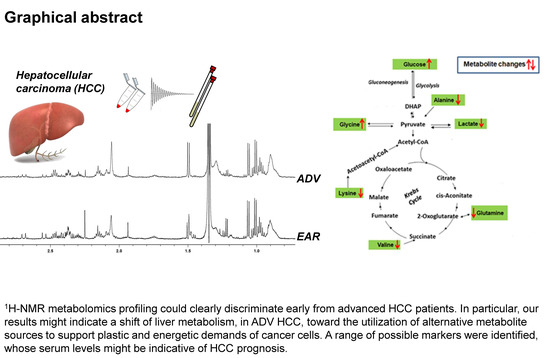
1H-NMR Based Serum Metabolomics Highlights Different Specific Biomarkers between Early and Advanced Hepatocellular Carcinoma Stages
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics and Clinical Outcomes
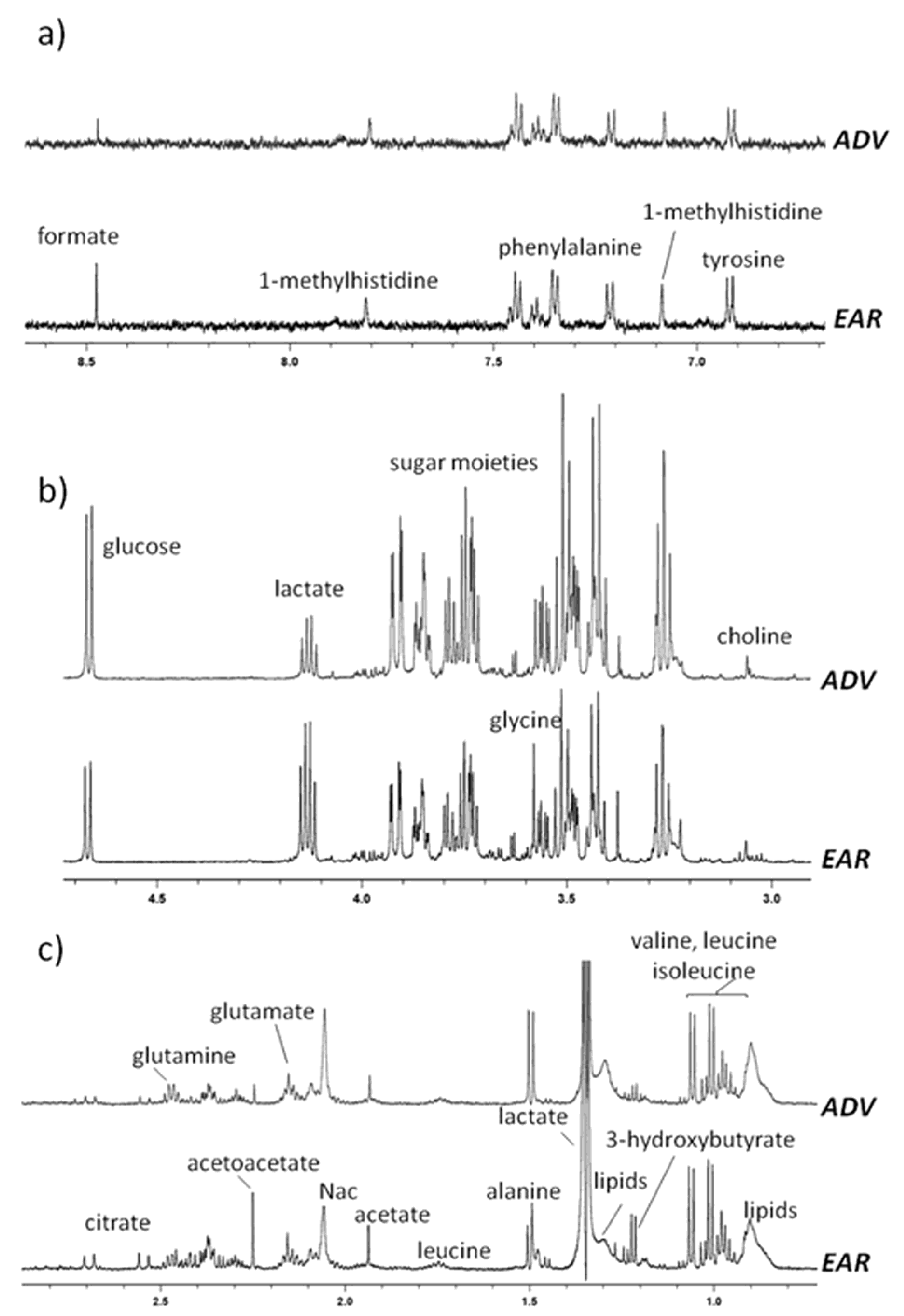
2.2. H-NMR Analysis of Serum Samples
2.3. Multivariate Analysis of NMR Data
2.4. Metabolic Pathway Analysis
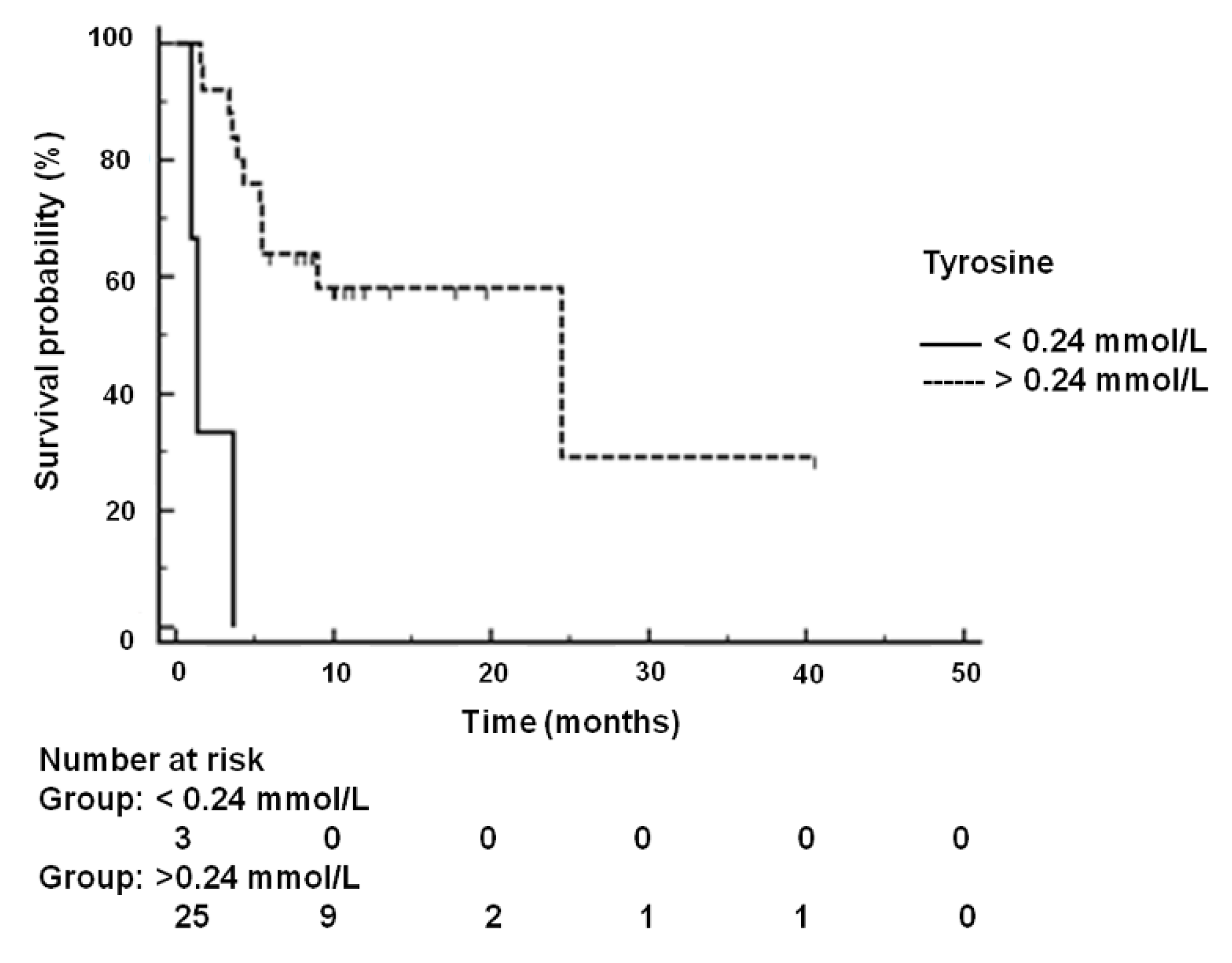
2.5. Kaplan-Meier Analysis of Disease-Free Survival and Overall Survival
3. Discussion
4. Materials and Methods
4.1. Patient Sampling
4.2. Sample Preparation and NMR Measurements
4.3. NMR Data Processing and Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Llovet, J.M.; Montal, R.; Sia, D.; Finn, R.S. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2018, 15, 599–616. [Google Scholar] [CrossRef]
- Llovet, J.M.; Villanueva, A.; Lachenmayer, A.; Finn, R.S. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat. Rev. Clin. Oncol. 2015, 12, 408–424. [Google Scholar] [CrossRef] [PubMed]
- De Matteis, S.; Ragusa, A.; Marisi, G.; De Domenico, S.; Casadei Gardini, A.; Bonafè, M.; Giudetti, A.M. Aberrant Metabolism in Hepatocellular Carcinoma Provides Diagnostic and Therapeutic Opportunities. Oxidative Med. Cell. Longev. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Vergara, D.; Casadei-Gardini, A.; Giudetti, A.M. Oxidative Molecular Mechanisms Underlying Liver Diseases: From Systems Biology to the Personalized Medicine. Oxidative Med. Cell. Longev. 2019, 2019, 7864316. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Sun, A.; Zhao, Y.; Ying, W.; Sun, H.; Yang, X.; Xing, B.; Sun, W.; Ren, L.; Hu, B. Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature 2019, 567, 257–261. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, J.; Li, J.; Feng, J.; Chen, Z.; Wang, X. Plasma metabolomic analysis of human hepatocellular carcinoma: Diagnostic and therapeutic study. Oncotarget 2016, 7, 47332. [Google Scholar] [CrossRef]
- Di Poto, C.; Ferrarini, A.; Zhao, Y.; Varghese, R.S.; Tu, C.; Zuo, Y.; Wang, M.; Ranjbar, M.R.N.; Luo, Y.; Zhang, C. Metabolomic characterization of hepatocellular carcinoma in patients with liver cirrhosis for biomarker discovery. Cancer Epidemiol. Biomark. Prev. 2017, 26, 675–683. [Google Scholar] [CrossRef]
- Fages, A.; Duarte-Salles, T.; Stepien, M.; Ferrari, P.; Fedirko, V.; Pontoizeau, C.; Trichopoulou, A.; Aleksandrova, K.; Tjønneland, A.; Olsen, A. Metabolomic profiles of hepatocellular carcinoma in a European prospective cohort. BMC Med. 2015, 13, 242. [Google Scholar] [CrossRef]
