Overexpression of Mitochondrial IF1 Prevents Metastatic Disease of Colorectal Cancer by Enhancing Anoikis and Tumor Infiltration of NK Cells
Abstract
:1. Introduction
2. Results
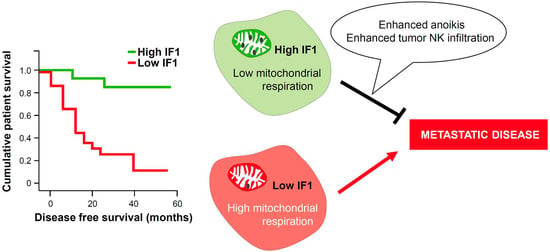
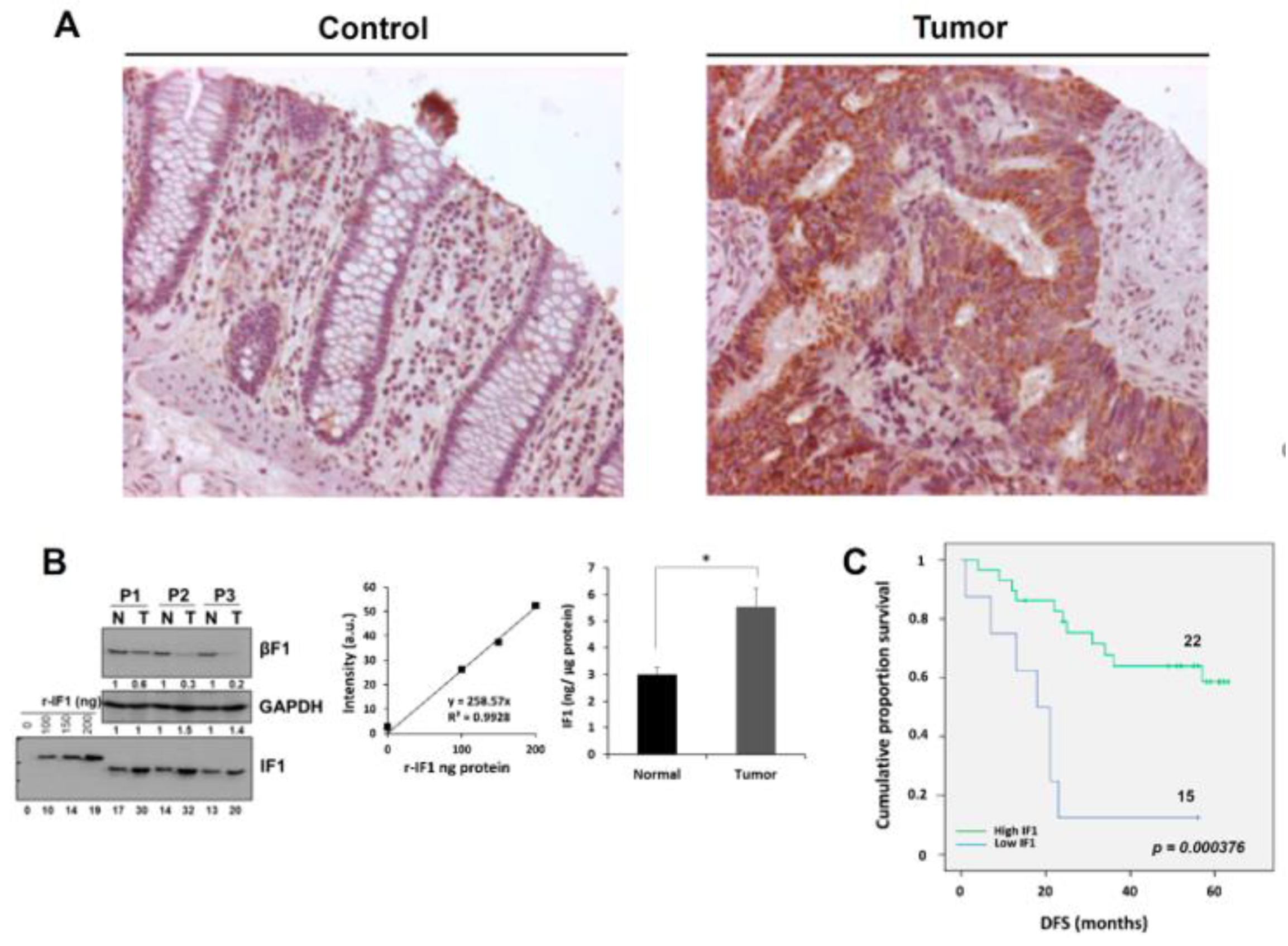
2.1. Increased Expression of IF1 Is Associated with Increased Survival of CRC Patients
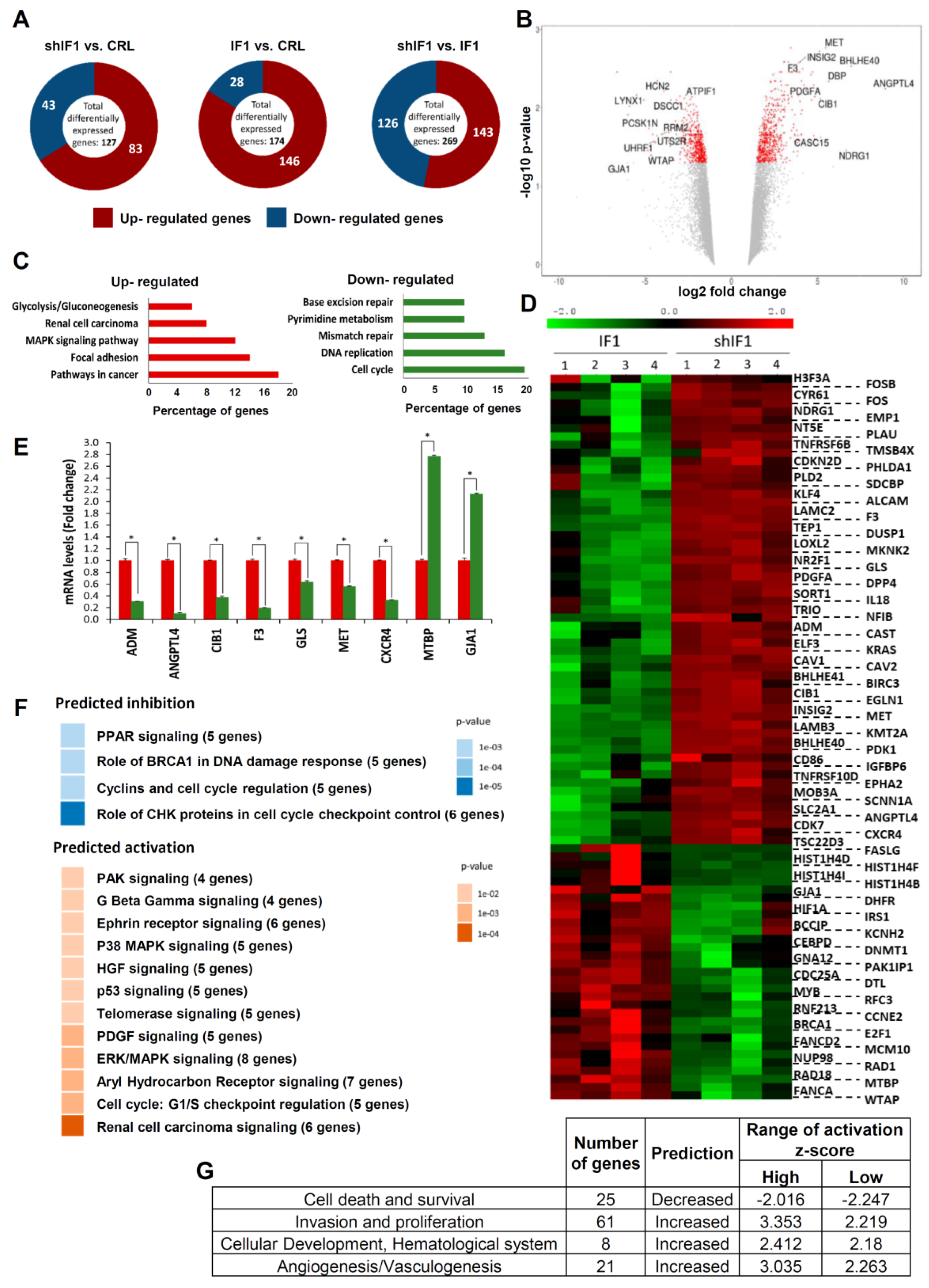
2.2. Transcriptomic Profiling of HCT116 Cells with Differential Expression of IF1
2.3. Proteomic Analysis of HCT116 Cells with Differential Expression of IF1
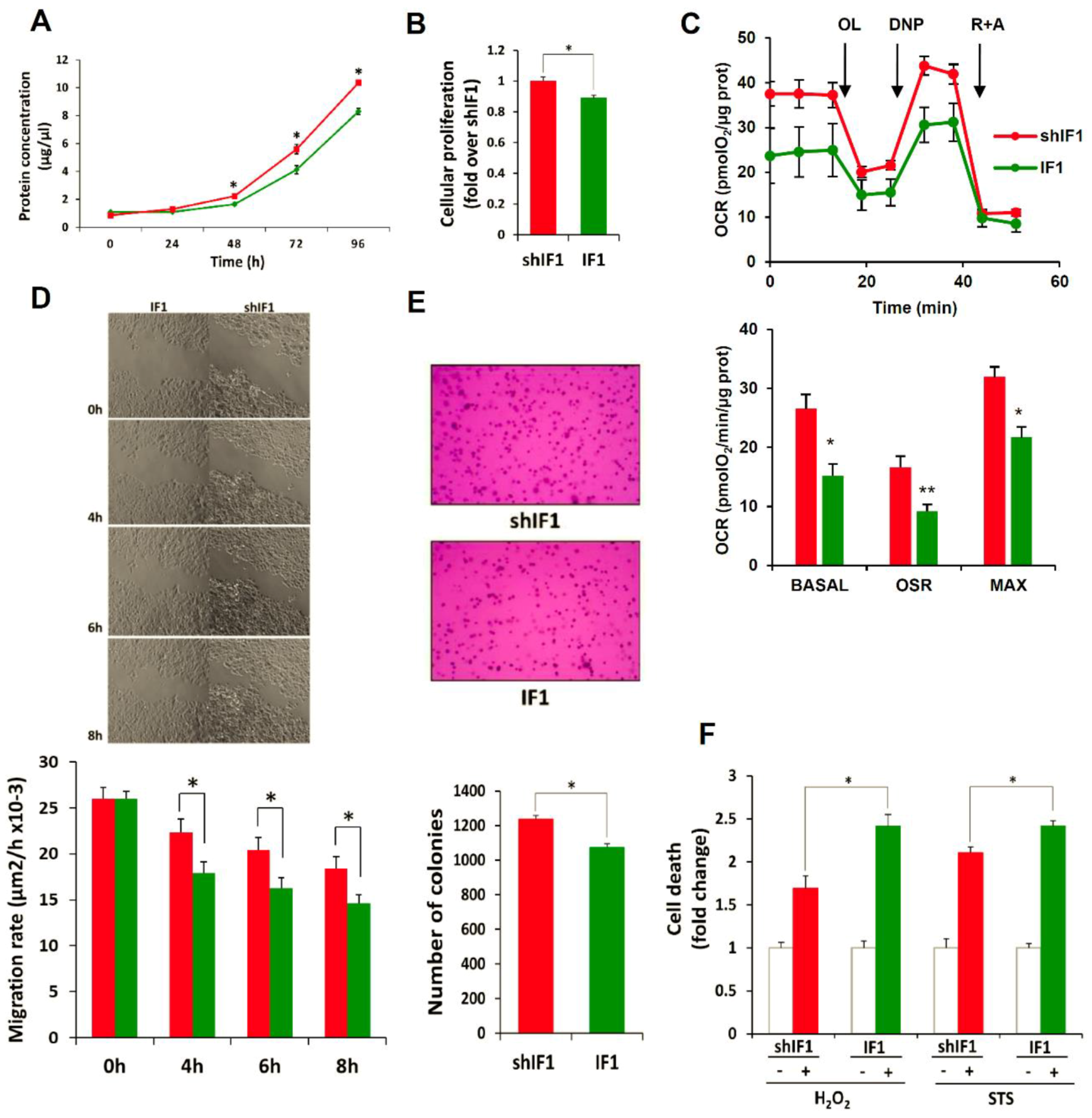
2.4. Phenotypic Analysis of HCT116 Cells with Differential Expression of IF1
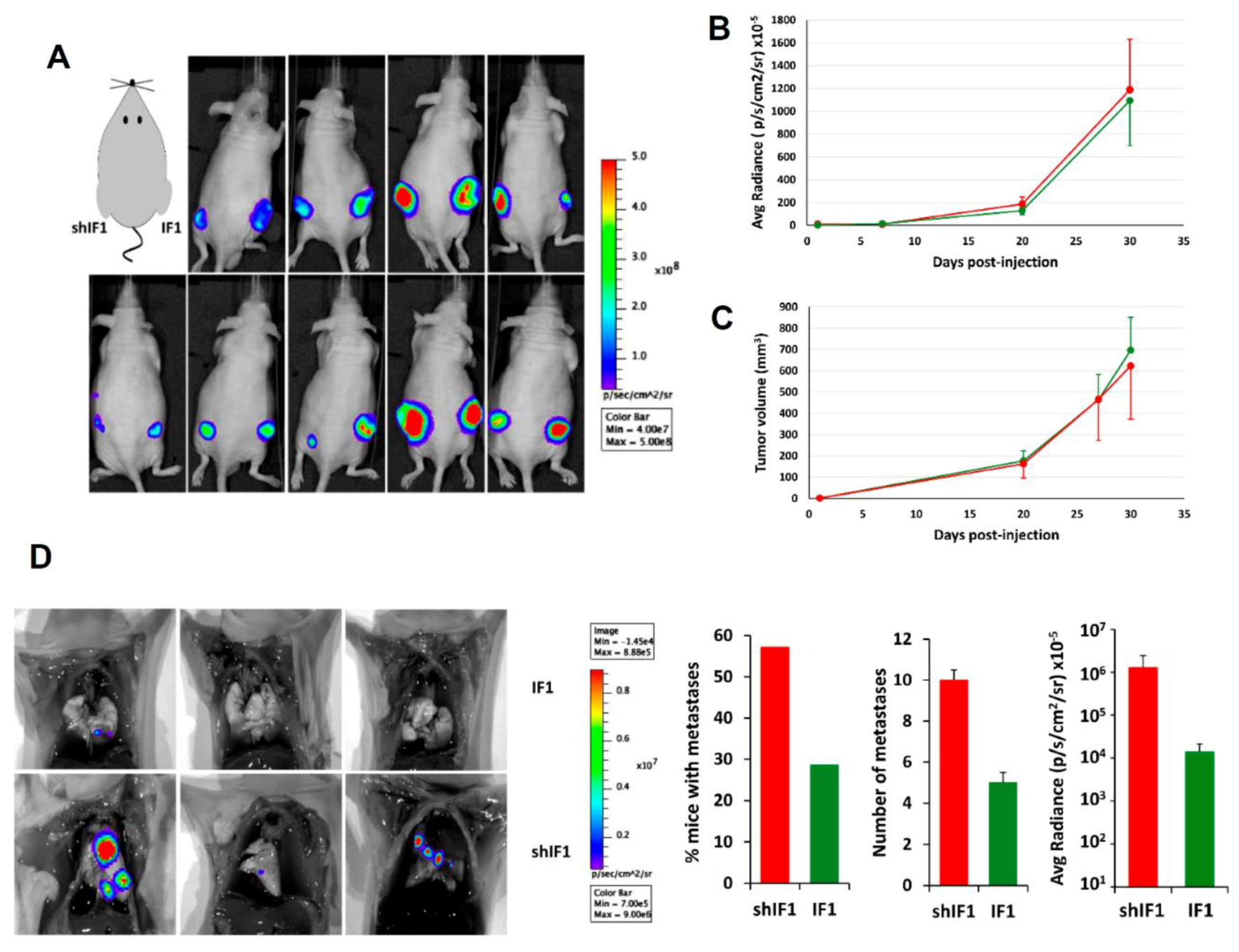
2.5. In Vivo Tumorigenesis of HCT116 Cells with Differential Expression of IF1
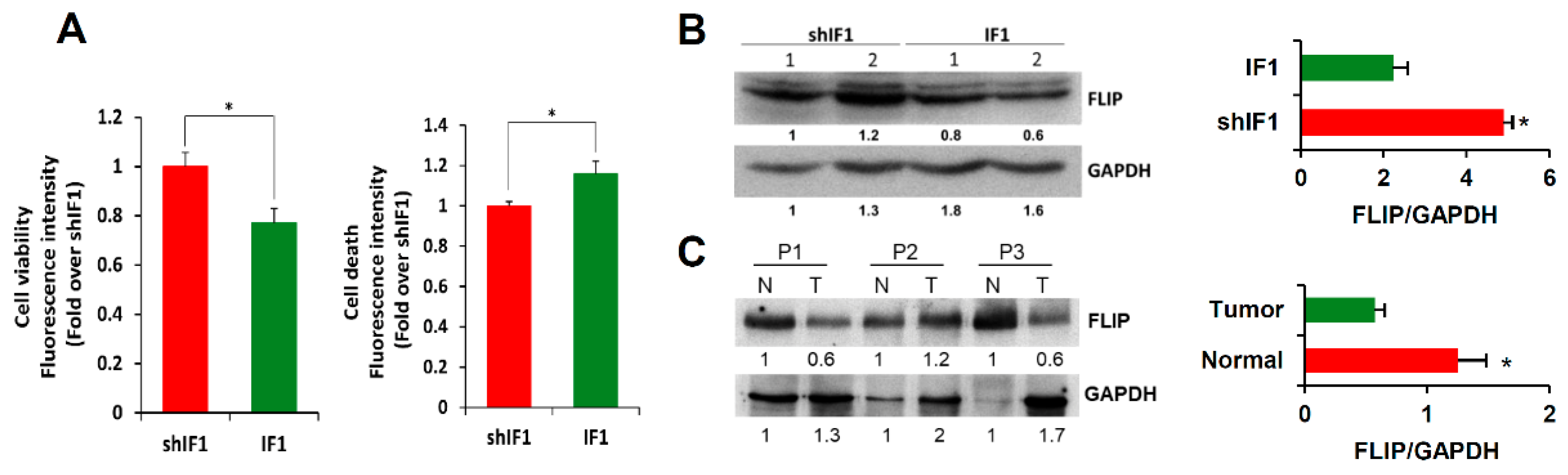
2.6. IF1 Favors Anoikis of Colon Cancer Cells
2.7. IF1 Favors Infiltration and Cytotoxicity of NK Cells
3. Discussion
4. Materials and Methods
4.1. Patient Specimens and IF1 Quantification
4.2. Generation of Cell Lines, Cell Cultures and Spheroids
4.3. NK Cell Isolation and Labeling
4.4. Cellular Lysis and Western Blotting
4.5. Gene Array Hybridization
4.6. Gene Data Analysis
4.7. Quantification of mRNA
4.8. Proteomic Analysis and ITRAQ Labeling
4.9. Reverse Phase-Liquid Chromatography RP-LC-MS/MS and Data Analysis
4.10. Immunohistochemistry
4.11. Cellular Proliferation, Cell Death Assays and Cellular Respiration
4.12. Soft-Agar, Wound Healing, Invasion and Anoikis Assays
4.13. In Vivo Assays
4.14. Flow Cytometry Analyses of Spheroid Invasion
4.15. In-Vivo Confocal Microscopy of NK cell Invasion and Cytotoxicity
4.16. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Miller, K.D.; Nogueira, L.; Mariotto, A.B.; Rowland, J.H.; Yabroff, K.R.; Alfano, C.M.; Jemal, A.; Kramer, J.L.; Siegel, R.L. Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 2019, 69, 363–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, P.P.; Sabatini, D.M. Cancer cell metabolism: Warburg and beyond. Cell 2008, 134, 703–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danhier, P.; Banski, P.; Payen, V.L.; Grasso, D.; Ippolito, L.; Sonveaux, P.; Porporato, P.E. Cancer metabolism in space and time: Beyond the Warburg effect. Biochim. Biophys. Acta 2017, 1858, 556–572. