Translating Biomarkers of Cholangiocarcinoma for Theranosis: A Systematic Review
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
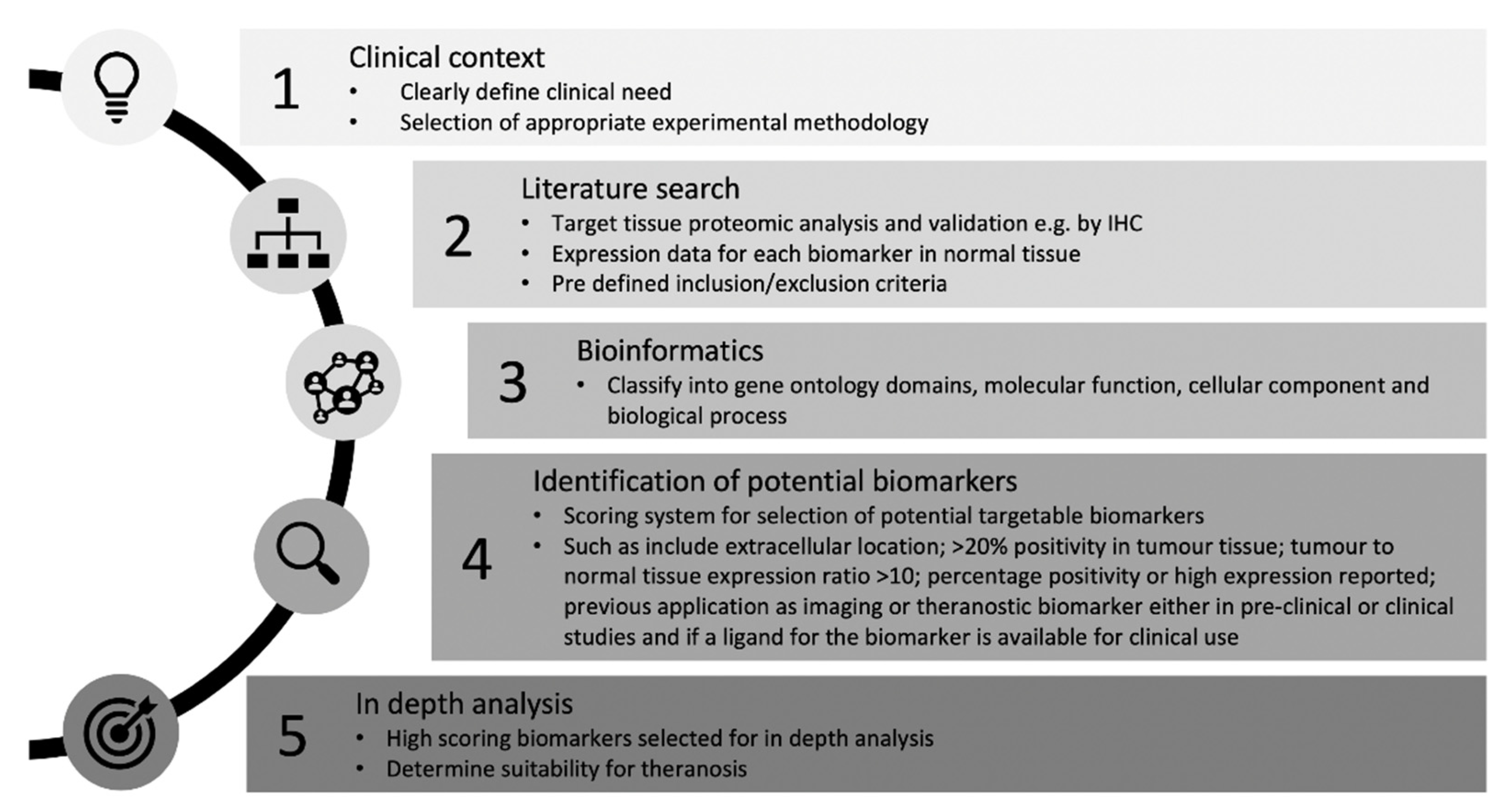
2.1. Systematic Search Strategy for Theranostic Biomarkers in CCA
2.2. Selection Criteria
2.3. Data Extraction
2.4. Bioinformatics
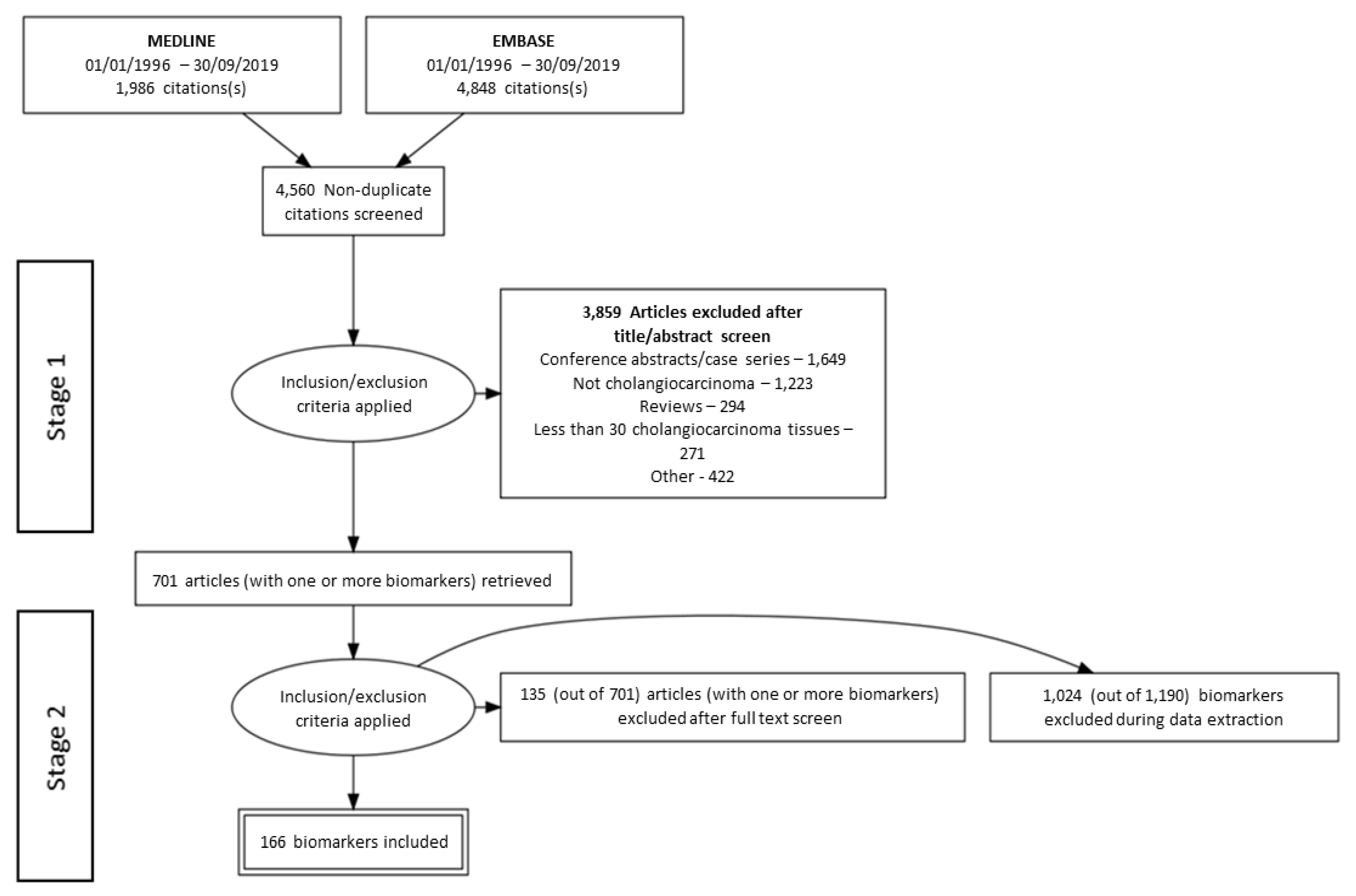
3. Results
3.1. Literature Search
3.2. Gene Ontology of Selected Candidate Biomarkers
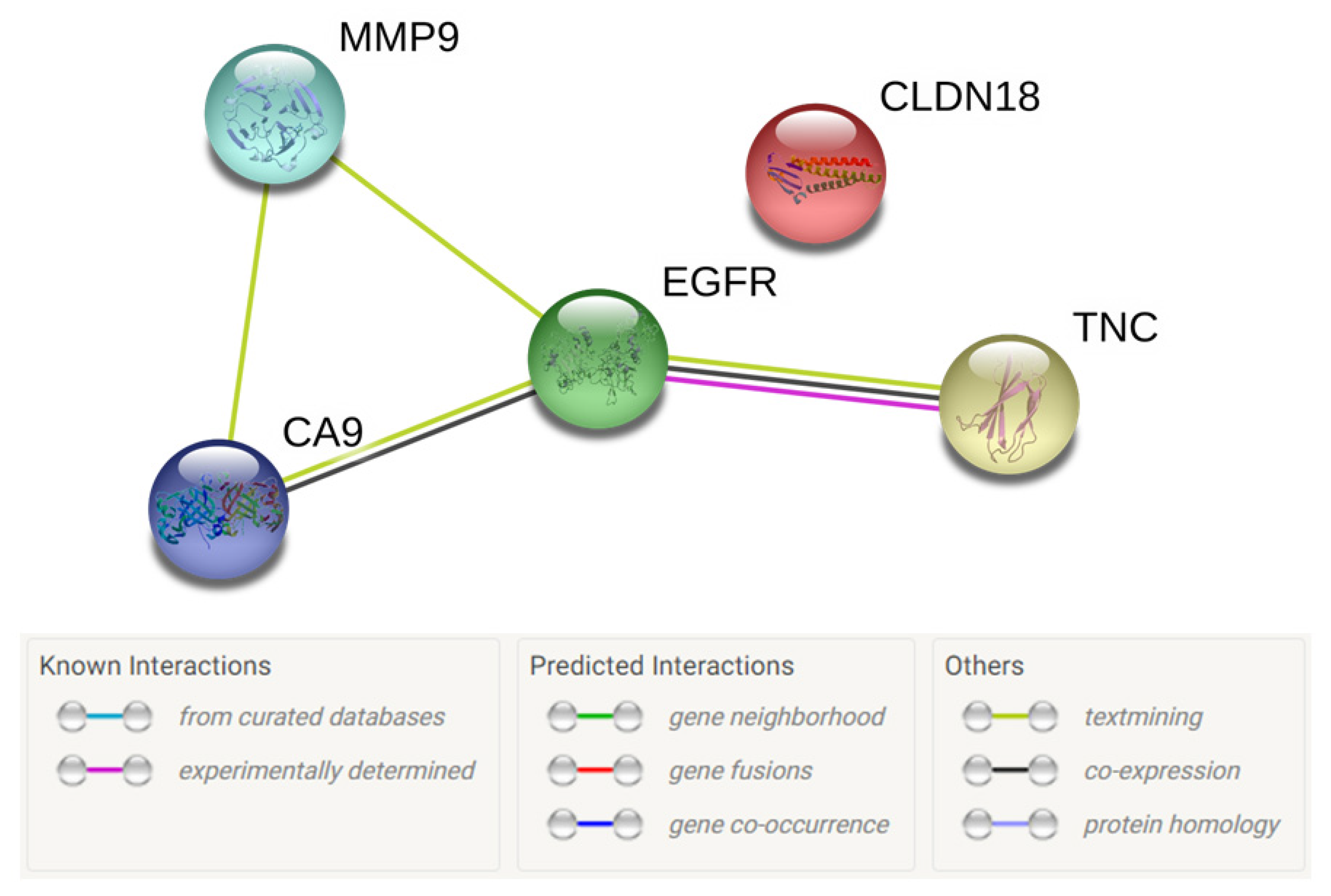
3.3. Selection of Biomarkers for Theranosis
| No. | Parameter | Score |
|---|---|---|
| 1 | Extracellular or membrane localisation of biomarker | 5 |
| 2 | >20% positivity in tumor tissue | 4 |
| 3 | Tumor to normal tissue ratio >10 | 3 |
| 4 | Percentage positivity or upregulation in tumor tissue | |
| >90% | 6 | |
| 70–90% | 5 | |
| 50–69% | 3 | |
| <49% | 0 | |
| 5 | Previous application to imaging | |
| Preclinical | 1 | |
| Clinical | 2 | |
| 6 | Ligand in human trials | 1 |
| Total | 22 | |
| No. | Tumor Biomarker | Extracellular | Membrane | Highest % Positivity/Upregulation Reported in CCA | Ligand in Human Trials | Previous Use in Pre-Clinical or Clinical Imaging | TASC-T (Max 22) |
|---|---|---|---|---|---|---|---|
| 1 | ANGPT1 | Yes | No | 43.7 | No | No | 9 |
| 2 | ANGPT2 | Yes | No | 57.6 | No | No | 9 |
| 3 | CA9 | No | Yes | 85 | Yes (Iodine-124 labeled cG250) | Yes [33] | 17 |
| 4 | CDH17 | No | Yes | 52.9 | No | No | 15 |
| 5 | CDX2 | No | No | 60 | No | No | 7 |
| 6 | CLDN18 | No | Yes | 90 | Yes (Claudiximab, Zolbetuximab) | No | 19 |
| 7 | EGFR | Yes | Yes | 75 | Yes (Cetuximab, Panitumumab) | Yes [34] | 17 |
| 8 | MMP7 | Yes | No | 80 | No | Yes [35] | 15 |
| 9 | MMP9 | Yes | No | 67 | Yes (Andecaliximab) | Yes [36] | 19 |
| 10 | SERPINA1 | Yes | No | 57 | No | No | 12 |
| 11 | SFRP1 | Yes | Yes | 60 | No | No | 12 |
| 12 | SLC2A1 | No | Yes | 52 | No | No | 12 |
| 13 | TFF1 | Yes | No | 98.4 | No | No | 15 |
| 14 | TGFB1 | Yes | No | 47 | Yes (Fresolimumab) | Yes [37] | 15 |
| 15 | TNC | Yes | No | 63.9 | Yes (Neuradiab, Tenatumomab) | Yes [38] | 18 |
| 16 | TP53 | No | No | 84 | No | Yes [39] | 13 |
3.4. Selected Biomarkers for Theranosis in CCA
3.4.1. Matrix Metalloproteinase 9 Presence in CCA
3.4.2. Claudin 18 Presence in CCA
3.4.3. Tenascin C Presence in CCA
3.4.4. Carbonic Anhydrase 9 Presence in CCA
3.4.5. Epidermal Growth Factor Receptor Presence in CCA
4. Discussion
4.1. Matrix Metalloproteinase 9
4.2. Claudin 18
4.3. Tenascin C
4.4. Carbonic Anhydrase 9
4.5. Epidermal Growth Factor Receptor
4.6. Challenges and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Patel, T. Cholangiocarcinoma. Nat. Clin. Pract. Gastroenterol. Hepatol. 2006, 3, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Razumilava, N.; Gores, G.J. Cholangiocarcinoma. Lancet 2014, 383, 2168–2179. [Google Scholar] [CrossRef]
- Buckholz, A.P.; Brown, R.S. Cholangiocarcinoma: Diagnosis and Management. Clin. Liver Dis. 2020, 24, 421–436. [Google Scholar] [CrossRef] [PubMed]
- Banales, J.M.; Marin, J.J.G.; Lamarca, A.; Rodrigues, P.M.; Khan, S.A.; Roberts, L.R.; Cardinale, V.; Carpino, G.; Andersen, J.B.; Braconi, C.; et al. Cholangiocarcinoma 2020: The next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Kamsa-ard, S.; Wiangnon, S.; Suwanrungruang, K.; Promthet, S.; Khuntikeo, N.; Kamsa-ard, S.; Mahaweerawat, S. Trends in liver cancer incidence between 1985 and 2009, Khon Kaen, Thailand: Cholangiocarcinoma. Asian Pac. J. Cancer Prev. 2011, 12, 2209–2213. [Google Scholar] [PubMed]