- Del Coco, L.; Vergara, D.; De Matteis, S.; Mensà, E.; Sabbatinelli, J.; Prattichizzo, F.; Bonfigli, A.R.; Storci, G.; Bravaccini, S.; Pirini, F. NMR-Based Metabolomic Approach Tracks Potential Serum Biomarkers of Disease Progression in Patients with Type 2 Diabetes Mellitus. J. Clin. Med. 2019, 8, 720. [Google Scholar] [CrossRef]
- Pinto, J.; Almeida, L.M.; Martins, A.S.; Duarte, D.; Barros, A.S.; Galhano, E.; Pita, C.; Almeida, M.d.C.; Carreira, I.M.; Gil, A.M. Prediction of gestational diabetes through NMR metabolomics of maternal blood. J. Proteome Res. 2015, 14, 2696–2706. [Google Scholar] [CrossRef]
- Goossens, C.; Nahon, P.; Le Moyec, L.; Triba, M.N.; Bouchemal, N.; Amathieu, R.; Ganne-Carrié, N.; Ziol, M.; Trinchet, J.-C.; Sellier, N. Sequential serum metabolomic profiling after radiofrequency ablation of hepatocellular carcinoma reveals different response patterns according to etiology. J. Proteome Res. 2016, 15, 1446–1454. [Google Scholar] [CrossRef] [PubMed]
- Lever, J.; Krzywinski, M.; Altman, N. Points of significance: Principal component analysis. Nat. Methods 2017, 14, 641–642. [Google Scholar] [CrossRef]
- Lu, Y.; Li, N.; Gao, L.; Xu, Y.-J.; Huang, C.; Yu, K.; Ling, Q.; Cheng, Q.; Chen, S.; Zhu, M. Acetylcarnitine is a candidate diagnostic and prognostic biomarker of hepatocellular carcinoma. Cancer Res. 2016, 76, 2912–2920. [Google Scholar] [CrossRef] [PubMed]
- El-Bacha, T.; Struchiner, C.J.; Cordeiro, M.T.; Almeida, F.C.; Marques, E.T.; Da Poian, A.T. 1H nuclear magnetic resonance metabolomics of plasma unveils liver dysfunction in dengue patients. J. Virol. 2016, 90, 7429–7443. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Cambiaghi, A.; Ferrario, M.; Masseroli, M. Analysis of metabolomic data: Tools, current strategies and future challenges for omics data integration. Brief. Bioinform. 2017, 18, 498–510. [Google Scholar] [CrossRef]
- Bharti, S.K.; Roy, R. Quantitative 1H NMR spectroscopy. TrAC Trends Anal. Chem. 2012, 35, 5–26. [Google Scholar] [CrossRef]
- Intlekofer, A.M.; Finley, L.W. Metabolic signatures of cancer cells and stem cells. Nat. Metab. 2019, 1, 177–188. [Google Scholar] [CrossRef]
- Ortmayr, K.; Dubuis, S.; Zampieri, M. Metabolic profiling of cancer cells reveals genome-wide crosstalk between transcriptional regulators and metabolism. Nat. Commun. 2019, 10, 1841. [Google Scholar] [CrossRef]