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Arago, M.; Formentini, L.; Cuezva, J.M. Mitochondria-mediated energy adaption in cancer: The H(+)-ATP synthase-geared switch of metabolism in human tumors. Antioxid. Redox Signal. 2013, 19, 285–298. [Google Scholar] [CrossRef] [Green Version]
- Chandel, N.S. Evolution of mitochondria as signaling organelles. Cell Metab. 2015, 22, 204–206. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Arago, M.; Garcia-Bermudez, J.; Martinez-Reyes, I.; Santacatterina, F.; Cuezva, J.M. Degradation of IF1 controls energy metabolism during osteogenic differentiation of stem cells. EMBO Rep. 2013, 14, 638–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Green, D.R. Metabolic reprogramming and metabolic dependency in T cells. Immunol. Rev. 2012, 249, 14–26. [Google Scholar] [CrossRef] [Green Version]
- Cuezva, J.M.; Ortega, A.D.; Willers, I.; Sanchez-Cenizo, L.; Aldea, M.; Sanchez-Arago, M. The tumor suppressor function of mitochondria: Translation into the clinics. Biochim. Biophys. Acta 2009, 1792, 1145–1158. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Formentini, L.; Sánchez-Aragó, M.; Sánchez-Cenizo, L.; Cuezva, J.M. The mitochondrial ATPase Inhibitory Factor 1 (IF1) triggers a ROS-mediated retrograde pro-survival and proliferative response. Mol. Cell 2012, 45, 731–742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Cenizo, L.; Formentini, L.; Aldea, M.; Ortega, A.D.; Garcia-Huerta, P.; Sanchez-Arago, M.; Cuezva, J.M. Up-regulation of the ATPase inhibitory factor 1 (IF1) of the mitochondrial H+-ATP synthase in human tumors mediates the metabolic shift of cancer cells to a Warburg phenotype. J. Biol. Chem. 2010, 285, 25308–25313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Arago, M.; Formentini, L.; Martinez-Reyes, I.; Garcia-Bermudez, J.; Santacatterina, F.; Sanchez-Cenizo, L.; Willers, I.M.; Aldea, M.; Najera, L.; Juarranz, A.; et al. Expression, regulation and clinical relevance of the ATPase inhibitory factor 1 in human cancers. Oncogenesis 2013, 2, e46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, J.; Zhang, L.; Zong, S.; Guo, R.; Liu, T.; Yi, J.; Wang, P.; Zhuo, W.; Yang, M. Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1. Science 2019, 364, 1068–1075. [Google Scholar] [CrossRef]
