Ex-Vivo Treatment of Tumor Tissue Slices as a Predictive Preclinical Method to Evaluate Targeted Therapies for Patients with Renal Carcinoma
Abstract
1. Introduction
2. Results
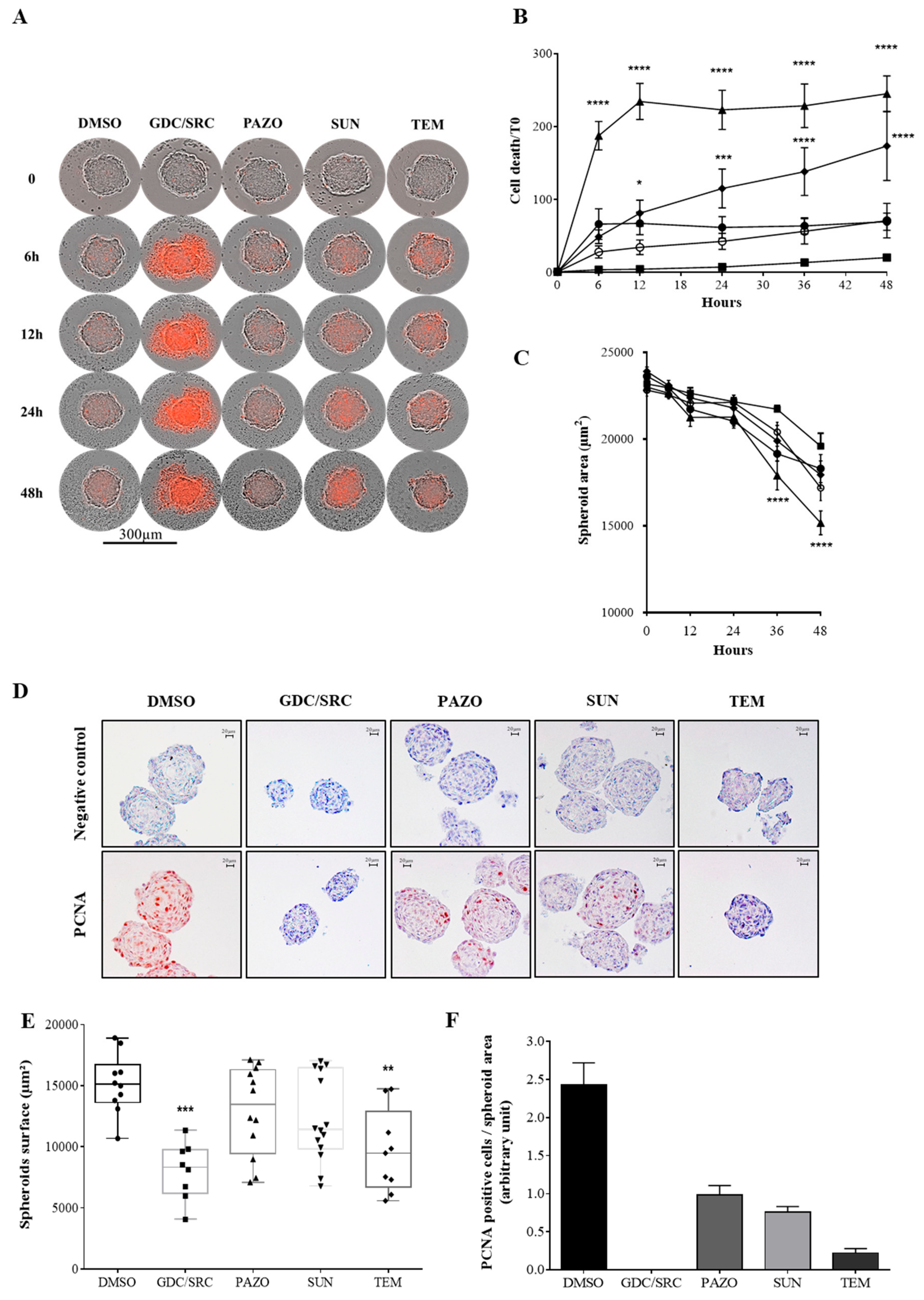
2.1. Evaluation of Drug Sensitivity on 786-0 Cell-Derived Spheroids
2.2. Tissue Slice Cultures of Renal Tumors
2.3. The Cytotoxic Effects of Drug Treatments Can Be Evaluated in Tissue-Slice Cultures
2.4. Acute Ex-Vivo Drug Treatments Identify Renal Tumor Subsets with Distinct Therapeutic Profiles
2.5. Predictive Biomarkers in Renal Tumor Slice Cultures
2.6. Prediction of Potential Correlations between Drug Sensitivity Responses and Tumor Immune Infiltration
3. Discussion
4. Materials and Methods
4.1. Reagents, Drugs and Antibodies
4.2. 3D-Spheroid Culture and Live Cell Tracking
4.3. Mice Orthotopic Tumor Xenograft Models
4.4. Patients and Clinical Samples
4.5. Preparation of Tissue Slices and Organotypic Culture
4.6. Slice Viability Assay
4.7. Immunohistochemistry Analysis
4.8. Statistical and Correlation Analyses
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Negrier, S.; Escudier, B.; Lasset, C.; Douillard, J.Y.; Savary, J.; Chevreau, C.; Ravaud, A.; Mercatello, A.; Peny, J.; Mousseau, M.; et al. Recombinant human interleukin-2, recombinant human interferon alfa-2a, or both in metastatic renal-cell carcinoma. Groupe Francais d’Immunotherapie. N. Engl. J. Med. 1998, 338, 1272–1278. [Google Scholar] [CrossRef]
- Figlin, R.; Sternberg, C.; Wood, C.G. Novel agents and approaches for advanced renal cell carcinoma. J. Urol. 2012, 188, 707–715. [Google Scholar] [CrossRef]
- Atkins, M.B.; Clark, J.I.; Quinn, D.I. Immune checkpoint inhibitors in advanced renal cell carcinoma: Experience to date and future directions. Ann. Oncol. Off. J. Eur. Soc. Med Oncol. 2017, 28, 1484–1494. [Google Scholar] [CrossRef]
- Motzer, R.J.; Penkov, K.; Haanen, J.; Rini, B.; Albiges, L.; Campbell, M.T.; Venugopal, B.; Kollmannsberger, C.; Negrier, S.; Uemura, M.; et al. Avelumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2019, 380, 1103–1115. [Google Scholar] [CrossRef] [PubMed]
- Rini, B.I.; Plimack, E.R.; Stus, V.; Gafanov, R.; Hawkins, R.; Nosov, D.; Pouliot, F.; Alekseev, B.; Soulieres, D.; Melichar, B.; et al. Pembrolizumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2019, 380, 1116–1127. [Google Scholar] [CrossRef] [PubMed]
- Cella, D.; Grunwald, V.; Escudier, B.; Hammers, H.J.; George, S.; Nathan, P.; Grimm, M.O.; Rini, B.I.; Doan, J.; Ivanescu, C.; et al. Patient-reported outcomes of patients with advanced renal cell carcinoma treated with nivolumab plus ipilimumab versus sunitinib (CheckMate 214): A randomised, phase 3 trial. Lancet Oncol. 2019, 20, 297–310. [Google Scholar] [CrossRef]