- Girolimetti, G.; Guerra, F.; Iommarini, L.; Kurelac, I.; Vergara, D.; Maffia, M.; Vidone, M.; Amato, L.B.; Leone, G.; Dusi, S. Platinum-induced mitochondrial DNA mutations confer lower sensitivity to paclitaxel by impairing tubulin cytoskeletal organization. Hum. Mol. Genet. 2017, 26, 2961–2974. [Google Scholar] [CrossRef] [PubMed]
- Giudetti, A.M.; De Domenico, S.; Ragusa, A.; Lunetti, P.; Gaballo, A.; Franck, J.; Simeone, P.; Nicolardi, G.; De Nuccio, F.; Santino, A. A specific lipid metabolic profile is associated with the epithelial mesenchymal transition program. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2019, 1864, 344–357. [Google Scholar] [CrossRef] [PubMed]
- Lunetti, P.; Di Giacomo, M.; Vergara, D.; De Domenico, S.; Maffia, M.; Zara, V.; Capobianco, L.; Ferramosca, A. Metabolic reprogramming in breast cancer results in distinct mitochondrial bioenergetics between luminal and basal subtypes. FEBS J. 2019, 286, 688–709. [Google Scholar] [CrossRef] [PubMed]
- De Castro, F.; Benedetti, M.; Del Coco, L.; Fanizzi, F.P. NMR-Based Metabolomics in Metal-Based Drug Research. Molecules 2019, 24, 2240. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Locasale, J.W.; Bielas, J.H.; O’sullivan, J.; Sheahan, K.; Cantley, L.C.; Vander Heiden, M.G.; Vitkup, D. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat. Biotechnol. 2013, 31, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Hui, S.; Ghergurovich, J.M.; Morscher, R.J.; Jang, C.; Teng, X.; Lu, W.; Esparza, L.A.; Reya, T.; Zhan, L.; Guo, J.Y. Glucose feeds the TCA cycle via circulating lactate. Nature 2017, 551, 115–118. [Google Scholar] [CrossRef]
- Beyoğlu, D.; Imbeaud, S.; Maurhofer, O.; Bioulac-Sage, P.; Zucman-Rossi, J.; Dufour, J.F.; Idle, J.R. Tissue metabolomics of hepatocellular carcinoma: Tumor energy metabolism and the role of transcriptomic classification. Hepatology 2013, 58, 229–238. [Google Scholar] [CrossRef]
- Fitian, A.I.; Nelson, D.R.; Liu, C.; Xu, Y.; Ararat, M.; Cabrera, R. Integrated metabolomic profiling of hepatocellular carcinoma in hepatitis C cirrhosis through GC/MS and UPLC/MS-MS. Liver Int. 2014, 34, 1428–1444. [Google Scholar] [CrossRef]
- Shao, Y.; Zhu, B.; Zheng, R.; Zhao, X.; Yin, P.; Lu, X.; Jiao, B.; Xu, G.; Yao, Z. Development of urinary pseudotargeted LC-MS-based metabolomics method and its application in hepatocellular carcinoma biomarker discovery. J. Proteome Res. 2014, 14, 906–916. [Google Scholar] [CrossRef]