- Esparza-Molto, P.B.; Nuevo-Tapioles, C.; Chamorro, M.; Najera, L.; Torresano, L.; Santacatterina, F.; Cuezva, J.M. Tissue-specific expression and post-transcriptional regulation of the ATPase inhibitory factor 1 (IF1) in human and mouse tissues. FASEB J. 2019, 33, 1836–1851. [Google Scholar] [CrossRef]
- Santacatterina, F.; Sanchez-Cenizo, L.; Formentini, L.; Mobasher, M.A.; Casas, E.; Rueda, C.B.; Martinez-Reyes, I.; Nunez de Arenas, C.; Garcia-Bermudez, J.; Zapata, J.M.; et al. Down-regulation of oxidative phosphorylation in the liver by expression of the ATPase inhibitory factor 1 induces a tumor-promoter metabolic state. Oncotarget 2016, 7, 490–508. [Google Scholar] [CrossRef] [Green Version]
- Formentini, L.; Pereira, M.P.; Sanchez-Cenizo, L.; Santacatterina, F.; Lucas, J.J.; Navarro, C.; Martinez-Serrano, A.; Cuezva, J.M. In vivo inhibition of the mitochondrial H+-ATP synthase in neurons promotes metabolic preconditioning. EMBO J. 2014, 33, 762–778. [Google Scholar] [CrossRef] [Green Version]
- Formentini, L.; Ryan, A.J.; Galvez-Santisteban, M.; Carter, L.; Taub, P.; Lapek, J.D., Jr.; Gonzalez, D.J.; Villarreal, F.; Ciaraldi, T.P.; Cuezva, J.M.; et al. Mitochondrial H+-ATP synthase in human skeletal muscle: Contribution to dyslipidaemia and insulin resistance. Diabetologia 2017, 60, 2052–2065. [Google Scholar] [CrossRef]
- Garcia-Bermudez, J.; Sanchez-Arago, M.; Soldevilla, B.; Del Arco, A.; Nuevo-Tapioles, C.; Cuezva, J.M. PKA Phosphorylates the ATPase Inhibitory Factor 1 and Inactivates Its Capacity to Bind and Inhibit the Mitochondrial H-ATP Synthase. Cell Rep. 2015, 12, 2143–2155. [Google Scholar] [CrossRef] [Green Version]
- Song, R.; Song, H.; Liang, Y.; Yin, D.; Zhang, H.; Zheng, T.; Wang, J.; Lu, Z.; Song, X.; Pei, T.; et al. Reciprocal activation between ATPase inhibitory factor 1 and NF-kappaB drives hepatocellular carcinoma angiogenesis and metastasis. Hepatology 2014, 60, 1659–1673. [Google Scholar] [CrossRef] [Green Version]