- Garnett, M.J.; Edelman, E.J.; Heidorn, S.J.; Greenman, C.D.; Dastur, A.; Lau, K.W.; Greninger, P.; Thompson, I.R.; Luo, X.; Soares, J.; et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012, 483, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. [Google Scholar] [CrossRef]
- Basu, A.; Bodycombe, N.E.; Cheah, J.H.; Price, E.V.; Liu, K.; Schaefer, G.I.; Ebright, R.Y.; Stewart, M.L.; Ito, D.; Wang, S.; et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 2013, 154, 1151–1161. [Google Scholar] [CrossRef]
- Holbeck, S.L.; Collins, J.M.; Doroshow, J.H. Analysis of Food and Drug Administration-approved anticancer agents in the NCI60 panel of human tumor cell lines. Mol. Cancer Ther. 2010, 9, 1451–1460. [Google Scholar] [CrossRef] [PubMed]
- Garnett, M.J.; McDermott, U. The evolving role of cancer cell line-based screens to define the impact of cancer genomes on drug response. Curr. Opin. Genet. Dev. 2014, 24, 114–119. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef] [PubMed]
- Iorio, F.; Knijnenburg, T.A.; Vis, D.J.; Bignell, G.R.; Menden, M.P.; Schubert, M.; Aben, N.; Goncalves, E.; Barthorpe, S.; Lightfoot, H.; et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016, 166, 740–754. [Google Scholar] [CrossRef] [PubMed]
- Koerfer, J.; Kallendrusch, S.; Merz, F.; Wittekind, C.; Kubick, C.; Kassahun, W.T.; Schumacher, G.; Moebius, C.; Gassler, N.; Schopow, N.; et al. Organotypic slice cultures of human gastric and esophagogastric junction cancer. Cancer Med. 2016, 5, 1444–1453. [Google Scholar] [CrossRef]
- Merz, F.; Gaunitz, F.; Dehghani, F.; Renner, C.; Meixensberger, J.; Gutenberg, A.; Giese, A.; Schopow, K.; Hellwig, C.; Schafer, M.; et al. Organotypic slice cultures of human glioblastoma reveal different susceptibilities to treatments. Neuro Oncol. 2013, 15, 670–681. [Google Scholar] [CrossRef]
- Senkowski, W.; Zhang, X.; Olofsson, M.H.; Isacson, R.; Hoglund, U.; Gustafsson, M.; Nygren, P.; Linder, S.; Larsson, R.; Fryknas, M. Three-Dimensional Cell Culture-Based Screening Identifies the Anthelmintic Drug Nitazoxanide as a Candidate for Treatment of Colorectal Cancer. Mol. Cancer Ther. 2015, 14, 1504–1516. [Google Scholar] [CrossRef]
- Sachs, N.; Clevers, H. Organoid cultures for the analysis of cancer phenotypes. Curr. Opin. Genet. Dev. 2014, 24, 68–73. [Google Scholar] [CrossRef]
- Weeber, F.; Ooft, S.N.; Dijkstra, K.K.; Voest, E.E. Tumor Organoids as a Pre-clinical Cancer Model for Drug Discovery. Cell Chem. Biol. 2017, 24, 1092–1100. [Google Scholar] [CrossRef]
