Effect of 1α,25(OH)2 Vitamin D3 in Mutant P53 Glioblastoma Cells: Involvement of Neutral Sphingomyelinase1
Abstract
:Simple Summary
Abstract
1. Introduction
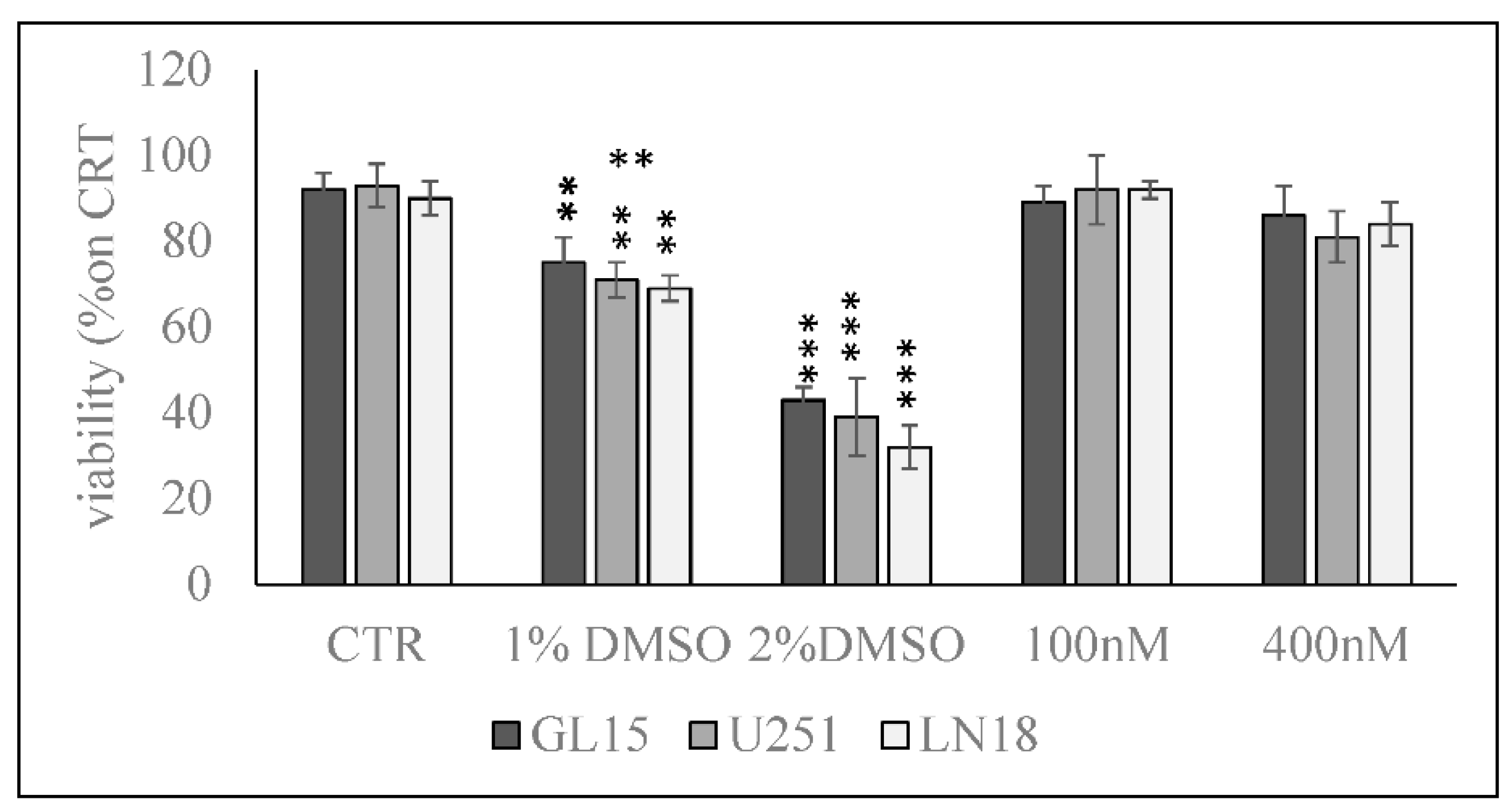
2. Results
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture
4.3. Cytotoxicity
4.4. Reverse Transcription Quantitative PCR (RTqPCR)
4.5. Protein Concentration and Western Blotting
4.6. Enzyme Activity Assay
4.7. Ultrafast Liquid Chromatography–Tandem Mass Spectrometry
4.8. Xenotrasnplantation in Chorioallantoic Membrane
4.9. Immunofluorescence
4.10. Immunohistochemistry
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nicholas, M.K.; Lukas, R.V.; Chmura, S.; Yamini, B.; Lesniak, M.; Pytel, P. Molecular heterogeneity in glioblastoma: Therapeutic opportunities and challenges. Semin. Oncol. 2011, 38, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.C.; Ashley, D.M.; López, G.L.; Malinzak, M.; Friedman, H.S.; Khasraw, M. Management of Glioblastoma: State of the Art and Future Directions. CA Cancer J. Clin. 2020, 70, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Badr, C.E.; da Hora, C.C.; Kirov, A.B.; Tabet, E.; Amante, R.; Maksoud, S.; Nibbs, A.E.; Fitzsimons, E.; Boukhali, M.; Chen, J.W.; et al. Obtusaquinone: A Cysteine-Modifying Compound That Targets Keap1 for Degradation. ACS Chem. Biol. 2020, 15, 1445–1454. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C. Anticancer activities and mechanism of action of the labdane diterpene coronarin D. Pathol. Res. Pract. 2020, 216, 152946. [Google Scholar] [CrossRef]
- Lo, W.L.; Hsu, T.I.; Yang, W.B.; Kao, T.J.; Wu, M.H.; Huang, Y.N.; Yeh, S.H.; Chuang, J.Y. Betulinic Acid-Mediated Tuning of PERK/CHOP Signaling by Sp1 Inhibition as a Novel Therapeutic Strategy for Glioblastoma. Cancers 2020, 12, 981. [Google Scholar] [CrossRef]
- Li, X.; Zhang, W.; Liang, L.; Duan, X.; Deng, J.; Zhou, Y. Natural Product-Derived Icaritin Exerts Anti-Glioblastoma Effects by Positively Modulating Estrogen Receptor β. Exp. Ther. Med. 2020, 19, 2841–2850. [Google Scholar] [CrossRef]
- Magrassi, L.; Butti, G.; Pezzotta, S.; Infuso, L.; Milanesi, G. Effects of Vitamin D and Retinoic Acid on Human Glioblastoma Cell Lines. Acta Neurochir. 1995, 133, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Diesel, B.; Radermacher, J.; Bureik, M.; Bernhardt, R.; Seifert, M.; Reichrath, J.; Fischer, U.; Meese, E. Vitamin D(3) metabolism in human glioblastoma multiforme: Functionality of CYP27B1 splice variants, metabolism of calcidiol, and effect of calcitriol. Clin. Cancer Res. 2005, 11, 5370–5380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reichrath, S.; Müller, C.S.L.; Gleissner, B.; Pfreundschuh, M.; Vogt, T.; Reichrath, J. Notch- And Vitamin D Signaling in 1,25(OH)2D3-resistant Glioblastoma Multiforme (GBM) Cell Lines. J. Steroid Biochem. Mol. Biol. 2010, 121, 420–424. [Google Scholar] [CrossRef]