- Kirstein, M.M.; Vogel, A. Epidemiology and Risk Factors of Cholangiocarcinoma. Visc. Med. 2016, 32, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Tavolari, S.; Brandi, G. Cholangiocarcinoma: Epidemiology and risk factors. Liver Int. 2019, 39, 19–31. [Google Scholar] [CrossRef]
- Belkouz, A.; Wilmink, J.W.; Haj Mohammad, N.; Hagendoorn, J.; de Vos-Geelen, J.; Dejong, C.H.C.; Homs, M.Y.V.; Groot Koerkamp, B.; van Gulik, T.M.; van Oijen, M.G.H.; et al. Advances in adjuvant therapy of biliary tract cancer: An overview of current clinical evidence based on phase II and III trials. Crit. Rev. Oncol. Hematol. 2020, 151, e102975. [Google Scholar] [CrossRef]
- Valle, J.; Wasan, H.; Palmer, D.H.; Cunningham, D.; Anthoney, A.; Maraveyas, A.; Madhusudan, S.; Iveson, T.; Hughes, S.; Pereira, S.P.; et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N. Engl. J. Med. 2010, 362, 1273–1281. [Google Scholar] [CrossRef]
- Tian, W.; Hu, W.; Shi, X.; Liu, P.; Ma, X.; Zhao, W.; Qu, L.; Zhang, S.; Shi, W.; Liu, A.; et al. Comprehensive genomic profile of cholangiocarcinomas in China. Oncol. Lett. 2020, 19, 3101–3110. [Google Scholar] [CrossRef]
- Ettrich, T.J.; Schwerdel, D.; Dolnik, A.; Beuter, F.; Blatte, T.J.; Schmidt, S.A.; Stanescu-Siegmund, N.; Steinacker, J.; Marienfeld, R.; Kleger, A.; et al. Genotyping of circulating tumor DNA in cholangiocarcinoma reveals diagnostic and prognostic information. Sci. Rep. 2019, 9, e13261. [Google Scholar] [CrossRef] [PubMed]
- Pene, F.; Courtine, E.; Cariou, A.; Mira, J.P. Toward theragnostics. Crit. Care Med. 2009, 37, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Idee, J.M.; Louguet, S.; Ballet, S.; Corot, C. Theranostics and contrast-agents for medical imaging: A pharmaceutical company viewpoint. Quant. Imaging Med. Surg. 2013, 3, 292–297. [Google Scholar] [CrossRef]
- Jeelani, S.; Reddy, R.C.; Maheswaran, T.; Asokan, G.S.; Dany, A.; Anand, B. Theranostics: A treasured tailor for tomorrow. J. Pharm. Bioallied. Sci. 2014, 6, S6–S8. [Google Scholar] [CrossRef] [PubMed]
- Pelaz, B.; Alexiou, C.; Alvarez-Puebla, R.A.; Alves, F.; Andrews, A.M.; Ashraf, S.; Balogh, L.P.; Ballerini, L.; Bestetti, A.; Brendel, C.; et al. Diverse Applications of Nanomedicine. ACS Nano. 2017, 11, 2313–2381. [Google Scholar] [CrossRef]
- Chen, G.; Roy, I.; Yang, C.; Prasad, P.N. Nanochemistry and Nanomedicine for Nanoparticle-based Diagnostics and Therapy. Chem. Rev. 2016, 116, 2826–2885. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Ehlerding, E.B.; Cai, W. Theranostic nanoparticles. J. Nucl. Med. 2014, 55, 1919–1922. [Google Scholar] [CrossRef] [PubMed]
- Kodiha, M.; Wang, Y.M.; Hutter, E.; Maysinger, D.; Stochaj, U. Off to the organelles - killing cancer cells with targeted gold nanoparticles. Theranostics 2015, 5, 357–370. [Google Scholar] [CrossRef]
- Kelloff, G.J.; Krohn, K.A.; Larson, S.M.; Weissleder, R.; Mankoff, D.A.; Hoffman, J.M.; Link, J.M.; Guyton, K.Z.; Eckelman, W.C.; Scher, H.I.; et al. The progress and promise of molecular imaging probes in oncologic drug development. Clin. Cancer Res. 2005, 11, 7967–7985. [Google Scholar] [CrossRef]
- Wu, T.T.; Zhou, S.H. Nanoparticle-Based Targeted Therapeutics in Head-And-Neck Cancer. Int. J. Med. Sci. 2015, 12, 187–200. [Google Scholar] [CrossRef]
- Wu, C.; Li, F.; Niu, G.; Chen, X. PET imaging of inflammation biomarkers. Theranostics. Theranostics 2013, 3, 448–466. [Google Scholar] [CrossRef]
