD-L1, CA9, PSMA, and Ki-67 tissue markers in localized renal cell carcinoma: A retrospective study of tissue microarrays using immunohistochemistry. PLoS ONE 2017. [Google Scholar] [CrossRef] [Green Version]
- Voss, M.H.; Hakimi, A.A.; Pham, C.G.; Brannon, A.R.; Chen, Y.-B.; Cunha, L.F.; Akin, O.; Liu, H.; Takeda, S.; Scott, S.N.; et al. Tumor genetic analyses of patients with metastatic renal cell carcinoma and extended benefit from mTOR inhibitor therapy. Clin. Cancer Res. 2014, 20, 1955–1964. [Google Scholar] [CrossRef] [Green Version]
- Kwiatkowski, D.J.; Choueiri, T.K.; Fay, A.P.; Rini, B.I.; Thorner, A.R.; de Velasco, G.; Tyburczy, M.E.; Hamieh, L.; Albiges, L.; Agarwal, N.; et al. Mutations in TSC1, TSC2, and MTOR are associated with response to rapalogs in patients with metastatic renal cell carcinoma. Clin. Cancer Res. 2016, 22, 2445–2452. [Google Scholar] [CrossRef] [Green Version]
- Ho, T.H.; Choueiri, T.K.; Wang, K.; Karam, J.A.; Chalmers, Z.; Frampton, G.; Elvin, J.A.; Johnson, A.; Liu, X.; Lin, Y.; et al. Correlation between molecular subclassifications of clear cell renal cell carcinoma and targeted therapy response. Eur. Urol. Focus 2016, 2, 204–209. [Google Scholar] [CrossRef]
- Hsieh, J.J.; Chen, D.; Wang, P.I.; Marker, M.; Redzematovic, A.; Chen, Y.B.; Selcuklu, S.D.; Weinhold, N.; Bouvier, N.; Huberman, K.H.; et al. Genomic biomarkers of a randomized trial comparing first-line everolimus and sunitinib in patients with metastatic renal cell carcinoma. Eur. Urol. 2017, 71, 405–414. [Google Scholar] [CrossRef] [Green Version]
- Miao, D.; Margolis, C.A.; Gao, W.; Voss, M.H.; Li, W.; Martini, D.J.; Norton, C.; Bossé, D.; Wankowicz, S.M.; Cullen, D.; et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 2018, 359, 801–806. [Google Scholar] [CrossRef] [Green Version]
- Braun, D.A.; Ishii, Y.; Walsh, A.M.; Van Allen, E.M.; Wu, C.J.; Shukla, S.A.; Choueiri, T.K. Clinical validation of PBRM1 alterations as a marker of immune checkpoint inhibitor response in renal cell carcinoma. JAMA Oncol. 2019, 5, 1631–1633. [Google Scholar] [CrossRef]
