Sequential Colocalization of ERa, PR, and AR Hormone Receptors Using Confocal Microscopy Enables New Insights into Normal Breast and Prostate Tissue and Cancers
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
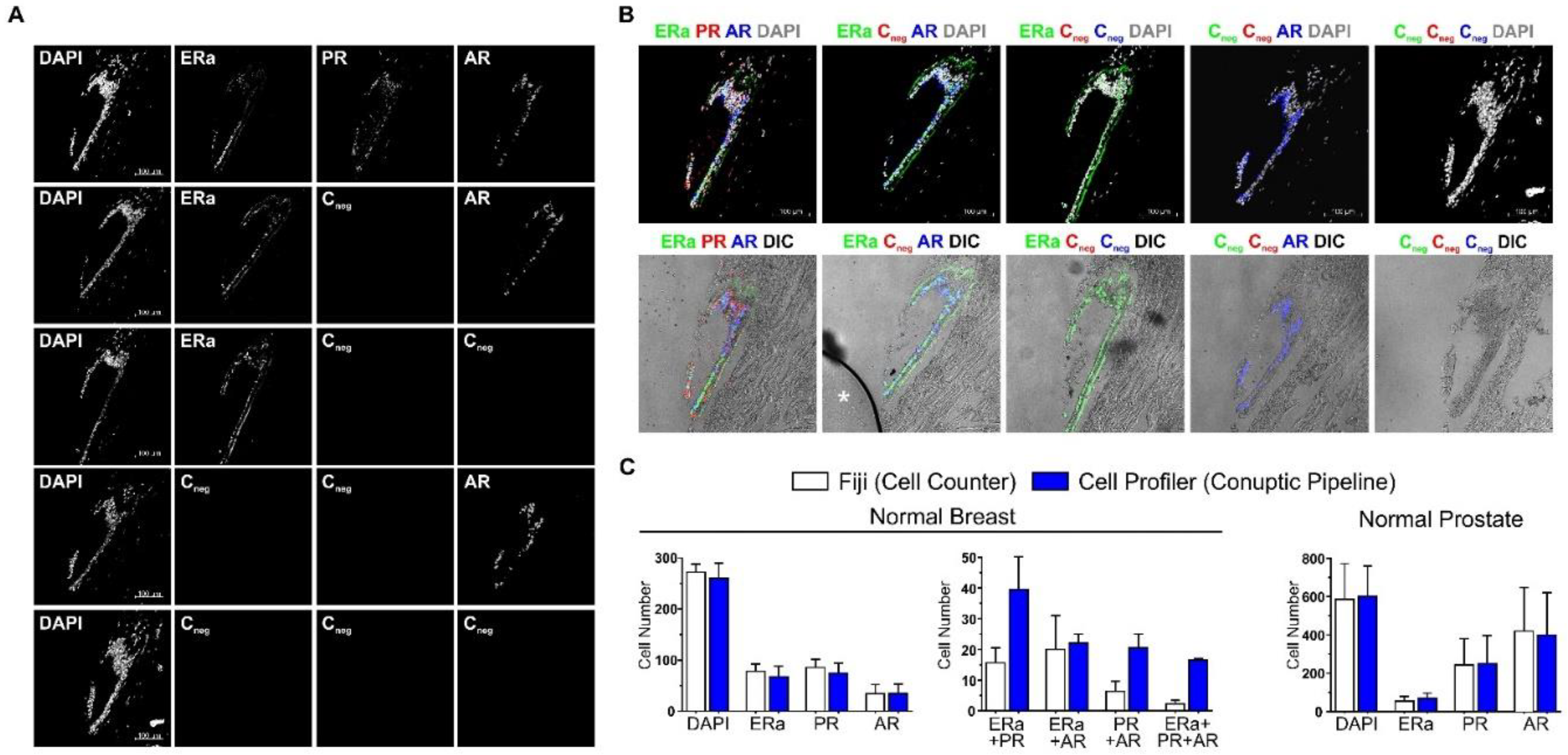
2.1. ColNu mIHCF Standardization
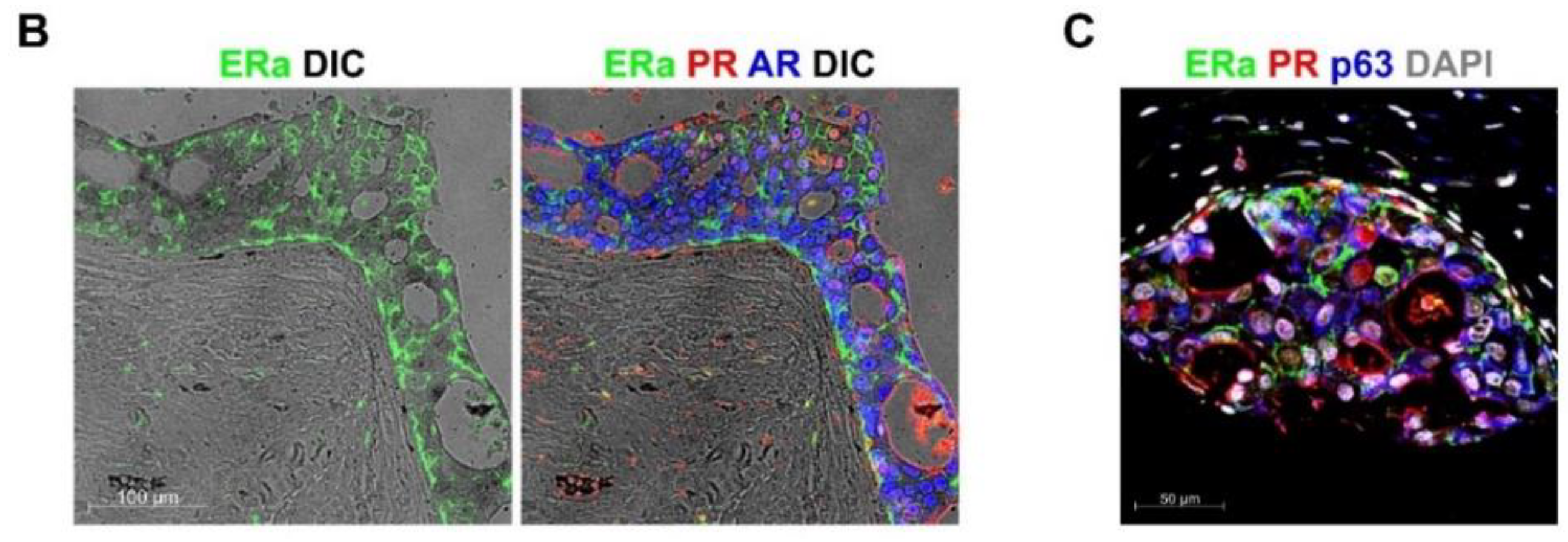
2.2. ColNu mIHCF in Normal Breast (NB) and Breast Cancer (BC)
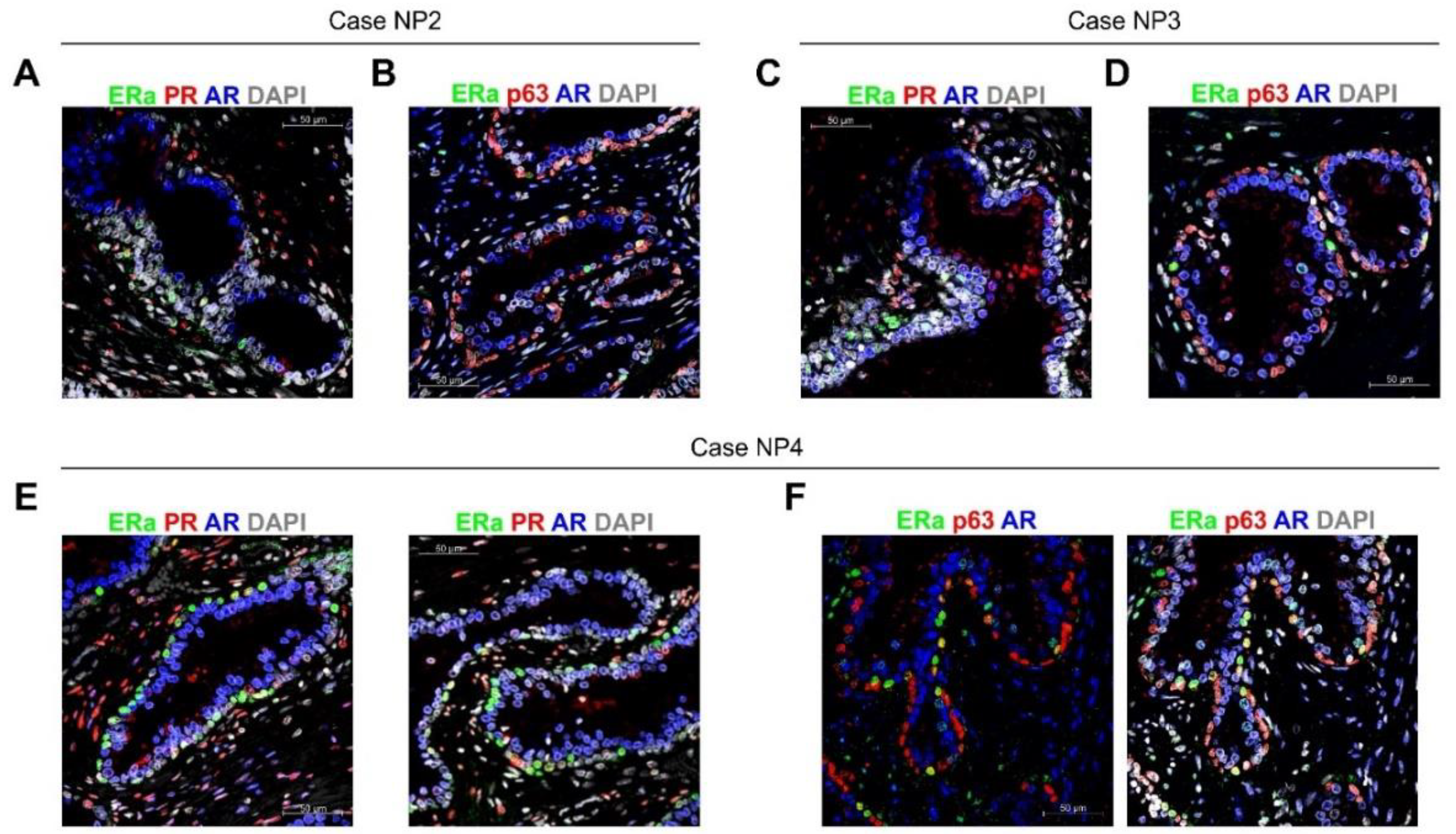
2.3. ColNu mIHCF in Normal Prostate (NP) from Paired Cancer Cases: ERa Marks a p63 + Basal Cell Subpopulation
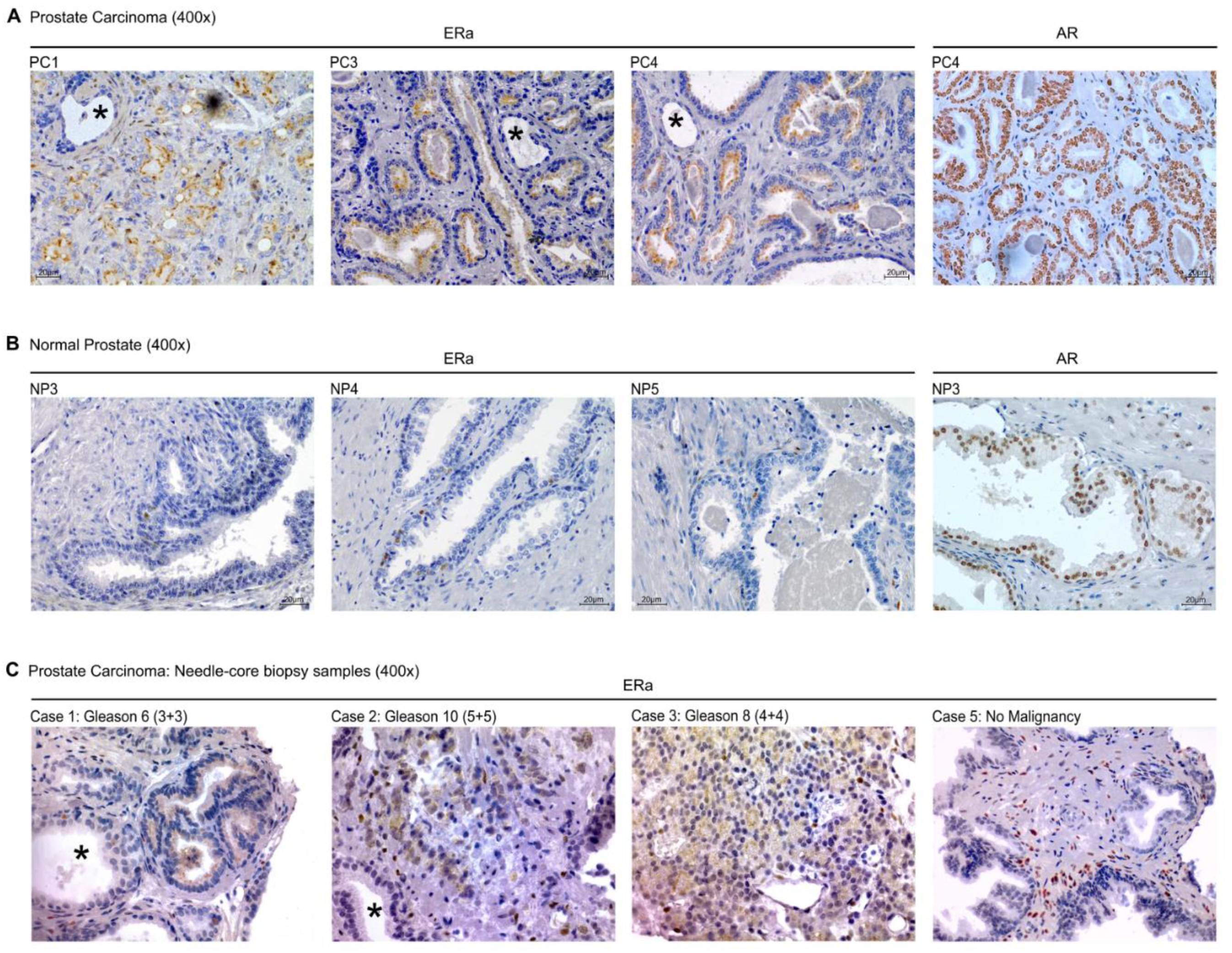
2.4. ColNu mIHCF in and Prostate Cancer (PC): Apical ERa is a Cancer Marker
2.5. Conventional IHC of ERa in Prostate Cancer in Tissue Sections and Needle-Core Biopsy
3. Discussion
4. Materials and Methods
4.1. Ethical Statement and Pathological and Clinical Data
4.2. Tissue Microarrays (TMA)
4.3. Prostate Needle-Core Biopsy
4.4. Immunohistochemistry (IHC)
4.5. ColNu mIHCF
4.6. Quantification of the Cell Populations
4.7. Graphs and Descriptive Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| TSA | Tyramide signal amplification |
| TMA | Tissue microarray |
| IVD | In vitro diagnostics |
| ERa | Estrogen receptor alpha |
| PR | Progesterone receptor |
| AR | Androgen receptor |
| NB | Normal breast |
| BC | Breast cancer |
| NP | Normal prostate |