- Huynh, N.T.; Passirani, C.; Saulnier, P.; Benoit, J.P. Lipid nanocapsules: A new platform for nanomedicine. Int. J. Pharm. 2009, 379, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Muhamad, N.; Plengsuriyakarn, T.; Na-Bangchang, K. Application of active targeting nanoparticle delivery system for chemotherapeutic drugs and traditional/herbal medicines in cancer therapy: A systematic review. Int. J. Nanomed. 2018, 13, 3921–3935. [Google Scholar] [CrossRef] [PubMed]
- Wiggers, J.K.; Ruys, A.T.; Groot Koerkamp, B.; Beuers, U.; ten Kate, F.J.; van Gulik, T.M. Differences in immunohistochemical biomarkers between intra- and extrahepatic cholangiocarcinoma: A systematic review and meta-analysis. J. Gastroenterol. Hepatol. 2014, 29, 1582–1594. [Google Scholar] [CrossRef] [PubMed]
- Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 30 November 2019).
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013, 41, D377–D386. [Google Scholar] [CrossRef]
- PANTHER. Available online: http://www.pantherdb.org/ (accessed on 30 November 2019).
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- STRING. Available online: http://string905.embl.de/ (accessed on 30 November 2019).
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef]
- van Oosten, M.; Crane, L.M.; Bart, J.; van Leeuwen, F.W.; van Dam, G.M. Selecting Potential Targetable Biomarkers for Imaging Purposes in Colorectal Cancer Using TArget Selection Criteria (TASC): A Novel Target Identification Tool. Transl. Oncol. 2011, 4, 71–82. [Google Scholar] [CrossRef]
- Divgi, C.R.; Pandit-Taskar, N.; Jungbluth, A.A.; Reuter, V.E.; Gonen, M.; Ruan, S.; Pierre, C.; Nagel, A.; Pryma, D.A.; Humm, J.; et al. Preoperative characterisation of clear-cell renal carcinoma using iodine-124-labelled antibody chimeric G250 (124I-cG250) and PET in patients with renal masses: A phase I trial. Lancet Oncol. 2007, 8, 304–310. [Google Scholar] [CrossRef]
- McKnight, B.N.; Kuda-Wedagedara, A.N.W.; Sevak, K.K.; Abdel-Atti, D.; Wiesend, W.N.; Ku, A.; Selvakumar, D.; Carlin, S.D.; Lewis, J.S.; Viola-Villegas, N.T. Imaging EGFR and HER3 through (89)Zr-labeled MEHD7945A (Duligotuzumab). Sci. Rep. 2018, 8, e9043. [Google Scholar] [CrossRef] [PubMed]
- Scherer, R.L.; VanSaun, M.N.; McIntyre, J.O.; Matrisian, L.M. Optical imaging of matrix metalloproteinase-7 activity in vivo using a proteolytic nanobeacon. Mol. Imaging 2008, 7, 118–131. [Google Scholar] [CrossRef] [PubMed]
- Hakimzadeh, N.; Pinas, V.A.; Molenaar, G.; de Waard, V.; Lutgens, E.; van Eck-Smit, B.L.F.; de Bruin, K.; Piek, J.J.; Eersels, J.L.H.; Booij, J.; et al. Novel molecular imaging ligands targeting matrix metalloproteinases 2 and 9 for imaging of unstable atherosclerotic plaques. PLoS ONE 2017, 12, e0187767. [Google Scholar] [CrossRef] [PubMed]
- den Hollander, M.W.; Bensch, F.; Glaudemans, A.W.; Oude Munnink, T.H.; Enting, R.H.; den Dunnen, W.F.; Heesters, M.A.; Kruyt, F.A.; Lub-de Hooge, M.N.; Cees de Groot, J.; et al. TGF-beta Antibody Uptake in Recurrent High-Grade Glioma Imaged with 89Zr-Fresolimumab PET. J. Nucl. Med. 2015, 56, 1310–1314. [Google Scholar] [CrossRef]
- Jacobson, O.; Yan, X.; Niu, G.; Weiss, I.D.; Ma, Y.; Szajek, L.P.; Shen, B.; Kiesewetter, D.O.; Chen, X. PET imaging of tenascin-C with a radiolabeled single-stranded DNA aptamer. J. Nucl. Med. 2015, 56, 616–621. [Google Scholar] [CrossRef]
- Li, D.; Bentley, C.; Anderson, A.; Wiblin, S.; Cleary, K.L.S.; Koustoulidou, S.; Hassanali, T.; Yates, J.; Greig, J.; Nordkamp, M.O.; et al. Development of a T-cell Receptor Mimic Antibody against Wild-Type p53 for Cancer Immunotherapy. Cancer Res. 2017, 77, 2699–2711. [Google Scholar] [CrossRef]