- Chen, T.; Xie, G.; Wang, X.; Fan, J.; Qiu, Y.; Zheng, X.; Qi, X.; Cao, Y.; Su, M.; Wang, X. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol. Cell. Proteom. 2011, 10, M110.004945. [Google Scholar] [CrossRef]
- Lee, D.-Y.; Kim, E.-H. Therapeutic Effects of Amino Acids in Liver Diseases: Current Studies and Future Perspectives. J. Cancer Prev. 2019, 24, 72. [Google Scholar] [CrossRef] [PubMed]
- Baniasadi, H.; Gowda, G.N.; Gu, H.; Zeng, A.; Zhuang, S.; Skill, N.; Maluccio, M.; Raftery, D. Targeted metabolic profiling of hepatocellular carcinoma and hepatitis C using LC-MS/MS. Electrophoresis 2013, 34, 2910–2917. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.-K.; Park, K.-G. Targeting glutamine metabolism for cancer treatment. Biomol. Ther. 2018, 26, 19–28. [Google Scholar] [CrossRef] [PubMed]
- AlSalloom, A.A.M. An update of biochemical markers of hepatocellular carcinoma. Int. J. Health Sci. 2016, 10, 121–136. [Google Scholar] [CrossRef]
- Hirayama, C.; Suyama, K.; Horie, Y.; Tanimoto, K.; Kato, S. Plasma amino acid patterns in hepatocellular carcinoma. Biochem. Med. Metab. Boil. 1987, 38, 127–133. [Google Scholar] [CrossRef]
- Amelio, I.; Cutruzzolá, F.; Antonov, A.; Agostini, M.; Melino, G. Serine and glycine metabolism in cancer. Trends Biochem. Sci. 2014, 39, 191–198. [Google Scholar] [CrossRef]
- Kampen, K.R.; Fancello, L.; Girardi, T.; Rinaldi, G.; Planque, M.; Sulima, S.O.; Loayza-Puch, F.; Verbelen, B.; Vereecke, S.; Verbeeck, J. Translatome analysis reveals altered serine and glycine metabolism in T-cell acute lymphoblastic leukemia cells. Nat. commun. 2019, 10, 2542. [Google Scholar] [CrossRef]
- Zhuang, H.; Li, Q.; Zhang, X.; Ma, X.; Wang, Z.; Liu, Y.; Yi, X.; Chen, R.; Han, F.; Zhang, N. Downregulation of glycine decarboxylase enhanced cofilin-mediated migration in hepatocellular carcinoma cells. Free. Radic. Biol. Med. 2018, 120, 1–12. [Google Scholar] [CrossRef]
- Tamanna, N.; Mahmood, N. Emerging roles of branched-chain amino acid supplementation in human diseases. Int. Sch. Res. Not. 2014, 2014, 235619. [Google Scholar] [CrossRef][Green Version]
- Chuang, J.; Yu, C.; Wang, S. Modulation of human lymphocyte proliferation by amino acids. Clin. Exp Immunol. 1990, 81, 173–176. [Google Scholar] [CrossRef]
- Kakazu, E.; Ueno, Y.; Kondo, Y.; Fukushima, K.; Shiina, M.; Inoue, J.; Tamai, K.; Ninomiya, M.; Shimosegawa, T. Branched chain amino acids enhance the maturation and function of myeloid dendritic cells ex vivo in patients with advanced cirrhosis. Hepatology 2009, 50, 1936–1945. [Google Scholar] [CrossRef] [PubMed]