- Bleijs, M.; van de Wetering, M.; Clevers, H.; Drost, J. Xenograft and organoid model systems in cancer research. EMBO J. 2019, 38, e101654. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Pauli, C.; Hopkins, B.D.; Prandi, D.; Shaw, R.; Fedrizzi, T.; Sboner, A.; Sailer, V.; Augello, M.; Puca, L.; Rosati, R.; et al. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discov. 2017, 7, 462–477. [Google Scholar] [CrossRef] [PubMed]
- Lang, H.; Beraud, C.; Bethry, A.; Danilin, S.; Lindner, V.; Coquard, C.; Rothhut, S.; Massfelder, T. Establishment of a large panel of patient-derived preclinical models of human renal cell carcinoma. Oncotarget 2016, 7, 59336–59359. [Google Scholar] [CrossRef] [PubMed]
- Morgan, K.M.; Riedlinger, G.M.; Rosenfeld, J.; Ganesan, S.; Pine, S.R. Patient-Derived Xenograft Models of Non-Small Cell Lung Cancer and Their Potential Utility in Personalized Medicine. Front. Oncol. 2017, 7, 2. [Google Scholar] [CrossRef]
- Maeda, H.; Khatami, M. Analyses of repeated failures in cancer therapy for solid tumors: Poor tumor-selective drug delivery, low therapeutic efficacy and unsustainable costs. Clin. Transl. Med. 2018, 7, 11. [Google Scholar] [CrossRef]
- Wong, C.C.; Cheng, K.W.; Rigas, B. Preclinical predictors of anticancer drug efficacy: Critical assessment with emphasis on whether nanomolar potency should be required of candidate agents. J. Pharmacol. Exp. Ther. 2012, 341, 572–578. [Google Scholar] [CrossRef]
- Ward, C.; Meehan, J.; Gray, M.; Kunkler, I.H.; Langdon, S.P.; Murray, A.; Argyle, D. Preclinical Organotypic Models for the Assessment of Novel Cancer Therapeutics and Treatment. Curr. Top. Microbiol. Immunol. 2019. [Google Scholar] [CrossRef]
- Altman, R.B. Predicting cancer drug response: Advancing the DREAM. Cancer Discov. 2015, 5, 237–238. [Google Scholar] [CrossRef][Green Version]
- Guyot, C.; Combe, C.; Clouzeau-Girard, H.; Moronvalle-Halley, V.; Desmouliere, A. Specific activation of the different fibrogenic cells in rat cultured liver slices mimicking in vivo situations. Virchows Arch. 2007, 450, 503–512. [Google Scholar] [CrossRef]
- Schmeichel, K.L.; Bissell, M.J. Modeling tissue-specific signaling and organ function in three dimensions. J. Cell. Sci. 2003, 116, 2377–2388. [Google Scholar] [CrossRef]
- Vaira, V.; Fedele, G.; Pyne, S.; Fasoli, E.; Zadra, G.; Bailey, D.; Snyder, E.; Faversani, A.; Coggi, G.; Flavin, R.; et al. Preclinical model of organotypic culture for pharmacodynamic profiling of human tumors. Proc. Natl. Acad. Sci. USA 2010, 107, 8352–8356. [Google Scholar] [CrossRef] [PubMed]
- De Hoogt, R.; Estrada, M.F.; Vidic, S.; Davies, E.J.; Osswald, A.; Barbier, M.; Santo, V.E.; Gjerde, K.; van Zoggel, H.; Blom, S.; et al. Protocols and characterization data for 2D, 3D, and slice-based tumor models from the PREDECT project. Sci. Data 2017, 4, 170170. [Google Scholar] [CrossRef] [PubMed]