- Itatsu, K.; Sasaki, M.; Yamaguchi, J.; Ohira, S.; Ishikawa, A.; Ikeda, H.; Sato, Y.; Harada, K.; Zen, Y.; Sato, H.; et al. Cyclooxygenase-2 is involved in the up-regulation of matrix metalloproteinase-9 in cholangiocarcinoma induced by tumor necrosis factor-alpha. Am. J. Pathol. 2009, 174, 829–841. [Google Scholar] [CrossRef]
- Onodera, M.; Zen, Y.; Harada, K.; Sato, Y.; Ikeda, H.; Itatsu, K.; Sato, H.; Ohta, T.; Asaka, M.; Nakanuma, Y. Fascin is involved in tumor necrosis factor-alpha-dependent production of MMP9 in cholangiocarcinoma. Lab. Invest. 2009, 89, 1261–1274. [Google Scholar] [CrossRef]
- Subimerb, C.; Pinlaor, S.; Khuntikeo, N.; Leelayuwat, C.; Morris, A.; McGrath, M.S.; Wongkham, S. Tissue invasive macrophage density is correlated with prognosis in cholangiocarcinoma. Mol. Med. Rep. 2010, 3, 597–605. [Google Scholar] [CrossRef]
- Shi, R.Y.; Yang, X.R.; Shen, Q.J.; Yang, L.X.; Xu, Y.; Qiu, S.J.; Sun, Y.F.; Zhang, X.; Wang, Z.; Zhu, K.; et al. High expression of Dickkopf-related protein 1 is related to lymphatic metastasis and indicates poor prognosis in intrahepatic cholangiocarcinoma patients after surgery. Cancer 2013, 119, 993–1003. [Google Scholar] [CrossRef]
- Sun, Q.; Zhao, C.; Xia, L.; He, Z.; Lu, Z.; Liu, C.; Jia, M.; Wang, J.; Niu, J. High expression of matrix metalloproteinase-9 indicates poor prognosis in human hilar cholangiocarcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 6157–6164. [Google Scholar] [PubMed]
- Tian, X.; Wang, Q.; Li, Y.; Hu, J.; Wu, L.; Ding, Q.; Zhang, C. The expression of S100A4 protein in human intrahepatic cholangiocarcinoma: Clinicopathologic significance and prognostic value. Pathol Oncol. Res. 2015, 21, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Li, F.; Sun, F.; Niu, J. Interleukin-8 is a prognostic indicator in human hilar cholangiocarcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 8376–8384. [Google Scholar] [PubMed]
- Park, Y.; Kim, K.; Paik, J.H.; Chie, E.K.; Jang, J.Y.; Kim, S.W.; Oh, D.Y. High expression of MMP-9 is associated with better prognosis in extrahepatic bile duct cancer patients. Eur. J. Surg. Oncol. 2018, 44, 638–643. [Google Scholar] [CrossRef]
- Keira, Y.; Takasawa, A.; Murata, M.; Nojima, M.; Takasawa, K.; Ogino, J.; Higashiura, Y.; Sasaki, A.; Kimura, Y.; Mizuguchi, T.; et al. An immunohistochemical marker panel including claudin-18, maspin, and p53 improves diagnostic accuracy of bile duct neoplasms in surgical and presurgical biopsy specimens. Virchows Arch. 2015, 466, 265–277. [Google Scholar] [CrossRef]
- Shinozaki, A.; Shibahara, J.; Noda, N.; Tanaka, M.; Aoki, T.; Kokudo, N.; Fukayama, M. Claudin-18 in biliary neoplasms. Its significance in the classification of intrahepatic cholangiocarcinoma. Virchows Arch. 2011, 459, 73–80. [Google Scholar] [CrossRef]
- Iguchi, T.; Yamashita, N.; Aishima, S.; Kuroda, Y.; Terashi, T.; Sugimachi, K.; Taguchi, K.; Taketomi, A.; Maehara, Y.; Tsuneyoshi, M. A comprehensive analysis of immunohistochemical studies in intrahepatic cholangiocarcinoma using the survival tree model. Oncology 2009, 76, 293–300. [Google Scholar] [CrossRef]
- Soejima, Y.; Takeuchi, M.; Akashi, T.; Sawabe, M.; Fukusato, T. beta4 and beta6 Integrin Expression Is Associated with the Subclassification and Clinicopathological Features of Intrahepatic Cholangiocarcinoma. Int. J. Mol. Sci. 2018, 19, 1004. [Google Scholar] [CrossRef]
- Bi, C.; Liu, M.; Rong, W.; Wu, F.; Zhang, Y.; Lin, S.; Liu, Y.; Wu, J.; Wang, L. High Beclin-1 and ARID1A expression corelates with poor survival and high recurrence in intrahepatic cholangiocarcinoma: A histopathological retrospective study. BMC Cancer 2019, 19, e213. [Google Scholar] [CrossRef]