- Tajiri, K.; Shimizu, Y. Branched-chain amino acids in liver diseases. World J. Gastroenterol. WJG 2013, 19, 7620. [Google Scholar] [CrossRef] [PubMed]
- Hakuno, D.; Hamba, Y.; Toya, T.; Adachi, T. Plasma amino acid profiling identifies specific amino acid associations with cardiovascular function in patients with systolic heart failure. PLoS ONE 2015, 10, e0117325. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.L.; Wu, S.T.; Yin, M.C.; Chen, H.T.; Chen, H.C. Protective effects from carnosine and histidine on acetaminophen-induced liver injury. J. Food Sci. 2009, 74, H259–H265. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Nakamura, Y.; Inaba, Y.; Matsumoto, M.; Kido, Y.; Asahara, S.; Matsuda, T.; Watanabe, H.; Maeda, A.; Inagaki, F. Histidine augments the suppression of hepatic glucose production by central insulin action. Diabetes 2013, 62, 2266–2277. [Google Scholar] [CrossRef] [PubMed]
- Mong, M.C.; Chao, C.Y.; Yin, M.C. Histidine and carnosine alleviated hepatic steatosis in mice consumed high saturated fat diet. Eur. J. Pharmacol. 2011, 653, 82–88. [Google Scholar] [CrossRef]
- Liu, T.; Shang, S.; Li, W.; Qin, X.; Sun, L.; Zhang, S.; Liu, Y. Assessment of hepatocellular carcinoma metastasis glycobiomarkers using advanced quantitative N-glycoproteome analysis. Front. Physiol. 2017, 8, 472. [Google Scholar] [CrossRef]
- Sonveaux, P.; Végran, F.; Schroeder, T.; Wergin, M.C.; Verrax, J.; Rabbani, Z.N.; De Saedeleer, C.J.; Kennedy, K.M.; Diepart, C.; Jordan, B.F. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J. Clin. Investig. 2008, 118, 3930–3942. [Google Scholar] [CrossRef]
- Kim, W.; Yoon, J.-H.; Jeong, J.-M.; Cheon, G.-J.; Lee, T.-S.; Yang, J.-I.; Park, S.-C.; Lee, H.-S. Apoptosis-inducing antitumor efficacy of hexokinase II inhibitor in hepatocellular carcinoma. Mol. Cancer Ther. 2007, 6, 2554–2562. [Google Scholar] [CrossRef]
- Palmieri, D.; Fitzgerald, D.; Shreeve, S.M.; Hua, E.; Bronder, J.L.; Weil, R.J.; Davis, S.; Stark, A.M.; Merino, M.J.; Kurek, R. Analyses of resected human brain metastases of breast cancer reveal the association between up-regulation of hexokinase 2 and poor prognosis. Mol. Cancer Res. 2009, 7, 1438–1445. [Google Scholar] [CrossRef]
- Goodwin, M.L.; Pennington, Z.; Westbroek, E.M.; Cottrill, E.; Ahmed, A.K.; Sciubba, D.M. Lactate and cancer: A “lactatic” perspective on spinal tumor metabolism (part 1). Ann. Transl. Med. 2019, 7, 220. [Google Scholar] [CrossRef] [PubMed]