- Ferronato, M.J.; Alonso, E.N.; Salomón, D.G.; Fermento, M.E.; Gandini, N.A.; Quevedo, M.A.; Mascaró, E.; Vitale, C.; Fall, Y.; Facchinetti, M.M.; et al. Antitumoral effects of the alkynylphosphonate analogue of calcitriol EM1 on glioblastoma multiforme cells. J. Steroid Biochem. Mol. Biol. 2018, 178, 22–35. [Google Scholar] [CrossRef]
- Emanuelsson, I.; Wikvall, K.; Friman, T.; Norlin, M. Vitamin D Analogues Tacalcitol and Calcipotriol Inhibit Proliferation and Migration of T98G Human Glioblastoma Cells. Basic Clin. Pharmacol. Toxicol. 2018, 123, 130–136. [Google Scholar] [CrossRef]
- McConnell, D.D.; McGreevy, J.W.; Williams, M.N.; Litofsky, N.S. Do Anti-Oxidants Vitamin D(3,) Melatonin, and Alpha-Lipoic Acid Have Synergistic Effects with Temozolomide on Cultured Glioblastoma Cells? Medicines 2018, 5, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elmaci, I.; Ozpinar, A.; Ozpinar, A.; Perez, J.L.; Altinoz, M.A. From epidemiology and neurometabolism to treatment: Vitamin D in pathogenesis of glioblastoma Multiforme (GBM) and a proposal for Vitamin D + all-trans retinoic acid + Temozolomide combination in treatment of GBM. Metab. Brain Dis. 2019, 34, 687–704. [Google Scholar] [CrossRef] [PubMed]
- Norlin, M. Effects of vitamin D in the nervous system: Special focus on interaction with steroid hormone signalling and a possible role in the treatment of brain cancer. J. Neuroendocrinol. 2020, 32, e12799. [Google Scholar] [CrossRef]
- Pike, J.W.; Meyer, M.B. The vitamin D receptor: New paradigms for the regulation of gene expression by 1,25-dihydroxyvitamin D(3). Endocrinol. Metab. Clin. N. Am. 2010, 39, 255–269. [Google Scholar]
- Bartoccini, E.; Marini, F.; Damaskopoulou, E.; Lazzarini, R.; Cataldi, S.; Cascianelli, G.; Gil Garcia, M.; Albi, E. Nuclear lipid microdomains regulate nuclear vitamin D3 uptake and influence embryonic hippocampal cell differentiation. Mol. Biol. Cell. 2011, 22, 3022–3031. [Google Scholar] [CrossRef]