- Gu, M.J. CA9 overexpression is an independent favorable prognostic marker in intrahepatic cholangiocarcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 862–866. [Google Scholar]
- Ogo, Y.; Nio, Y.; Yano, S.; Toga, T.; Koike, M.; Hashimoto, K.; Itakura, M.; Maruyama, R. Immunohistochemical expression of HER-1 and HER-2 in extrahepatic biliary carcinoma. Anticancer Res. 2006, 26, 763–770. [Google Scholar]
- Schmitz, K.J.; Lang, H.; Wohlschlaeger, J.; Sotiropoulos, G.C.; Reis, H.; Schmid, K.W.; Baba, H.A. AKT and ERK1/2 signaling in intrahepatic cholangiocarcinoma. World J. Gastroenterol. 2007, 13, 6470–6477. [Google Scholar] [CrossRef]
- Yoshikawa, D.; Ojima, H.; Iwasaki, M.; Hiraoka, N.; Kosuge, T.; Kasai, S.; Hirohashi, S.; Shibata, T. Clinicopathological and prognostic significance of EGFR, VEGF, and HER2 expression in cholangiocarcinoma. Br. J. Cancer 2008, 98, 418–425. [Google Scholar] [CrossRef]
- Pignochino, Y.; Sarotto, I.; Peraldo-Neia, C.; Penachioni, J.Y.; Cavalloni, G.; Migliardi, G.; Casorzo, L.; Chiorino, G.; Risio, M.; Bardelli, A.; et al. Targeting EGFR/HER2 pathways enhances the antiproliferative effect of gemcitabine in biliary tract and gallbladder carcinomas. BMC Cancer 2010, 10, e631. [Google Scholar] [CrossRef]
- Shafizadeh, N.; Grenert, J.P.; Sahai, V.; Kakar, S. Epidermal growth factor receptor and HER-2/neu status by immunohistochemistry and fluorescence in situ hybridization in adenocarcinomas of the biliary tree and gallbladder. Hum. Pathol. 2010, 41, 485–492. [Google Scholar] [CrossRef]
- Miyamoto, M.; Ojima, H.; Iwasaki, M.; Shimizu, H.; Kokubu, A.; Hiraoka, N.; Kosuge, T.; Yoshikawa, D.; Kono, T.; Furukawa, H.; et al. Prognostic significance of overexpression of c-Met oncoprotein in cholangiocarcinoma. Br. J. Cancer 2011, 105, 131–138. [Google Scholar] [CrossRef]
- Gu, M.J.; Choi, J.H. Clinicopathological significance of E-cadherin, beta-catenin and epidermal growth factor receptor expression in intrahepatic cholangiocarcinoma. Hepatogastroenterology 2012, 59, 1241–1244. [Google Scholar] [CrossRef]
- Simbolo, M.; Fassan, M.; Ruzzenente, A.; Mafficini, A.; Wood, L.D.; Corbo, V.; Melisi, D.; Malleo, G.; Vicentini, C.; Malpeli, G.; et al. Multigene mutational profiling of cholangiocarcinomas identifies actionable molecular subgroups. Oncotarget 2014, 5, 2839–2852. [Google Scholar] [CrossRef]
- Yang, X.; Wang, W.; Wang, C.; Wang, L.; Yang, M.; Qi, M.; Su, H.; Sun, X.; Liu, Z.; Zhang, J.; et al. Characterization of EGFR family gene aberrations in cholangiocarcinoma. Oncol. Rep. 2014, 32, 700–708. [Google Scholar] [CrossRef] [PubMed]
- Moon, A.; Chin, S.; Kim, H.K.; Kwak, J.J.; Koh, E.S.; Kim, Y.W.; Jang, K.T. EGFR, COX2, p-AKT expression and PIK3CA mutation in distal extrahepatic bile duct carcinoma. Pathology 2016, 48, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Padthaisong, S.; Thanee, M.; Techasen, A.; Namwat, N.; Yongvanit, P.; Liwatthakun, A.; Hankla, K.; Sangkhamanon, S.; Loilome, W. Nimotuzumab Inhibits Cholangiocarcinoma Cell Metastasis via Suppression of the Epithelial-Mesenchymal Transition Process. Anticancer Res. 2017, 37, 3591–3597. [Google Scholar] [CrossRef]