- Heberer, M.; Talke, H.; Maier, K.; Gerok, W. Metabolism of phenylalanine in liver diseases. Klin. Wochenschr. 1980, 58, 1189–1196. [Google Scholar] [CrossRef] [PubMed]
- Tessari, P.; Vettore, M.; Millioni, R.; Puricelli, L.; Orlando, R. Effect of liver cirrhosis on phenylalanine and tyrosine metabolism. Curr. Opin. Clin. Nutr. Metab. Care 2010, 13, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Beckonert, O.; Keun, H.C.; Ebbels, T.M.; Bundy, J.; Holmes, E.; Lindon, J.C.; Nicholson, J.K. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat. Protoc. 2007, 2, 2692. [Google Scholar] [CrossRef]
- Guleria, A.; Pratap, A.; Dubey, D.; Rawat, A.; Chaurasia, S.; Sukesh, E.; Phatak, S.; Ajmani, S.; Kumar, U.; Khetrapal, C.L. NMR based serum metabolomics reveals a distinctive signature in patients with Lupus Nephritis. Sci. Rep. 2016, 6, 35309. [Google Scholar] [CrossRef]
- Kostidis, S.; Addie, R.D.; Morreau, H.; Mayboroda, O.A.; Giera, M. Quantitative NMR analysis of intra-and extracellular metabolism of mammalian cells: A tutorial. Anal. Chim. Acta 2017, 980, 1–24. [Google Scholar] [CrossRef]
- Eastment, H.; Krzanowski, W. Cross-validatory choice of the number of components from a principal component analysis. Technometrics 1982, 24, 73–77. [Google Scholar] [CrossRef]
- Trygg, J.; Wold, S. Orthogonal projections to latent structures (O-PLS). J. Chemom. J. Chemom. Soc. 2002, 16, 119–128. [Google Scholar] [CrossRef]
- Bro, R.; Kjeldahl, K.; Smilde, A.; Kiers, H. Cross-validation of component models: A critical look at current methods. Anal. Bioanal. Chem. 2008, 390, 1241–1251. [Google Scholar] [CrossRef]
- Consonni, R.; Cagliani, L.; Benevelli, F.; Spraul, M.; Humpfer, E.; Stocchero, M. NMR and chemometric methods: A powerful combination for characterization of balsamic and traditional balsamic vinegar of Modena. Anal. Chim. Acta 2008, 611, 31–40. [Google Scholar] [CrossRef]
- Girelli, C.R.; Del Coco, L.; Fanizzi, F.P. 1H NMR spectroscopy and multivariate analysis as possible tool to assess cultivars, from specific geographical areas, in EVOOs. Eur. J. Lipid Sci. Technol. 2016, 118, 1380–1388. [Google Scholar] [CrossRef]
- Liu, Y.; Hong, Z.; Tan, G.; Dong, X.; Yang, G.; Zhao, L.; Chen, X.; Zhu, Z.; Lou, Z.; Qian, B. NMR and LC/MS-based global metabolomics to identify serum biomarkers differentiating hepatocellular carcinoma from liver cirrhosis. Int. J. Cancer 2014, 135, 658–668. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. MetPA: A web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 2010, 26, 2342–2344. [Google Scholar] [CrossRef] [PubMed]