- Wei, S.; Fukuhara, H.; Kawada, C.; Kurabayashi, A.; Furihata, M.; Ogura, S.; Inoue, K.; Shuin, T. Silencing of ATPase Inhibitory Factor 1 Inhibits Cell Growth via Cell Cycle Arrest in Bladder Cancer. Pathobiology 2015, 82, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Yin, T.; Lu, L.; Xiong, Z.; Wei, S.; Cui, D. ATPase inhibitory factor 1 is a prognostic marker and contributes to proliferation and invasion of human gastric cancer cells. Biomed. Pharmacother. 2015, 70, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Shan, Q.; Li, P.; Wu, Y.; Xie, J.; Wang, X. ATPase inhibitory factor 1 is a potential prognostic marker for the migration and invasion of glioma. Oncol. Lett. 2015, 10, 2075–2080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Ledo, L.; Nuevo-Tapioles, C.; Cuevas-Martin, C.; Martinez-Reyes, I.; Soldevilla, B.; Gonzalez-Llorente, L.; Cuezva, J.M. Overexpression of the ATPase inhibitory factor 1 favors a non-metastatic phenotype in breast cancer. Front. Oncol. 2017, 7, 69. [Google Scholar] [CrossRef] [PubMed]
- Paoli, P.; Giannoni, E.; Chiarugi, P. Anoikis molecular pathways and its role in cancer progression. Biochim. Biophys. Acta 2013, 1833, 3481–3498. [Google Scholar] [CrossRef] [Green Version]
- Krueger, A.; Baumann, S.; Krammer, P.H.; Kirchhoff, S. FLICE-inhibitory proteins: Regulators of death receptor-mediated apoptosis. Mol. Cell. Biol. 2001, 21, 8247–8254. [Google Scholar] [CrossRef] [Green Version]
- Wilson, T.R.; McLaughlin, K.M.; McEwan, M.; Sakai, H.; Rogers, K.M.; Redmond, K.M.; Johnston, P.G.; Longley, D.B. c-FLIP: A key regulator of colorectal cancer cell death. Cancer Res. 2007, 67, 5754–5762. [Google Scholar] [CrossRef] [Green Version]
- Shadel, G.S.; Horvath, T.L. Mitochondrial ROS signaling in organismal homeostasis. Cell 2015, 163, 560–569. [Google Scholar] [CrossRef] [Green Version]
- Formentini, L.; Santacatterina, F.; Nunez de Arenas, C.; Stamatakis, K.; Lopez-Martinez, D.; Logan, A.; Fresno, M.; Smits, R.; Murphy, M.P.; Cuezva, J.M. Mitochondrial ROS production protects the Intestine from Inflammation through Functional M2 macrophage polarization. Cell Rep. 2017, 19, 1202–1213. [Google Scholar] [CrossRef] [Green Version]