- Gomes, R.V.; Rodrigues, M.A.; Rodrigues, J.; Vidigal, P.T.; Damasceno, K.A.; Lima, H.A.; Gomes, D.A.; Machado, C.J.; and Resende, V. Expression of epidermal growth factor receptor (EGFR) in cholangiocarcinomas: Predictive factors and survival. Rev. Col. Bras. Cir. 2018, 45, e1826. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.F.; Liu, Z.L.; Pan, C.; Yang, X.Q.; Ning, S.L.; Liu, H.D.; Guo, S.; Yu, J.M.; Zhang, Z.L. HMGB1 correlates with angiogenesis and poor prognosis of perihilar cholangiocarcinoma via elevating VEGFR2 of vessel endothelium. Oncogene 2019, 38, 868–880. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Ma, X.; Liang, M.; Li, D.; Ma, P.; Wang, S.; Wu, Z.; Zhao, X. Prediction for early recurrence of intrahepatic mass-forming cholangiocarcinoma: Quantitative magnetic resonance imaging combined with prognostic immunohistochemical markers. Cancer Imaging 2019, 19, e49. [Google Scholar] [CrossRef]
- Kumar, V.; Soni, P.; Garg, M.; Kamholz, S.; Chandra, A.B. Emerging Therapies in the Management of Advanced-Stage Gastric Cancer. Front. Pharmacol. 2018, 9, e404. [Google Scholar] [CrossRef]
- Nair, A.; Ingram, N.; Verghese, E.T.; Wijetunga, I.; Markham, A.F.; Wyatt, J.; Prasad, K.R.; Coletta, P.L. Neutrophil Gelatinase-associated Lipocalin as a Theragnostic Marker in Perihilar Cholangiocarcinoma. Anticancer Res. 2018, 38, 6737–6744. [Google Scholar] [CrossRef]
- Garcia-Hernandez, V.; Quiros, M.; Nusrat, A. Intestinal epithelial claudins: Expression and regulation in homeostasis and inflammation. Ann. N. Y. Acad. Sci. 2017, 1397, 66–79. [Google Scholar] [CrossRef]
- Niimi, T.; Nagashima, K.; Ward, J.M.; Minoo, P.; Zimonjic, D.B.; Popescu, N.C.; Kimura, S. Claudin-18, a novel downstream target gene for the T/EBP/NKX2.1 homeodomain transcription factor, encodes lung- and stomach-specific isoforms through alternative splicing. Mol. Cell Biol. 2001, 21, 7380–7390. [Google Scholar] [CrossRef]
- Sahin, U.; Koslowski, M.; Dhaene, K.; Usener, D.; Brandenburg, G.; Seitz, G.; Huber, C.; Tureci, O. Claudin-18 splice variant 2 is a pan-cancer target suitable for therapeutic antibody development. Clin. Cancer Res. 2008, 14, 7624–7634. [Google Scholar] [CrossRef]
- Dottermusch, M.; Kruger, S.; Behrens, H.M.; Halske, C.; Rocken, C. Expression of the potential therapeutic target claudin-18.2 is frequently decreased in gastric cancer: Results from a large Caucasian cohort study. Virchows Arch. 2019, 475, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Chiquet-Ehrismann, R. Tenascins. Int. J. Biochem. Cell Biol. 2004, 36, 986–990. [Google Scholar] [CrossRef]
- Chiquet-Ehrismann, R.; Mackie, E.J.; Pearson, C.A.; Sakakura, T. Tenascin: An extracellular matrix protein involved in tissue interactions during fetal development and oncogenesis. Cell 1986, 47, 131–139. [Google Scholar] [CrossRef]
- Terada, T.; Nakanuma, Y. Expression of tenascin, type IV collagen and laminin during human intrahepatic bile duct development and in intrahepatic cholangiocarcinoma. Histopathology 1994, 25, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Midwood, K.S.; Orend, G. The role of tenascin-C in tissue injury and tumorigenesis. J. Cell Commun. Signal. 2009, 3, 287–310. [Google Scholar] [CrossRef]
- Leppanen, J.; Lindholm, V.; Isohookana, J.; Haapasaari, K.M.; Karihtala, P.; Lehenkari, P.P.; Saarnio, J.; Kauppila, J.H.; Karttunen, T.J.; Helminen, O.; et al. Tenascin C, Fibronectin, and Tumor-Stroma Ratio in Pancreatic Ductal Adenocarcinoma. Pancreas 2019, 48, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.Z.; Jiang, P.; Wu, G.; Chen, K.; Zhang, X.; Li, X. The co-expression of MMP-9 and Tenascin-C is significantly associated with the progression and prognosis of pancreatic cancer. Diagn. Pathol. 2015, 10, e211. [Google Scholar] [CrossRef] [PubMed]