- Cataldi, S.; Arcuri, C.; Hunot, S.; Mecca, C.; Codini, M.; Laurenti, M.E.; Ferri, I.; Loreti, E.; Garcia-Gil, M.; Traina, G.; et al. Effect of Vitamin D in HN9.10e Embryonic Hippocampal Cells and in Hippocampus from MPTP-Induced Parkinson’s Disease Mouse Model. Front. Cell Neurosci. 2018, 12, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marini, F.; Bartoccini, E.; Cascianelli, G.; Voccoli, V.; Baviglia, M.G.; Magni, M.V.; Garcia-Gil, M.; Albi, E. Effect of 1alpha,25-dihydroxyvitamin D3 in embryonic hippocampal cells. Hippocampus 2010, 20, 696–705. [Google Scholar]
- Christakos, S.; Raval-Pandya, M.; Wernyi, R.P.; Yang, W. Genomic mechanisms involved in pleiotropic actions of 1,25-dihydroxyvitamin D3. Biochem. J. 1996, 316, 361–371. [Google Scholar]
- Okazaki, T.; Bielawska, A.; Domae, N.; Bell, R.M.; Hannun, Y.A. Characteristics and partial purification of a novel cytosolic, magnesium-independent, neutral sphingomyelinase activated in the early signal transduction of 1 alpha,25-dihydroxyvitamin D3-induced HL-60 cell differentiation. J. Biol. Chem. 1994, 269, 4070–4077. [Google Scholar] [PubMed]
- Okazaki, T.; Bielawska, A.; Bell, R.M.; Hannun, Y.A. Role of ceramide as a lipid mediator of 1 alpha,25-dihydroxyvitamin D3-induced HL-60 cell differentiation. J. Biol. Chem. 1990, 265, 15823–15831. [Google Scholar] [PubMed]
- Airola, M.V.; Hannun, Y.A. Sphingolipidmetabolism and neutralsphingomyelinases. Handb. Exp. Pharmacol. 2013, 215, 57–76. [Google Scholar]
- Perrotta, C.; Cervia, D.; De Palma, C.; Assi, E.; Pellegrino, P.; Bassi, M.T.; Clementi, E. The emergingrole of acid sphingomyelinase in autophagy. Apoptosis 2015, 20, 635–644. [Google Scholar] [PubMed]
- Shamseddine, A.A.; Airola, M.V.; Hannun, Y. A Roles and regulation of neutral sphingomyelinase-2 in cellular and pathological processes. Adv. Biol. Regul. 2015, 57, 24–41. [Google Scholar] [CrossRef] [Green Version]
- Cataldi, S.; Borrelli, A.; Ceccarini, M.R.; Nakashidze, I.; Codini, M.; Belov, O.; Ivanov, A.; Krasavin, E.; Ferri, I.; Conte, C.; et al. Acid and Neutral Sphingomyelinase Behavior in Radiation-Induced Liver Pyroptosis and in the Protective/Preventive Role of rMnSOD. Int. J. Mol. Sci. 2020, 21, 3281. [Google Scholar] [CrossRef]
- Assi, E.; Cervia, D.; Bizzozero, L.; Capobianco, A.; Pambianco, S.; Morisi, F.; De Palma, C.; Moscheni, C.; Pellegrino, P.; Clementi, E.; et al. Modulation of Acid Sphingomyelinase in Melanoma Reprogrammes the Tumour Immune Microenvironment. Mediat. Inflamm. 2015, 2015, 370482. [Google Scholar] [CrossRef]