- Orange, J.S. Formation and function of the lytic NK-cell immunological synapse. Nat. Rev. Immunol. 2008, 8, 713–725. [Google Scholar] [CrossRef] [Green Version]
- Esparza-Moltó, P.B.; Cuezva, J.M. The role of mitochondrial H+-ATP synthase in cancer. Front. Oncol. 2018, 8, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Aguilar, A.; Martinez-Reyes, I.; Cuezva, J.M. Changes in the turnover of the cellular proteome during metabolic reprogramming: A role for mtROS in proteostasis. J. Proteome Res. 2019, 18, 3142–3155. [Google Scholar] [CrossRef] [PubMed]
- Jover, R.; Zapater, P.; Castells, A.; Llor, X.; Andreu, M.; Cubiella, J.; Balaguer, F.; Sempere, L.; Xicola, R.M.; Bujanda, L.; et al. The efficacy of adjuvant chemotherapy with 5-fluorouracil in colorectal cancer depends on the mismatch repair status. Eur. J. Cancer 2009, 45, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Casimiro, M.C.; Crosariol, M.; Loro, E.; Li, Z.; Pestell, R.G. Cyclins and cell cycle control in cancer and disease. Genes Cancer 2012, 3, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.M.; Shah, Y.M.; Gonzalez, F.J. The role of peroxisome proliferator-activated receptors in carcinogenesis and chemoprevention. Nat. Rev. Cancer 2012, 12, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Wang, W.H. Peroxisome proliferator-activated receptor gamma and colorectal cancer. World J. Gastrointest. Oncol. 2010, 2, 159–164. [Google Scholar] [CrossRef]
- Cairns, R.A.; Harris, I.S.; Mak, T.W. Regulation of cancer cell metabolism. Nat. Rev. Cancer 2011, 11, 85–95. [Google Scholar] [CrossRef] [Green Version]
- Kondoh, H.; Lleonart, M.E.; Nakashima, Y.; Yokode, M.; Tanaka, M.; Bernard, D.; Gil, J.; Beach, D. A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid. Redox Signal. 2007, 9, 293–299. [Google Scholar] [CrossRef]
- Xu, X.; Duan, S.; Yi, F.; Ocampo, A.; Liu, G.H.; Izpisua Belmonte, J.C. Mitochondrial regulation in pluripotent stem cells. Cell Metab. 2013, 18, 325–332. [Google Scholar] [CrossRef] [Green Version]