- Leppanen, J.; Bogdanoff, S.; Lehenkari, P.P.; Saarnio, J.; Kauppila, J.H.; Karttunen, T.J.; Huhta, H.; Helminen, O. Tenascin-C and fibronectin in normal esophageal mucosa, Barrett’s esophagus, dysplasia and adenocarcinoma. Oncotarget 2017, 8, 66865–66877. [Google Scholar] [CrossRef][Green Version]
- Qi, W.; Yang, Z.; Li, H.; Cui, Y.; Xuan, Y. The role of Tenascin-C and Twist1 in gastric cancer: Cancer progression and prognosis. APMIS 2019, 127, 64–71. [Google Scholar] [CrossRef]
- Lundin, M.; Nordling, S.; Lundin, J.; Haglund, C. Tenascin-C expression and its prognostic significance in colorectal cancer. Oncology 2007, 72, 403–409. [Google Scholar] [CrossRef]
- Hicke, B.J.; Stephens, A.W.; Gould, T.; Chang, Y.F.; Lynott, C.K.; Heil, J.; Borkowski, S.; Hilger, C.S.; Cook, G.; Warren, S.; et al. Tumor targeting by an aptamer. J. Nucl. Med. 2006, 47, 668–678. [Google Scholar]
- He, X.; Chen, X.; Liu, L.; Zhang, Y.; Lu, Y.; Zhang, Y.; Chen, Q.; Ruan, C.; Guo, Q.; Li, C.; et al. Sequentially Triggered Nanoparticles with Tumor Penetration and Intelligent Drug Release for Pancreatic Cancer Therapy. Adv. Sci. (Weinh) 2018, 5, e1701070. [Google Scholar] [CrossRef] [PubMed]
- Tiede, C.; Tang, A.A.; Deacon, S.E.; Mandal, U.; Nettleship, J.E.; Owen, R.L.; George, S.E.; Harrison, D.J.; Owens, R.J.; Tomlinson, D.C.; et al. Adhiron: A stable and versatile peptide display scaffold for molecular recognition applications. Protein Eng. Des. Sel. 2014, 27, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Yang, Y.P.; Dikici, E.; Deo, S.K.; Daunert, S. Beyond Antibodies as Binding Partners: The Role of Antibody Mimetics in Bioanalysis. Annu. Rev. Anal. Chem. (Palo Alto Calif.) 2017, 10, 293–320. [Google Scholar] [CrossRef] [PubMed]
- Hilvo, M.; Baranauskiene, L.; Salzano, A.M.; Scaloni, A.; Matulis, D.; Innocenti, A.; Scozzafava, A.; Monti, S.M.; Di Fiore, A.; De Simone, G.; et al. Biochemical characterization of CA IX, one of the most active carbonic anhydrase isozymes. J. Biol. Chem. 2008, 283, 27799–27809. [Google Scholar] [CrossRef]
- Alterio, V.; Hilvo, M.; Di Fiore, A.; Supuran, C.T.; Pan, P.; Parkkila, S.; Scaloni, A.; Pastorek, J.; Pastorekova, S.; Pedone, C.; et al. Crystal structure of the catalytic domain of the tumor-associated human carbonic anhydrase IX. Proc. Natl. Acad. Sci. USA 2009, 106, 16233–16238. [Google Scholar] [CrossRef]
- Driessen, A.; Landuyt, W.; Pastorekova, S.; Moons, J.; Goethals, L.; Haustermans, K.; Nafteux, P.; Penninckx, F.; Geboes, K.; Lerut, T.; et al. Expression of carbonic anhydrase IX (CA IX), a hypoxia-related protein, rather than vascular-endothelial growth factor (VEGF), a pro-angiogenic factor, correlates with an extremely poor prognosis in esophageal and gastric adenocarcinomas. Ann. Surg. 2006, 243, 334–340. [Google Scholar] [CrossRef]
- Swietach, P.; Hulikova, A.; Vaughan-Jones, R.D.; Harris, A.L. New insights into the physiological role of carbonic anhydrase IX in tumour pH regulation. Oncogene 2010, 29, 6509–6521. [Google Scholar] [CrossRef]
- De Simone, G.; Supuran, C.T. Carbonic anhydrase IX: Biochemical and crystallographic characterization of a novel antitumor target. Biochim. Biophys. Acta 2010, 1804, 404–409. [Google Scholar] [CrossRef]
- Bayat-Mokhtari, R.; Homayouni, T.S.; Baluch, N.; Morgatskaya, E.; Kumar, S.; Das, B.; Yeger, H. Combination therapy in combating cancer. Oncotarget 2017, 8, 38022–38043. [Google Scholar] [CrossRef]
- Safety Study of SLC-0111 in Subjects with Advanced Solid Tumours. Available online: https://clinicaltrials.gov/ct2/show/NCT02215850 (accessed on 29 September 2020).