| PC | Prostate cancer |
| DIC | Differential interfering contrast |
| DAPI | 4′,6-Diamidino-2-phenylindole |
| FFPE | Formalin-fixed paraffin-embedded |
References
- Stack, E.C.; Wang, C.; Roman, K.A.; Hoyt, C.C. Multiplexed Immunohistochemistry, Imaging, and Quantitation: A Review, with an Assessment of Tyramide Signal Amplification, Multispectral Imaging and Multiplex Analysis. Methods 2014, 70, 46–58. [Google Scholar] [CrossRef]
- Gerdes, M.J.; Sevinsky, C.J.; Sood, A.; Adak, S.; Bello, M.O.; Bordwell, A.; Can, A.; Corwin, A.; Dinn, S.; Filkins, R.J.; et al. Highly Multiplexed Single-Cell Analysis of Formalin-Fixed, Paraffin-Embedded Cancer Tissue. Proc. Natl. Acad. Sci. USA 2013, 110, 11982–11987. [Google Scholar] [CrossRef] [Green Version]
- Parra, E.R.; Uraoka, N.; Jiang, M.; Cook, P.; Gibbons, D.; Forget, M.; Bernatchez, C.; Haymaker, C.; Wistuba, I.I.; Rodriguez-Canales, J. Validation of Multiplex Immunofluorescence Panels using Multispectral Microscopy for Immune-Profiling of Formalin-Fixed and Paraffin-Embedded Human Tumor Tissues. Sci. Rep. 2017, 7, 13380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blom, S.; Paavolainen, L.; Bychkov, D.; Turkki, R.; Mäki-Teeri, P.; Hemmes, A.; Välimäki, K.; Lundin, J.; Kallioniemi, O.; Pellinen, T. Systems Pathology by Multiplexed Immunohistochemistry and Whole-Slide Digital Image Analysis. Sci. Rep. 2017, 7, 15580. [Google Scholar] [CrossRef] [PubMed]