- LeBleu, V.S.; O’Connell, J.T.; Gonzalez Herrera, K.N.; Wikman, H.; Pantel, K.; Haigis, M.C.; de Carvalho, F.M.; Damascena, A.; Domingos Chinen, L.T.; Rocha, R.M.; et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 2014, 16, 992–1003. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.N.; Zeng, Z.L.; Lu, J.; Wang, Y.; Liu, Z.X.; He, M.M.; Zhao, Q.; Wang, Z.X.; Li, T.; Lu, Y.X.; et al. CPT1A-mediated fatty acid oxidation promotes colorectal cancer cell metastasis by inhibiting anoikis. Oncogene 2018, 37, 6025–6040. [Google Scholar] [CrossRef] [PubMed]
- Riihimaki, M.; Hemminki, A.; Sundquist, J.; Hemminki, K. Patterns of metastasis in colon and rectal cancer. Sci. Rep. 2016, 6, 29765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morvan, M.G.; Lanier, L.L. NK cells and cancer: You can teach innate cells new tricks. Nat. Rev. Cancer 2016, 16, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef] [Green Version]
- Ohtani, H. Focus on TILs: Prognostic significance of tumor infiltrating lymphocytes in human colorectal cancer. Cancer Immun. Arch. 2007, 7, 4. [Google Scholar]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and granzymes: Function, dysfunction and human pathology. Nat. Rev. Immunol. 2015, 15, 388–400. [Google Scholar] [CrossRef]
- Takeda, K.; Hayakawa, Y.; Smyth, M.J.; Kayagaki, N.; Yamaguchi, N.; Kakuta, S.; Iwakura, Y.; Yagita, H.; Okumura, K. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in surveillance of tumor metastasis by liver natural killer cells. Nat. Med. 2001, 7, 94–100. [Google Scholar] [CrossRef]
- D’Alterio, C.; Buoncervello, M.; Ierano, C.; Napolitano, M.; Portella, L.; Rea, G.; Barbieri, A.; Luciano, A.; Scognamiglio, G.; Tatangelo, F.; et al. Targeting CXCR4 potentiates anti-PD-1 efficacy modifying the tumor microenvironment and inhibiting neoplastic PD-1. J. Exp. Clin. Cancer Res. 2019, 38, 432. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Wang, D.; Wang, X.; Sun, S.; Zhang, Y.; Wang, S.; Miao, R.; Xu, X.; Qu, X. CXCL12/CXCR4 promotes inflammation-driven colorectal cancer progression through activation of RhoA signaling by sponging miR-133a-3p. J. Exp. Clin. Cancer Res. 2019, 38, 32. [Google Scholar] [CrossRef]