- Dunne, R.F.; Loh, K.P.; Williams, G.R.; Jatoi, A.; Mustian, K.M.; Mohile, S.G. Cachexia and Sarcopenia in Older Adults with Cancer: A Comprehensive Review. Cancers 2019, 11, 1861. [Google Scholar] [CrossRef] [PubMed]


| Patient’s Characteristics | Patients Recommended to Radiofrequency (Early Stage) (n = 28) | Patients Recommended to Sorafenib (Advanced Stage) (n = 36) |
|---|---|---|
| Median age (range) | 65 (38–86) | 70 (67–71) |
| Gender | ||
| Male | 25 (89.3%) | 32 (88.7%) |
| Female | 3 (10.7%) | 4 (11.3%) |
| Diabetes | ||
| Yes | 6 (21.4%) | 15 (41.7%) |
| No | 22 (78.6%) | 21 (58.3%) |
| Metformin treatment | ||
| Yes | 4 (14.3%) | 9 (25%) |
| No | 24 (85.7%) | 27 (75%) |
| Etiology | ||
| HCV | 13 (46.4%) | 16 (44.4%) |
| HBV | 4 (14.3%) | 5 (13.9%) |
| NASH | 2 (7.1%) | 8 (22.2%) |
| Others | 9 (32.1%) | 7 (19.5%) |
| BCLC stage | ||
| 0/A | 28 (100%) | 0 (0%) |
| B | 0 (0%) | 16 (44.4%) |
| C | 0 (0%) | 20 (55.6%) |
| Child pugh | ||
| A | 25 (89.3%) | 32 (88.9%) |
| B | 3 (10.7%) | 4 (11.1%) |
| ECOG | ||
| 0 | 28 (100%) | 27 (75.0%) |
| >0 | 0 (0%) | 9 (25.0%) |
| Extrahepatic disease | ||
| Yes | 0 (0%) | 19 (52.8%) |
| No | 28 (100%) | 17 (47.2%) |
| Portal Vein Thrombosis | ||
| Yes | 0 (0%) | 13 (26.1%) |
| No | 28 (100%) | 23 (63.9%) |
| Metabolite | Chemical Shift (ppm) | ADV Integrals (Mean ± SD) | EAR Integrals (Mean ± SD) | Ratio ADV/EAR | p-Value |
|---|---|---|---|---|---|
| Alanine | 1.49 | 9.40 × 10−3 ± 3.61 × 10-3 | 1.55 × 10−2 ± 5.74 × 10−3 | 0.6 | 2.69 × 10−6 |
| Glycine | 3.60 | 2.42 × 10−2 ± 7.80 × 10−3 | 1.42 × 10−2 ± 8.85 × 10−3 | 1.7 | 9.94 × 10−6 |
| Glutamine | 2.47 | 8.23 × 10−3 ± 3.36 × 10−3 | 1.21 × 10−2 ± 3.52 × 10−3 | 0.7 | 1.22 × 10−5 |
| β-Glucose | 4.66 | 2.41 × 10−2 ± 9.20 × 10−3 | 1.29 × 10−2 ± 9.85 × 10−3 | 1.9 | 1.47 × 10−5 |
| α-Glucose | 5.25 | 1.75 × 10−2 ± 6.42 × 10−3 | 9.56 × 10−3 ± 7.31 × 10−3 | 1.8 | 2.11 × 10−5 |
| Galactose | 3.94 | 1.97 × 10−2 ± 5.66 × 10−3 | 1.28 × 10−2 ± 6.58 × 10−3 | 1.5 | 2.83 × 10−5 |
| 1-Methylhistidine | 7.78 | 1.67 × 10−4 ± 8.25 × 10−4 | 8.91 × 10−4 ± 3.56 × 10−4 | 0.2 | 4.49 × 10−5 |
| Lactate | 1.34 | 1.25 × 10−1 ± 7.70 × 10−2 | 2.15 × 10−1 ± 1.21 × 10−1 | 0.6 | 9.08 × 10−5 |
| Lysine | 1.74 | 4.08 × 10−4 ± 5.87 × 10−4 | 1.07 × 10−3 ± 6.74 × 10−4 | 0.4 | 1.92 × 10−4 |
| N-acetylglycoproteins | 2.06 | 2.85 × 10−2 ± 7.64 × 10−3 | 2.24 × 10−2 ± 4.88 × 10−3 | 1.3 | 4.39 × 10−4 |
| Valine | 1.04 | 1.07 × 10−2 ± 2.77 × 10−3 | 1.27 × 10−2 ± 2.37 × 10−3 | 0.8 | 3.11 × 10−3 |