- Tsujikawa, T.; Kumar, S.; Borkar, R.N.; Azimi, V.; Thibault, G.; Chang, Y.H.; Balter, A.; Kawashima, R.; Choe, G.; Sauer, D.; et al. Quantitative Multiplex Immunohistochemistry Reveals Myeloid-Inflamed Tumor-Immune Complexity Associated with Poor Prognosis. Cell Rep. 2017, 19, 203–217. [Google Scholar] [CrossRef] [PubMed]
- Morrison, L.E.; Lefever, M.R.; Behman, L.J.; Leibold, T.; Roberts, E.A.; Horchner, U.B.; Bauer, D.R. Brightfield Multiplex Immunohistochemistry with Multispectral Imaging. Lab. Investig. 2020, 100, 1124–1136. [Google Scholar] [CrossRef]
- Akturk, G.; Sweeney, R.; Remark, R.; Merad, M.; Gnjatic, S. Multiplexed Immunohistochemical Consecutive Staining on Single Slide (MICSSS): Multiplexed Chromogenic IHC Assay for High-Dimensional Tissue Analysis. Methods Mol. Biol. 2020, 2055, 497–519. [Google Scholar]
- Wang, G.; Achim, C.L.; Hamilton, R.L.; Wiley, C.A.; Soontornniyomkij, V. Tyramide Signal Amplification Method in Multiple-Label Immunofluorescence Confocal Microscopy. Methods 1999, 18, 459–464. [Google Scholar] [CrossRef]
- Tóth, Z.E.; Mezey, E. Simultaneous Visualization of Multiple Antigens with Tyramide Signal Amplification using Antibodies from the Same Species. J. Histochem. Cytochem. 2007, 55, 545–554. [Google Scholar] [CrossRef] [Green Version]