- Cheal, S.M.; Punzalan, B.; Doran, M.G.; Evans, M.J.; Osborne, J.R.; Lewis, J.S.; Zanzonico, P.; Larson, S.M. Pairwise comparison of 89Zr- and 124I-labeled cG250 based on positron emission tomography imaging and nonlinear immunokinetic modeling: In vivo carbonic anhydrase IX receptor binding and internalization in mouse xenografts of clear-cell renal cell carcinoma. Eur. J. Nucl. Med. Mol. Imaging 2014, 41, 985–994. [Google Scholar] [CrossRef]
- Pastorekova, S.; Parkkila, S.; Parkkila, A.K.; Opavsky, R.; Zelnik, V.; Saarnio, J.; Pastorek, J. Carbonic anhydrase IX, MN/CA IX: Analysis of stomach complementary DNA sequence and expression in human and rat alimentary tracts. Gastroenterology 1997, 112, 398–408. [Google Scholar] [CrossRef] [PubMed]
- Price, T.J.; Tang, M.; Gibbs, P.; Haller, D.G.; Peeters, M.; Arnold, D.; Segelov, E.; Roy, A.; Tebbutt, N.; Pavlakis, N.; et al. Targeted therapy for metastatic colorectal cancer. Expert Rev. Anticancer Ther. 2018, 18, 991–1006. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, E.B.; Hanson, R.N. Imaging EGFR and HER2 by PET and SPECT: A review. Med. Res. Rev. 2014, 34, 596–643. [Google Scholar] [CrossRef] [PubMed]
- Merla, A.; Liu, K.G.; Rajdev, L. Targeted Therapy in Biliary Tract Cancers. Curr. Treat. Options Oncol. 2015, 16, e48. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Schmitz, K.R.; Jeffrey, P.D.; Wiltzius, J.J.; Kussie, P.; Ferguson, K.M. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell 2005, 7, 301–311. [Google Scholar] [CrossRef]
- Sickmier, E.A.; Kurzeja, R.J.; Michelsen, K.; Vazir, M.; Yang, E.; Tasker, A.S. The Panitumumab EGFR Complex Reveals a Binding Mechanism That Overcomes Cetuximab Induced Resistance. PLoS ONE 2016, 11, e0163366. [Google Scholar] [CrossRef]
- van Helden, E.J.; Elias, S.G.; Gerritse, S.L.; van Es, S.C.; Boon, E.; Huisman, M.C.; van Grieken, N.C.T.; Dekker, H.; van Dongen, G.; Vugts, D.J.; et al. [(89)Zr]Zr-cetuximab PET/CT as biomarker for cetuximab monotherapy in patients with RAS wild-type advanced colorectal cancer. Eur. J. Nucl. Med. Mol. Imaging 2019. [Google Scholar] [CrossRef]
- Hughes, N.R.; Pairojkul, C.; Royce, S.G.; Clouston, A.; Bhathal, P.S. Liver fluke-associated and sporadic cholangiocarcinoma: An immunohistochemical study of bile duct, peribiliary gland and tumour cell phenotypes. J. Clin. Pathol. 2006, 59, 1073–1078. [Google Scholar] [CrossRef]
- Stang, A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 2010, 25, 603–605. [Google Scholar] [CrossRef]
- Sauerbrei, W.; Taube, S.E.; McShane, L.M.; Cavenagh, M.M.; Altman, D.G. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): An Abridged Explanation and Elaboration. J. Natl. Cancer Inst. 2018, 110, 803–811. [Google Scholar] [CrossRef]
- Espinoza, J.A.; Riquelme, I.; Sagredo, E.A.; Rosa, L.; Garcia, P.; Bizama, C.; Apud-Bell, M.; Leal, P.; Weber, H.; Benavente, F.; et al. Mucin 5B, carbonic anhydrase 9 and claudin 18 are potential theranostic markers of gallbladder carcinoma. Histopathology 2019, 74, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Franz, M.; Jung, C.; Lauten, A.; Figulla, H.R.; Berndt, A. Tenascin-C in cardiovascular remodeling: Potential impact for diagnosis, prognosis estimation and targeted therapy. Cell Adhes. Migr. 2015, 9, 90–95. [Google Scholar] [CrossRef] [PubMed]


| Biomarker | Country | Site of Tumor | No. | % High/+ve Presence | Normal Tissue Control | Presence in Normal Tissues | Risk of Bias | References |
|---|---|---|---|---|---|---|---|---|
| MMP9 * | Multiple (mainly Eastern) | iCCA | 302 | 45.6–62.5% +ve | Normal liver tissue | Weak or −ve | Low-High | [40,41,42,43,44,45,46,47] |
| eCCA | 214 | 47.3–58% +ve | ||||||
| pCCA | 120 | 67.2–67.7% +ve | ||||||
| CLDN18 | Japan | iCCA | 27 | NR | Biliary epithelium | NR | High | [48] |
| eCCA | 32 | NR | ||||||
| Japan | iCCA | 83 | 43% +ve | Biliary epithelium | −ve | High | [49] | |
| eCCA | 99 | 90% +ve | ||||||
| TNC | Japan | iCCA | 61 | 63.9% +ve | NR | NR | High | [50] |
| Japan | iCCA | 48 | 37.5% high | NR | NR | High | [51] | |
| CA9 | China | iCCA | 113 | 85.0% high | NR | NR | Low | [52] |
| Korea | iCCA | 85 | 44.7% +ve | n = 4 normal liver | NR | High | [53] | |
| EGFR * | Multiple (mainly Eastern) | iCCA | 634 | 26.1–100% +ve | Normal liver tissue | +ve membrane | Low-High | [50,54,55,56,57,58,59,60,61,62,63,64,65,66,67] |
| eCCA | 402 | 18–79% +ve | ||||||
| pCCA | 121 | 45.5% high | ||||||
| CCA | 173 | 28.6–55% high |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wijetunga, I.; McVeigh, L.E.; Charalambous, A.; Antanaviciute, A.; Carr, I.M.; Nair, A.; Prasad, K.R.; Ingram, N.; Coletta, P.L. Translating Biomarkers of Cholangiocarcinoma for Theranosis: A Systematic Review. Cancers 2020, 12, 2817. https://doi.org/10.3390/cancers12102817
Wijetunga I, McVeigh LE, Charalambous A, Antanaviciute A, Carr IM, Nair A, Prasad KR, Ingram N, Coletta PL. Translating Biomarkers of Cholangiocarcinoma for Theranosis: A Systematic Review. Cancers. 2020; 12(10):2817. https://doi.org/10.3390/cancers12102817
Chicago/Turabian StyleWijetunga, Imeshi, Laura E. McVeigh, Antonia Charalambous, Agne Antanaviciute, Ian M. Carr, Amit Nair, K. Raj Prasad, Nicola Ingram, and P. Louise Coletta. 2020. "Translating Biomarkers of Cholangiocarcinoma for Theranosis: A Systematic Review" Cancers 12, no. 10: 2817. https://doi.org/10.3390/cancers12102817
APA StyleWijetunga, I., McVeigh, L. E., Charalambous, A., Antanaviciute, A., Carr, I. M., Nair, A., Prasad, K. R., Ingram, N., & Coletta, P. L. (2020). Translating Biomarkers of Cholangiocarcinoma for Theranosis: A Systematic Review. Cancers, 12(10), 2817. https://doi.org/10.3390/cancers12102817