- McDermott, D.F.; Huseni, M.A.; Atkins, M.B.; Motzer, R.J.; Rini, B.I.; Escudier, B.; Fong, L.; Joseph, R.W.; Pal, S.K.; Reeves, J.A.; et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat. Med. 2018, 24, 749–757. [Google Scholar] [CrossRef]
- Fay, A.P.; de Velasco, G.; Ho, T.H.; Van Allen, E.M.; Murray, B.; Albiges, L.; Signoretti, S.; Hakimi, A.A.; Stanton, M.L.; Bellmunt, J.; et al. Whole-exome sequencing in two extreme phenotypes of response to VEGF-targeted therapies in patients with metastatic clear cell renal cell carcinoma. J. Natl. Compr. Cancer Netw. JNCCN 2016, 14, 820–824. [Google Scholar] [CrossRef] [Green Version]
- Motzer, R.J.; Tannir, N.M.; McDermott, D.F.; ArénFrontera, O.; Melichar, B.; Choueiri, T.K.; Plimack, E.R.; Barthélémy, P.; Porta, C.; George, S.; et al. Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N. Engl. J. Med. 2018, 378, 1277–1290. [Google Scholar] [CrossRef]
- Motzer, R.J.; Escudier, B.; McDermott, D.F.; George, S.; Hammers, H.J.; Srinivas, S.; Tykodi, S.S.; Sosman, J.A.; Procopio, G.; Plimack, E.R.; et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N. Engl. J. Med. 2015, 373, 1803–1813. [Google Scholar] [CrossRef]
- Maia, M.C.; Almeida, L.; Bergerot, P.G.; Dizman, N.; Pal, S.K. Relationship of tumor mutational burden (TMB) to immunotherapy response in metastatic renal cell carcinoma (mRCC). J. Clin. Oncol. 2018, 36 (Suppl. 6), 662. [Google Scholar] [CrossRef]
- Pan, D.; Kobayashi, A.; Jiang, P.; Ferrari de Andrade, L.; Tay, R.E.; Luoma, A.M.; Tsoucas, D.; Qiu, X.; Lim, K.; Rao, P.; et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 2018, 359, 770–775. [Google Scholar] [CrossRef] [Green Version]
- Soussi, T.; Wiman, K.G. TP53: An oncogene in disguise. Cell Death Differ. 2015, 22, 1239–1249. [Google Scholar] [CrossRef]
- Ding, H.; Zhao, J.; Zhang, Y.; Yu, J.; Liu, M.; Li, X.; Xu, L.; Lin, M.; Liu, C.; He, Z.; et al. Systematic analysis of drug vulnerabilities conferred by tumor suppressor loss. Cell Rep. 2019, 27, 3331.e6–3344.e6. [Google Scholar] [CrossRef] [Green Version]

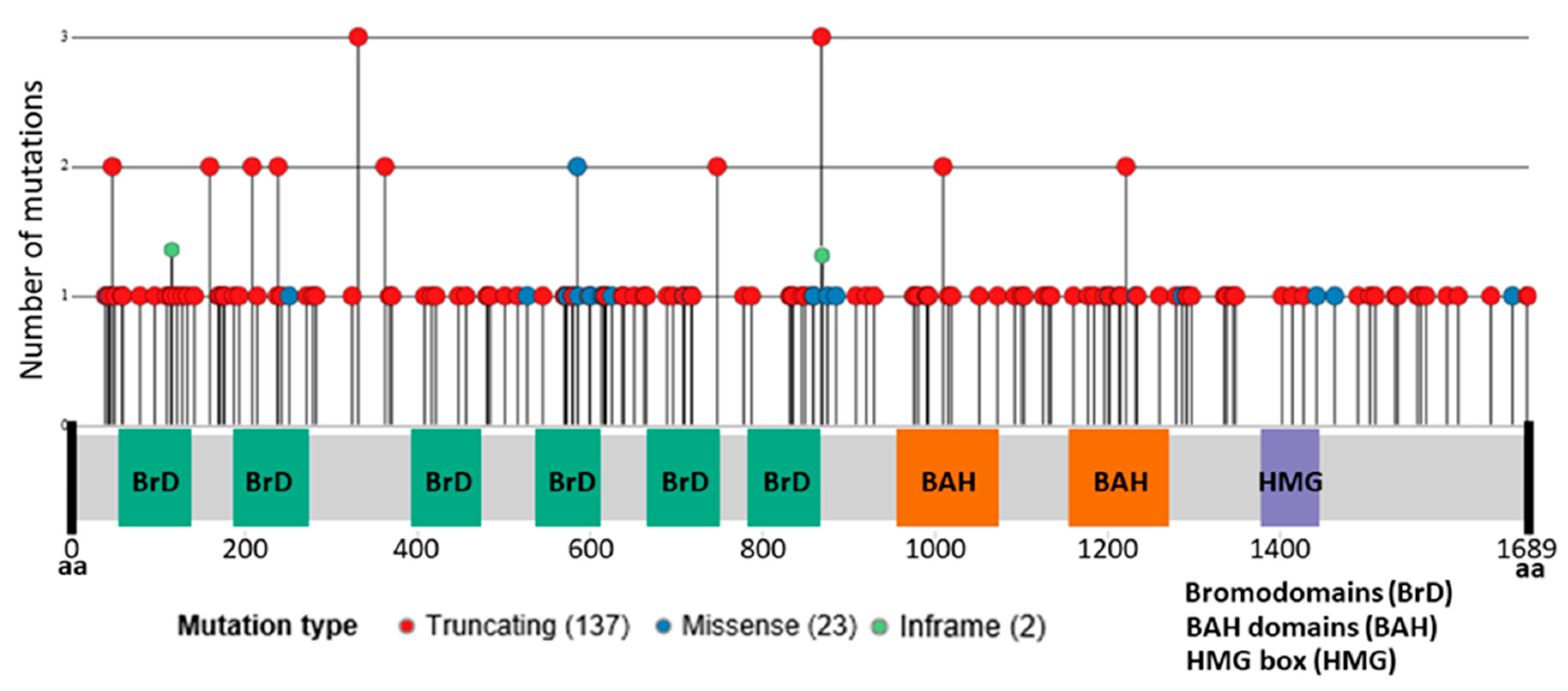