- Pivetta, E.; Spessotto, P. Multispectral Imaging Technology: Visualize, Analyze, Phenotyping, and Quantify Immune Cells in Situ. Int. J. Biol. Markers 2020, 35, 26–30. [Google Scholar] [CrossRef]
- Fassler, D.J.; Abousamra, S.; Gupta, R.; Chen, C.; Zhao, M.; Paredes, D.; Batool, S.A.; Knudsen, B.S.; Escobar-Hoyos, L.; Shroyer, K.R.; et al. Deep Learning-Based Image Analysis Methods for Brightfield-Acquired Multiplex Immunohistochemistry Images. Diagn. Pathol. 2020, 15, 1–11. [Google Scholar]
- Taube, J.M.; Akturk, G.; Angelo, M.; Engle, E.L.; Gnjatic, S.; Greenbaum, S.; Greenwald, N.F.; Hedvat, C.V.; Hollmann, T.J.; Juco, J.; et al. The Society for Immunotherapy of Cancer Statement on Best Practices for Multiplex Immunohistochemistry (IHC) and Immunofluorescence (IF) Staining and Validation. J. Immunother. Cancer 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Stein, J.E.; Rimm, D.L.; Wang, D.W.; Bell, J.M.; Johnson, D.B.; Sosman, J.A.; Schalper, K.A.; Anders, R.A.; Wang, H.; et al. Comparison of Biomarker Modalities for Predicting Response to PD-1/PD-L1 Checkpoint Blockade: A Systematic Review and Meta-Analysis. JAMA Oncol. 