- Ji, Q.K.; Ma, J.W.; Liu, R.H.; Li, X.S.; Shen, F.Z.; Huang, L.Y.; Hui, L.; Ma, Y.J.; Jin, B.Z. CDCA7L promotes glioma proliferation by targeting CCND1 and predicts an unfavorable prognosis. Mol. Med. Rep. 2019, 20, 1149–1156. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Zhang, Y.; Liu, X.; Li, Z.; Xu, W.; He, S.; Huang, Y.; Zhang, H. Acid sphingomyelinase contributes to evodiamine-induced apoptosis in human gastric cancer SGC-7901 cells. DNA Cell Biol. 2011, 30, 407–412. [Google Scholar] [CrossRef]
- Albi, E.; Cataldi, S.; Ceccarini, M.R.; Conte, C.; Ferri, I.; Fettucciari, K.; Patria, F.F.; Beccari, T.; Codini, M. Gentamicin Targets Acid Sphingomyelinase in Cancer: The Case of the Human Gastric Cancer NCI-N87 Cells. Int. J. Mol. Sci. 2019, 20, 4375. [Google Scholar] [CrossRef] [Green Version]
- Clarke, C.J. Neutral Sphingomyelinases in Cancer: Friend or Foe? Adv. Cancer Res. 2018, 140, 97–119. [Google Scholar]
- Magrassi, L.; Adorni, L.; Montorfano, G.; Rapelli, S.; Butti, G.; Berra, B.; Milanesi, G. Vitamin D metabolites activate the sphingomyelin pathway and induce death of glioblastoma cells. Acta Neurochir. 1998, 140, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Meek, D.W. Regulation of the p53 response and its relationship to cancer. Biochem. J. 2015, 469, 325–346. [Google Scholar] [CrossRef] [PubMed]
- Hewison, M.; Freeman, L.; Hughes, S.V.; Evans, K.N.; Bland, R.; Eliopoulos, A.G.; Kilby, M.D.; Moss, P.A.; Chakraverty, R. Differential regulation of vitamin D receptor and its ligand in human monocyte-derived dendritic cells. J. Immunol. 2003, 170, 5382–5389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhang, Y.; Qian, C.; Cai, M.; Li, Y.; Li, Z.; You, Q.; Wang, Q.; Hu, R.; Guo, Q. GSK3β/β-catenin signaling is correlated with the differentiation of glioma cells induced by wogonin. Toxicol. Lett. 2013, 222, 212–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bien-Möller, S.; Balz, E.; Herzog, S.; Plantera, L.; Vogelgesang, S.; Weitmann, K.; Seifert, C.; Fink, M.A.; Marx, S.; Bialke, A.; et al. Association of Glioblastoma Multiforme Stem Cell Characteristics, Differentiation, and Microglia Marker Genes with Patient Survival. Stem Cells Int. 2018, 2018, 9628289. [Google Scholar] [CrossRef] [Green Version]
- Sousa, J.R.; Rosa, É.P.C.; Nunes, I.F.O.C.; Carvalho, C.M.R.G. Effect of vitamin D supplementation on patients with systemic lupus erythematosus: A systematic review. Rev. Bras. Reumatol. Engl. Ed. 2017, 57, 466–471. [Google Scholar] [CrossRef]
- Valle-Leal, J.; Limón-Armenta, J.; Serrano-Osuna, R.; López-Morales, C.M.; Alvárez-Bastidas, L. Active form of vitamin D in overweight and obese pediatric patients in northwest Mexico. Bol. Med. Hosp. Infant. M. 2017, 74, 413–418. [Google Scholar]