- Misra, S.; Moro, C.F.; Del Chiaro, M.; Pouso, S.; Sebestyen, A.; Lohr, M.; Bjornstedt, M.; Verbeke, C.S. Ex vivo organotypic culture system of precision-cut slices of human pancreatic ductal adenocarcinoma. Sci. Rep. 2019, 9, 2133. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, M.M.; Merz, F.; Wichmann, G.; Kubick, C.; Wittekind, C.; Lordick, F.; Dietz, A.; Bechmann, I. Slice cultures from head and neck squamous cell carcinoma: A novel test system for drug susceptibility and mechanisms of resistance. Br. J. Cancer 2014, 110, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Marciniak, A.; Cohrs, C.M.; Tsata, V.; Chouinard, J.A.; Selck, C.; Stertmann, J.; Reichelt, S.; Rose, T.; Ehehalt, F.; Weitz, J.; et al. Using pancreas tissue slices for in situ studies of islet of Langerhans and acinar cell biology. Nat. Protoc. 2014, 9, 2809–2822. [Google Scholar] [CrossRef]
- Rebours, V.; Albuquerque, M.; Sauvanet, A.; Ruszniewski, P.; Levy, P.; Paradis, V.; Bedossa, P.; Couvelard, A. Hypoxia pathways and cellular stress activate pancreatic stellate cells: Development of an organotypic culture model of thick slices of normal human pancreas. PLoS ONE 2013, 8, e76229. [Google Scholar] [CrossRef]
- Kang, C.; Qiao, Y.; Li, G.; Baechle, K.; Camelliti, P.; Rentschler, S.; Efimov, I.R. Human Organotypic Cultured Cardiac Slices: New Platform For High Throughput Preclinical Human Trials. Sci. Rep. 2016, 6, 28798. [Google Scholar] [CrossRef]
- Jiang, T.; Zhou, C.; Ren, S. Role of IL-2 in cancer immunotherapy. Oncoimmunology 2016, 5, e1163462. [Google Scholar] [CrossRef]
- Roelants, C.; Giacosa, S.; Pillet, C.; Bussat, R.; Champelovier, P.; Bastien, O.; Guyon, L.; Arnoux, V.; Cochet, C.; Filhol, O. Combined inhibition of PI3K and Src kinases demonstrates synergistic therapeutic efficacy in clear-cell renal carcinoma. Oncotarget 2018, 9, 30066–30078. [Google Scholar] [CrossRef]
- Ricketts, C.J.; Linehan, W.M. Multi-regional Sequencing Elucidates the Evolution of Clear Cell Renal Cell Carcinoma. Cell 2018, 173, 540–542. [Google Scholar] [CrossRef]
- Kaelin, W.G., Jr. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat. Rev. Cancer 2008, 8, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Ricketts, C.J.; Crooks, D.R.; Linehan, W.M. Targeting HIF2alpha in Clear-Cell Renal Cell Carcinoma. Cancer Cell 2016, 30, 515–517. [Google Scholar] [CrossRef] [PubMed]