2019, 5, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Gorris, M.A.J.; Halilovic, A.; Rabold, K.; van Duffelen, A.; Wickramasinghe, I.N.; Verweij, D.; Wortel, I.M.N.; Textor, J.C.; de Vries, I.J.M.; Figdor, C.G. Eight-Color Multiplex Immunohistochemistry for Simultaneous Detection of Multiple Immune Checkpoint Molecules within the Tumor Microenvironment. J. Immunol. 2018, 200, 347–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeong, J.; Tan, T.; Chow, Z.L.; Cheng, Q.; Lee, B.; Seet, A.; Lim, J.X.; Lim, J.C.T.; Ong, C.C.H.; Thike, A.A.; et al. Multiplex immunohistochemistry/immunofluorescence (mIHC/IF) for PD-L1 Testing in Triple-Negative Breast Cancer: A Translational Assay Compared with Conventional IHC. J. Clin. Pathol. 2020, 73, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Beaulande, M.; Ben Hadj, S.; Chamorey, E.; Schiappa, R.; Long-Mira, E.; Lassalle, S.; Butori, C.; Cohen, C.; Leroy, S.; et al. Chromogenic Multiplex Immunohistochemistry Reveals Modulation of the Immune Microenvironment Associated with Survival in Elderly Patients with Lung Adenocarcinoma. Cancers 2018, 10, 326. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.K.; Wang, M.; Sun, Y.; Di Costanzo, N.; Mitchell, C.; Achuthan, A.; Hamilton, J.A.; Busuttil, R.A.; Boussioutas, A. Macrophage Spatial Heterogeneity in Gastric Cancer Defined by Multiplex Immunohistochemistry. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, Y.; Masugi, Y.; Abe, T.; Yamazaki, K.; Ueno, A.; Fujii-Nishimura, Y.; Hori, S.; Yagi, H.; Abe, Y.; Kitago, M.; et al. Three Distinct Stroma Types in Human Pancreatic Cancer Identified by Image Analysis of Fibroblast Subpopulations and Collagen. Clin. Cancer Res. 2020. [Google Scholar] [CrossRef]
- Karihtala, K.; Leivonen, S.K.; Bruck, O.; Karjalainen-Lindsberg, M.L.; Mustjoki, S.; Pellinen, T.; Leppa, S. Prognostic Impact of Tumor-Associated Macrophages on Survival is Checkpoint Dependent in Classical Hodgkin Lymphoma. Cancers 2020, 12, 877. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.Y.; Lim, J.Q.; Yeong, J.; Ravi, V.; Guan, P.; Boot, A.; Tay, T.K.Y.; Selvarajan, S.; Md Nasir, N.D.; Loh, J.H.; et al. Multiomic Analysis and Immunoprofiling Reveal Distinct Subtypes of Human Angiosarcoma. J. Clin. Investig. 2020, 130, 5833–5846. [Google Scholar] [CrossRef]
- Mitchell, R.T.; Camacho-Moll, M.; Macdonald, J.; Anderson, R.A.; Kelnar, C.J.; O’Donnell, M.; Sharpe, R.M.; Smith, L.B.; Grigor, K.M.; Wallace, W.H.; et al. Intratubular Germ Cell Neoplasia of the Human Testis: Heterogeneous Protein Expression and Relation to Invasive Potential. Mod. Pathol. 2014, 27, 1255–1266. [Google Scholar] [CrossRef] [PubMed]
- Moch, H.; Humphrey, P.A.; Ulbright, T.M.; Reuter, V.E. WHO Classification of Tumours of the Urinary System and Male Genital Organs; International Agency for Research on Cancer (IARC): Lyon, France, 2016.