| Author [Ref.] | Year | Country | Gender M(%)/F(%) | Age (y) | N | BAP1 MT(%) | PBRM1 MT(%) | Tech | Survival Efficacy Parameters | |
|---|---|---|---|---|---|---|---|---|---|---|
| RFS (HR/ p) | OS (HR/ p) | |||||||||
| Kapur [40] | 2013 | USA | 80(55)/65(45) | 62 | 145 | 21(14) | 78(54) | NGS | NA | BAP1 MT: 4.6; PBRM1 MT: 10.6 (2.7/0.044) |
| Gossage [41] | 2014 | UK | 83(63)/49(37) | 62 | 132 | 14(11) | 42(33) | NGS | BAP1 MT: 1.2 a; PBRM1 MT: 4.9 a (n.sp./0.059) | n.sp. (n.sp./NS) |
| Disease Type | Author [Ref.] | Year | Country | Gender M(%)/F(%) | Age (y) | N | PBRM1 -/Low (%) | Tech | Ab (Dilution) | PBRM1 Cut-off | Survival Efficacy Parameters | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RFS/PFS (HR/p) | OS/CSS (HR/p) | |||||||||||
| Loc | da Costa [43] | 2013 | Brazil | 66(59)/46(41) | 56 | 112 | 34(30) | IHC | PB1 Ab b (1:25) | Presence | PBRM1 -: 66.7% vs. PBRM1 +: 87.3% d (n.sp./0.048) | PBRM1 -: 70.6 vs. PBRM1 +: 89.7% f (n.sp./0.017) |
| Hakimi [46] | 2013 | USA | 408(67)/200(33) | 61 | 609 | 198(33) | NGS | - | - | NA | n.sp. (n.sp./NS) | |
| Pawlowski [42] | 2013 | Switzerland | NA | NA | 279 | 175(64) a. | IHC | PB1Ab c (1:25) | >5% | NA | PBRM1 -: 80 vs. PBRM1 +: not reach (n.sp./0.025) | |
| Nam [45] | 2015 | Korea | 485(74)/172(26) | NA | 657 | NA | IHC | PB1 Ab c (1:100) | >50% | PBRM1 LE: 109.5 vs. PBRM1 HE: 156.2 (n.sp./<0.001) | PBRM1 LE: 145.5 vs. PBRM1 HE: 171.7 (n.sp./<0.001) | |
| Joseph [36] | 2016 | USA | 823(62)/435(33) | 64 | 1330 | 674(51) | IHC | PB1 Ab c | Presence | PBRM1 -: higher mtx. risk (1.46/0.001) | n.sp. (n.sp/NS) | |
| Kim [52] | 2017 | Korea | 244(70)/107(30) | 54 | 351 | 208(59) | IHC | PB1 Ab | Score > 2 | n.sp. (0.81/0.64) | n.sp. (1.86/0.1) | |
| Loc and Mtx | Jiang [47] | 2017 | USA | 118(74)/42(26) | 60 | 160 | 49(31) | IHC | PB1 Abc (1:50) | >5% | n.sp. (0.79/0.21) | n.sp. (0.42/2.45e–05) |
| Carlo [49] | 2017 | USA | 77(73)/28(27) | 57 | 105 | 53(51) | NGS | - | - | PBRM1 MT: 12.0 vs. PBRM1 WT: 6.9 e (n.sp./0.01) | PBRM1 MT: not reach vs. PBRM1 WT: 36.3 (n.sp./0.12) | |