- Webster, W.S.; Lohse, C.M.; Thompson, R.H.; Dong, H.; Frigola, X.; Dicks, D.L.; Sengupta, S.; Frank, I.; Leibovich, B.C.; Blute, M.L.; et al. Mononuclear cell infiltration in clear-cell renal cell carcinoma independently predicts patient survival. Cancer 2006, 107, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Vuong, L.; Kotecha, R.R.; Voss, M.H.; Hakimi, A.A. Tumor Microenvironment Dynamics in Clear-Cell Renal Cell Carcinoma. Cancer Discov. 2019, 9, 1349–1357. [Google Scholar] [CrossRef]
- Kraus, V.B. Biomarkers as drug development tools: Discovery, validation, qualification and use. Nat. Rev. Rheumatol. 2018, 14, 354–362. [Google Scholar] [CrossRef]
- Motzer, R.J.; Tannir, N.M.; McDermott, D.F.; Aren Frontera, O.; Melichar, B.; Choueiri, T.K.; Plimack, E.R.; Barthelemy, P.; Porta, C.; George, S.; et al. Nivolumab plus Ipilimumab versus Sunitinib in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2018, 378, 1277–1290. [Google Scholar] [CrossRef]
- Rheinlander, A.; Schraven, B.; Bommhardt, U. CD45 in human physiology and clinical medicine. Immunol. Lett. 2018, 196, 22–32. [Google Scholar] [CrossRef]
- Wu, P.; Wu, D.; Li, L.; Chai, Y.; Huang, J. PD-L1 and Survival in Solid Tumors: A Meta-Analysis. PLoS ONE 2015, 10, e0131403. [Google Scholar] [CrossRef]
- Thompson, R.H.; Gillett, M.D.; Cheville, J.C.; Lohse, C.M.; Dong, H.; Webster, W.S.; Chen, L.; Zincke, H.; Blute, M.L.; Leibovich, B.C.; et al. Costimulatory molecule B7-H1 in primary and metastatic clear cell renal cell carcinoma. Cancer 2005, 104, 2084–2091. [Google Scholar] [CrossRef]
- Thompson, R.H.; Dong, H.; Lohse, C.M.; Leibovich, B.C.; Blute, M.L.; Cheville, J.C.; Kwon, E.D. PD-1 is expressed by tumor-infiltrating immune cells and is associated with poor outcome for patients with renal cell carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2007, 13, 1757–1761. [Google Scholar] [CrossRef]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.; Robert, L.; Chmielowski, B.; Spasic, M.; Henry, G.; Ciobanu, V.; et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Guertl, B.; Senanayake, U.; Nusshold, E.; Leuschner, I.; Mannweiler, S.; Ebner, B.; Hoefler, G. Lim1, an embryonal transcription factor, is absent in multicystic renal dysplasia, but reactivated in nephroblastomas. Pathobiology 2011, 78, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Hamaidi, I.; Coquard, C.; Danilin, S.; Dormoy, V.; Beraud, C.; Rothhut, S.; Barthelmebs, M.; Benkirane-Jessel, N.; Lindner, V.; Lang, H.; et al. The Lim1 oncogene as a new therapeutic target for metastatic human renal cell carcinoma. Oncogene 2019, 38, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Schnizlein-Bick, C.T.; Mandy, F.F.; O’Gorman, M.R.; Paxton, H.; Nicholson, J.K.; Hultin, L.E.; Gelman, R.S.; Wilkening, C.L.; Livnat, D. Use of CD45 gating in three and four-color flow cytometric immunophenotyping: Guideline from the National Institute of Allergy and Infectious Diseases, Division of AIDS. Cytometry 2002, 50, 46–52. [Google Scholar] [CrossRef]