| Pathway Name | Matched Metabolites | Raw p (*10−6) | = −log(p) | Holm Adjust (*10−5) | FDR (*10−5) | Impact |
|---|---|---|---|---|---|---|
| Alanine, aspartate and glutamate metabolism | alanine, glutamine (2/24) | 0.39 | 14.76 | 1.2 | 0.48 | 0.26401 |
| Glycine, serine and threonine metabolism | glycine (1/48) | 9.94 | 11.52 | 23.2 | 1.99 | 0.18774 |
| Lysine degradation | lysine, glycine (2/47) | 9.66 | 11.55 | 23.2 | 1.99 | 0.14675 |
| Aminoacyl-tRNA biosynthesis | glutamine, glycine, valine, alanine, lysine (5/75) | 0.45 | 14.62 | 1.3 | 0.48 | 0.05634 |
| Amino sugar and nucleotide sugar metabolism | N-acetyl-d-glucosamine, α-glucose (2/88) | 0.08 | 16.37 | 0.3 | 0.25 | 0.01122 |
| Pyruvate metabolism | lactate (1/32) | 0.59 | 7.44 | 328 | 64.80 | 0.13756 |
| Lysine biosynthesis | lysine (1/32) | 94.80 | 9.26 | 75.8 | 11.70 | 0.09993 |
| Taurine and hypotaurine metabolism | alanine (1/20) | 2.69 | 12.83 | 7.3 | 1.07 | 0.03237 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casadei-Gardini, A.; Del Coco, L.; Marisi, G.; Conti, F.; Rovesti, G.; Ulivi, P.; Canale, M.; Frassineti, G.L.; Foschi, F.G.; Longo, S.; et al. 1H-NMR Based Serum Metabolomics Highlights Different Specific Biomarkers between Early and Advanced Hepatocellular Carcinoma Stages. Cancers 2020, 12, 241. https://doi.org/10.3390/cancers12010241
Casadei-Gardini A, Del Coco L, Marisi G, Conti F, Rovesti G, Ulivi P, Canale M, Frassineti GL, Foschi FG, Longo S, et al. 1H-NMR Based Serum Metabolomics Highlights Different Specific Biomarkers between Early and Advanced Hepatocellular Carcinoma Stages. Cancers. 2020; 12(1):241. https://doi.org/10.3390/cancers12010241
Chicago/Turabian StyleCasadei-Gardini, Andrea, Laura Del Coco, Giorgia Marisi, Fabio Conti, Giulia Rovesti, Paola Ulivi, Matteo Canale, Giovanni Luca Frassineti, Francesco Giuseppe Foschi, Serena Longo, and et al. 2020. "1H-NMR Based Serum Metabolomics Highlights Different Specific Biomarkers between Early and Advanced Hepatocellular Carcinoma Stages" Cancers 12, no. 1: 241. https://doi.org/10.3390/cancers12010241
APA StyleCasadei-Gardini, A., Del Coco, L., Marisi, G., Conti, F., Rovesti, G., Ulivi, P., Canale, M., Frassineti, G. L., Foschi, F. G., Longo, S., Fanizzi, F. P., & Giudetti, A. M. (2020). 1H-NMR Based Serum Metabolomics Highlights Different Specific Biomarkers between Early and Advanced Hepatocellular Carcinoma Stages. Cancers, 12(1), 241. https://doi.org/10.3390/cancers12010241