- Queiroz, D.J.M.; Silva, A.S.; Diniz, A.D.S.; Carvalho, A.T.; Araújo, E.P.D.S.; Neves, J.P.R.; Lacerda, L.M.; Toscano, L.T.; Gonçalves, M.D.C.R. Vitamin D insufficiency/deficiency and its association with cardiometabolic risk factors in Brazilian adolescents. Nutr. Hosp. 2019, 36, 142–148. [Google Scholar]
- Zhang, Y.; Dube, C.; Gibert, M., Jr.; Cruickshanks, N.; Wang, B.; Coughlan, M.; Yang, Y.; Setiady, I.; Deveau, C.; Saoud, K.; et al. The p53 Pathway in Glioblastoma. Cancers 2018, 10, 297. [Google Scholar]
- Wang, S.Y.; Zhang, J.L.; Zhang, D.; Bao, X.Q.; Sun, H. Recent advances in study of sphingolipids on liver diseases. Yao Xue Xue Bao 2015, 50, 1551–1558. [Google Scholar]
- Molino, S.; Tate, E.; McKillop, W.M.; Medin, J.A. Sphingolipid pathway enzymes modulate cell fate and immune responses. Immunotherapy 2017, 9, 1185–1198. [Google Scholar] [CrossRef]
- Ma, Y.Y.; Mou, X.Z.; Ding, Y.H.; Zou, H.; Huang, D.S. Delivery systems of ceramide in targeted cancer therapy: Ceramide alone or in combination with other anti-tumor agents. Expert Opin. Drug Deliv. 2016, 13, 1397–1406. [Google Scholar] [CrossRef]
- Li, F.; Zhang, N. Ceramide: Therapeutic Potential in Combination Therapy for Cancer Treatment. Curr. Drug Metab. 2015, 17, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Khiste, S.K.; Liu, Z.; Roy, K.R.; Uddin, M.B.; Hosain, S.B.; Gu, X.; Nazzal, S.; Hill, R.A.; Liu, Y.Y. Ceramide-RubusosideNanomicelles, a Potential Therapeutic Approach to Target Cancers Carrying p53 Missense Mutations. Mol. Cancer Ther. 2020, 19, 564–574. [Google Scholar] [CrossRef] [Green Version]
- Madigan, J.P.; Robey, R.W.; Poprawski, J.E.; Huang, H.; Clarke, C.J.; Gottesman, M.M.; Cabot, M.C.; Rosenberg, D.W. A role for ceramide glycosylation in resistance to oxaliplatin in colorectal cancer. Exp. Cell Res. 2020, 388, 111860. [Google Scholar] [CrossRef]
- Barón-Mendoza, I.; González-Arenas, A. Relationship between the effect of polyunsaturated fatty acids (PUFAs) on brain plasticity and the improvement on cognition and behavior in individuals with autism spectrum disorder. Nutr. Neurosci. 2020, 25, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Bocchini, V.; Casalone, R.; Collini, P.; Rebel, G.; Lo Curto, F. Changes in glial fibrillary acidic protein and karyotype during culturing of two cell lines established from human glioblastoma multiforme. Cell Tissue Res. 1991, 265, 73–81. [Google Scholar] [CrossRef]