- Zerdes, I.; Matikas, A.; Bergh, J.; Rassidakis, G.Z.; Foukakis, T. Genetic, transcriptional and post-translational regulation of the programmed death protein ligand 1 in cancer: Biology and clinical correlations. Oncogene 2018, 37, 4639–4661. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.Y.; Subjeck, J.R.; Shrikant, P.A.; Kim, H.L. Temsirolimus, an mTOR inhibitor, enhances anti-tumour effects of heat shock protein cancer vaccines. Br. J. Cancer 2011, 104, 643–652. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria; Available online: https://www.R-project.org/ (accessed on 17 February 2018).
- Dienstmann, R.; Tabernero, J. Cancer: A precision approach to tumour treatment. Nature 2017, 548, 40–41. [Google Scholar] [CrossRef]
- Jiang, X.; Seo, Y.D.; Chang, J.H.; Coveler, A.; Nigjeh, E.N.; Pan, S.; Jalikis, F.; Yeung, R.S.; Crispe, I.N.; Pillarisetty, V.G. Long-lived pancreatic ductal adenocarcinoma slice cultures enable precise study of the immune microenvironment. Oncoimmunology 2017, 6, e1333210. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roelants, C.; Pillet, C.; Franquet, Q.; Sarrazin, C.; Peilleron, N.; Giacosa, S.; Guyon, L.; Fontanell, A.; Fiard, G.; Long, J.-A.; et al. Ex-Vivo Treatment of Tumor Tissue Slices as a Predictive Preclinical Method to Evaluate Targeted Therapies for Patients with Renal Carcinoma. Cancers 2020, 12, 232. https://doi.org/10.3390/cancers12010232
Roelants C, Pillet C, Franquet Q, Sarrazin C, Peilleron N, Giacosa S, Guyon L, Fontanell A, Fiard G, Long J-A, et al. Ex-Vivo Treatment of Tumor Tissue Slices as a Predictive Preclinical Method to Evaluate Targeted Therapies for Patients with Renal Carcinoma. Cancers. 2020; 12(1):232. https://doi.org/10.3390/cancers12010232
Chicago/Turabian StyleRoelants, Caroline, Catherine Pillet, Quentin Franquet, Clément Sarrazin, Nicolas Peilleron, Sofia Giacosa, Laurent Guyon, Amina Fontanell, Gaëlle Fiard, Jean-Alexandre Long, and et al. 2020. "Ex-Vivo Treatment of Tumor Tissue Slices as a Predictive Preclinical Method to Evaluate Targeted Therapies for Patients with Renal Carcinoma" Cancers 12, no. 1: 232. https://doi.org/10.3390/cancers12010232
APA StyleRoelants, C., Pillet, C., Franquet, Q., Sarrazin, C., Peilleron, N., Giacosa, S., Guyon, L., Fontanell, A., Fiard, G., Long, J.-A., Descotes, J.-L., Cochet, C., & Filhol, O. (2020). Ex-Vivo Treatment of Tumor Tissue Slices as a Predictive Preclinical Method to Evaluate Targeted Therapies for Patients with Renal Carcinoma. Cancers, 12(1), 232. https://doi.org/10.3390/cancers12010232