- Kordass, T.; Osen, W.; Eichmuller, S.B. Controlling the Immune Suppressor: Transcription Factors and MicroRNAs Regulating CD73/NT5E. Front. Immunol. 2018, 9, 813. [Google Scholar] [CrossRef] [Green Version]
- Sek, K.; Molck, C.; Stewart, G.D.; Kats, L.; Darcy, P.K.; Beavis, P.A. Targeting adenosine receptor signaling in cancer immunotherapy. Int. J. Mol. Sci. 2018, 19, 3837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.X.; Zhao, Y.Y.; Shen, J.; Sun, X.; Liu, Y.; Liu, H.; Wang, Y.; Wang, J. Nanoenabled Modulation of Acidic Tumor Microenvironment Reverses Anergy of Infiltrating T Cells and Potentiates Anti-PD-1 Therapy. Nano. Lett. 2019, 19, 2774–2783. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Arago, M.; Chamorro, M.; Cuezva, J.M. Selection of cancer cells with repressed mitochondria triggers colon cancer progression. Carcinogenesis 2010, 31, 567–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Univariate Analysis | Overall Survival | Disease-Free Survival | ||
| Variable | HR (95% CI) | p-Value | HR (95% CI) | p-Value |
| Age (year) | ||||
| ≤70 | 1 (Reference) | 1 (Reference) | ||
| >70 | 3.01 (0.97–9.27) | 0.055 | 3.67 (1.20–11.23) | 0.022 |
| Gender | ||||
| Female | 1 (Reference) | 1 (Reference) | ||
| Male | 0.59 (0.22–1.53) | 0.280 | 0.58 (0.22–1.47) | 0.252 |
| Grade | ||||
| I | 1 (Reference) | 1 (Reference) | ||
| II | 2.94 (0.38–22.62) | 0.300 | 3.56 (0.46–27.27) | 0.221 |
| III | 7.094 (0.78–63.99) | 0.081 | 8.62 (0.95–78.21) | 0.055 |
| Lymph nodes | ||||
| No | 1 (Reference) | 1 (Reference) | ||
| Yes | 4.62 (1.49–14.30) | 0.008 | 3.61 (1.28–10.23) | 0.015 |
| CEA | ||||
| ≤5 µg/L | 1 (Reference) | 1 (Reference) | ||
| >5 µg/L | 2.79 (1.06–7.36) | 0.038 | 2.64 (1.02–6.86) | 0.045 |
| Metastasis | ||||
| No | 1 (Reference) | 1 (Reference) | ||
| Yes | 4.34 (1.57–11.94) | 0.004 | 3.87 (1.42–10.52) | 0.008 |
| IF1 | ||||
| Low | 1 (Reference) | 1 (Reference) | ||
| High | 0.176 (0.06–0.49) | 0.001 | 0.19 (0.07–0.52) | 0.001 |
| Multivariate Analysis | Overall Survival | Disease-Free Survival | ||