- Ceccarini, M.R.; Codini, M.; Cataldi, S.; Vannini, S.; Lazzarini, A.; Floridi, A.; Moretti, M.; Villarini, M.; Fioretti, B.; Beccari, T.; et al. Acid sphingomyelinaseas target of LyciumChinense: Promising new action for cellhealth. Lipids Health Dis. 2016, 15, 570. [Google Scholar] [CrossRef] [Green Version]
- Lazzarini, A.; Macchiarulo, A.; Floridi, A.; Coletti, A.; Cataldi, S.; Codini, M.; Lazzarini, R.; Bartoccini, E.; Cascianelli, G.; Ambesi-Impiombato, F.S.; et al. Very-long-chain fatty acid sphingomyelin in nuclear lipid microdomains of hepatocytes and hepatoma cells: Can the exchange from C24:0 to C16:0 affect signal proteins and vitamin D receptor? Mol. Biol. Cell. 2015, 26, 2418–2425. [Google Scholar] [CrossRef] [PubMed]
- Traina, G. Mast cells in the brain—Old cells, new target. J. Integr. Neurosci. 2017, 16, S69–S83. [Google Scholar] [CrossRef] [PubMed]
- Uematsu, E.; Takino, S.; Okajima, H.; Tong, B.; Sugiyama, T.; Yamada, T.; Niimura, S.; Yamashiro, H. Use of in OvoChorioallantoic Membrane Engraftment to Culture Testes From Neonatal Mice. Comp. Med. 2014, 64, 264–269. [Google Scholar] [PubMed]
- Tubaro, C.; Arcuri, C.; Giambanco, I.; Donato, R. S100B in myoblasts regulates the transition from activation to quiescence and from quiescence to activation and reduces apoptosis. Biochim. Biophys. Acta 2011, 1813, 1092–1104. [Google Scholar] [CrossRef] [Green Version]
- Curcio, F.; Lazzarini, A.; Floridi, A.; Cataldi, S.; Lazzarini, R.; Loreti, E.; Ferri, I.; Ambesi-Impiombato, F.S. How microgravitychanges galectin-3 in thyroidfollicles. Biomed. Res. Int. 2014, 2014, 652863. [Google Scholar]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cataldi, S.; Arcuri, C.; Lazzarini, A.; Nakashidze, I.; Ragonese, F.; Fioretti, B.; Ferri, I.; Conte, C.; Codini, M.; Beccari, T.; et al. Effect of 1α,25(OH)2 Vitamin D3 in Mutant P53 Glioblastoma Cells: Involvement of Neutral Sphingomyelinase1. Cancers 2020, 12, 3163. https://doi.org/10.3390/cancers12113163
Cataldi S, Arcuri C, Lazzarini A, Nakashidze I, Ragonese F, Fioretti B, Ferri I, Conte C, Codini M, Beccari T, et al. Effect of 1α,25(OH)2 Vitamin D3 in Mutant P53 Glioblastoma Cells: Involvement of Neutral Sphingomyelinase1. Cancers. 2020; 12(11):3163. https://doi.org/10.3390/cancers12113163
Chicago/Turabian StyleCataldi, Samuela, Cataldo Arcuri, Andrea Lazzarini, Irina Nakashidze, Francesco Ragonese, Bernard Fioretti, Ivana Ferri, Carmela Conte, Michela Codini, Tommaso Beccari, and et al. 2020. "Effect of 1α,25(OH)2 Vitamin D3 in Mutant P53 Glioblastoma Cells: Involvement of Neutral Sphingomyelinase1" Cancers 12, no. 11: 3163. https://doi.org/10.3390/cancers12113163