| Mtx | Tennenbaum [50] | 2017 | USA | 116(69)/51(31) | 60 | 167 | 64(38) | NGS | - | - | NA | n.sp. (0.87/0.49) |
| Kim [48] | 2015 | Korea | 44(83)/9(17) | 62 | 53 | 28(52) | IHC | PB1 Ab c (1:100) | Score >2.5 | NA | PBRM1 LE: 46 vs. PBRM1 HE: 24 (n.sp./0.022) | |
| Voss [51] | 2018 | USA | 279(74)/98(26) | 61 | 357 | 160(45) | NGS | - | - | PBRM1 MT: 11.1 vs. PBRM1 WT: 8.2 (0.67/0.004) | PBRM1 MT: 35.5 vs. PBRM1 WT: 23.8 (0.63/0.002) | |
| Author [Ref.] | Year | Country | Gender M(%)/F(%) | Age (y) | N | BAP1 MT (%) | PBRM1 MT (%) | Tech | Ab(Dilution) | PBRM1 Cut-off | Everolimus PFS | Sunitinib PFS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PBRM1 MT or LE vs. PBRM1 WT or HE (HR/ p) | PBRM1 MT or LE vs. PBRM1 WT or HE (HR/ p) | |||||||||||
| Hsieh [56] | 2016 | USA | 168 (76)/52(24) | 62 | 220 | 42(19) | 101(46) | NGS | - | - | 12.8 vs. 5.5 (0.53/ 0.004) | 11.0 vs. 8.3 (0.79/ 0.4) |
| Kim [48] | 2015 | Korea | 44(83)/9(17) | 62 | 53 | NA | 25(47) | IHC | PB1 Ab a (1:100) | 2.5 | 3.0 vs. 1.9 (NA/ 0.1) | 7.3 vs. 9 (NA/ 0.8) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carril-Ajuria, L.; Santos, M.; Roldán-Romero, J.M.; Rodriguez-Antona, C.; de Velasco, G. Prognostic and Predictive Value of PBRM1 in Clear Cell Renal Cell Carcinoma. Cancers 2020, 12, 16. https://doi.org/10.3390/cancers12010016
Carril-Ajuria L, Santos M, Roldán-Romero JM, Rodriguez-Antona C, de Velasco G. Prognostic and Predictive Value of PBRM1 in Clear Cell Renal Cell Carcinoma. Cancers. 2020; 12(1):16. https://doi.org/10.3390/cancers12010016
Chicago/Turabian StyleCarril-Ajuria, Lucía, María Santos, Juan María Roldán-Romero, Cristina Rodriguez-Antona, and Guillermo de Velasco. 2020. "Prognostic and Predictive Value of PBRM1 in Clear Cell Renal Cell Carcinoma" Cancers 12, no. 1: 16. https://doi.org/10.3390/cancers12010016
APA StyleCarril-Ajuria, L., Santos, M., Roldán-Romero, J. M., Rodriguez-Antona, C., & de Velasco, G. (2020). Prognostic and Predictive Value of PBRM1 in Clear Cell Renal Cell Carcinoma. Cancers, 12(1), 16. https://doi.org/10.3390/cancers12010016