| Variable | HR (95% CI) | p-value | HR (95% CI) | p-value |
| Age (year) | ||||
| ≤70 | ------- | ------- | 1 (Reference) | |
| >70 | ------- | ------- | 2.50 (0.76–8.21) | 0.131 |
| Lymph nodes | ||||
| No | 1 (Reference) | 1 (Reference) | ||
| Yes | 3.18 (0.95–10.61) | 0.059 | 2.25 (0.71–7.13) | 0.165 |
| CEA | ||||
| ≤5 µg/L | 1 (Reference) | 1 (Reference) | ||
| >5 µg/L | 0.96 (0.17–5.20) | 0.960 | 1.31 (0.21–7.98) | 0.767 |
| Metastasis | ||||
| No | 1 (Reference) | 1 (Reference) | ||
| Yes | 3.23 (0.55–18.77) | 0.191 | 1.94 (0.31–12.08) | 0.476 |
| IF1 | ||||
| Low | 1 (Reference) | 1 (Reference) | ||
| High | 0.250 (0.08–0.76) | 0.015 | 0.32 (0.10–0.99) | 0.049 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Llorente, L.; Santacatterina, F.; García-Aguilar, A.; Nuevo-Tapioles, C.; González-García, S.; Tirpakova, Z.; Toribio, M.L.; Cuezva, J.M. Overexpression of Mitochondrial IF1 Prevents Metastatic Disease of Colorectal Cancer by Enhancing Anoikis and Tumor Infiltration of NK Cells. Cancers 2020, 12, 22. https://doi.org/10.3390/cancers12010022
González-Llorente L, Santacatterina F, García-Aguilar A, Nuevo-Tapioles C, González-García S, Tirpakova Z, Toribio ML, Cuezva JM. Overexpression of Mitochondrial IF1 Prevents Metastatic Disease of Colorectal Cancer by Enhancing Anoikis and Tumor Infiltration of NK Cells. Cancers. 2020; 12(1):22. https://doi.org/10.3390/cancers12010022
Chicago/Turabian StyleGonzález-Llorente, Lucía, Fulvio Santacatterina, Ana García-Aguilar, Cristina Nuevo-Tapioles, Sara González-García, Zuzana Tirpakova, María Luisa Toribio, and José M. Cuezva. 2020. "Overexpression of Mitochondrial IF1 Prevents Metastatic Disease of Colorectal Cancer by Enhancing Anoikis and Tumor Infiltration of NK Cells" Cancers 12, no. 1: 22. https://doi.org/10.3390/cancers12010022
APA StyleGonzález-Llorente, L., Santacatterina, F., García-Aguilar, A., Nuevo-Tapioles, C., González-García, S., Tirpakova, Z., Toribio, M. L., & Cuezva, J. M. (2020). Overexpression of Mitochondrial IF1 Prevents Metastatic Disease of Colorectal Cancer by Enhancing Anoikis and Tumor Infiltration of NK Cells. Cancers, 12(1), 22. https://doi.org/10.3390/cancers12010022