- Allison, K.H.; Hammond, M.E.H.; Dowsett, M.; McKernin, S.E.; Carey, L.A.; Fitzgibbons, P.L.; Hayes, D.F.; Lakhani, S.R.; Chavez-MacGregor, M.; Perlmutter, J.; et al. Estrogen and Progesterone Receptor Testing in Breast Cancer: ASCO/CAP Guideline Update. J. Clin. Oncol. 2020, 38, 1346–1366. [Google Scholar] [CrossRef] [PubMed]
- Hammond, M.E.; Hayes, D.F.; Dowsett, M.; Allred, D.C.; Hagerty, K.L.; Badve, S.; Fitzgibbons, P.L.; Francis, G.; Goldstein, N.S.; Hayes, M.; et al. American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for Immunohistochemical Testing of Estrogen and Progesterone Receptors in Breast Cancer. J. Clin. Oncol. 2010, 28, 2784–2795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baschong, W.; Suetterlin, R.; Laeng, R.H. Control of Autofluorescence of Archival Formaldehyde-Fixed, Paraffin-Embedded Tissue in Confocal Laser Scanning Microscopy (CLSM). J. Histochem. Cytochem. 2001, 49, 1565–1572. [Google Scholar] [CrossRef] [Green Version]
- Schnell, S.A.; Staines, W.A.; Wessendorf, M.W. Reduction of Lipofuscin-Like Autofluorescence in Fluorescently Labeled Tissue. J. Histochem. Cytochem. 1999, 47, 719–730. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Yu, H.; Zheng, D.; Cao, Q.; Wang, Y.; Harris, D.; Wang, Y. Sudan Black B Reduces Autofluorescence in Murine Renal Tissue. Arch. Pathol. Lab. Med. 2011, 135, 1335–1342. [Google Scholar] [CrossRef]
- Viegas, M.S.; Martins, T.C.; Seco, F.; do Carmo, A. An Improved and Cost-Effective Methodology for the Reduction of Autofluorescence in Direct Immunofluorescence Studies on Formalin-Fixed Paraffin-Embedded Tissues. Eur. J. Histochem. 2007, 51, 59–66. [Google Scholar]
- Garcia-Lavandeira, M.; Quereda, V.; Flores, I.; Saez, C.; Diaz-Rodriguez, E.; Japon, M.A.; Ryan, A.K.; Blasco, M.A.; Dieguez, C.; Malumbres, M.; et al. A GRFa2/Prop1/stem (GPS) Cell Niche in the Pituitary. PLoS ONE 2009, 4, e4815. [Google Scholar] [CrossRef] [Green Version]
- Diaz-Rodriguez, E.; Garcia-Lavandeira, M.; Perez-Romero, S.; Senra, A.; Canibano, C.; Palmero, I.; Borrello, M.G.; Dieguez, C.; Alvarez, C.V. Direct Promoter Induction of p19Arf by Pit-1 Explains the Dependence Receptor RET/Pit-1/p53-Induced Apoptosis in the Pituitary Somatotroph Cells. Oncogene 2012, 31, 2824–2835. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Lavandeira, M.; Diaz-Rodriguez, E.; Bahar, D.; Garcia-Rendueles, A.R.; Rodrigues, J.S.; Dieguez, C.; Alvarez, C.V. Pituitary Cell Turnover: From Adult Stem Cell Recruitment through Differentiation to Death. Neuroendocrinology 2015, 101, 175–192. [Google Scholar] [CrossRef]
- Bravo, S.B.; Garcia-Rendueles, M.E.; Garcia-Rendueles, A.R.; Rodrigues, J.S.; Perez-Romero, S.; Garcia-Lavandeira, M.; Suarez-Farina, M.; Barreiro, F.; Czarnocka, B.; Senra, A.; et al. Humanized Medium (h7H) Allows Long-Term Primary Follicular Thyroid Cultures from Human Normal Thyroid, Benign Neoplasm, and Cancer. J. Clin. Endocrinol. Metab. 2013, 98, 2431–2441. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rendueles, A.; Rodrigues, J.S.; Garcia-Rendueles, M.; Suarez-Farina, M.; Perez-Romero, S.; Barreiro, F.; Bernabeu, I.; Rodriguez-Garcia, J.; Fugazzola, L.; Sakai, T.; et al. Rewiring of the Apoptotic TGF-Beta-SMAD/NFkappaB Pathway through an Oncogenic Function of p27 in Human Papillary Thyroid Cancer. Oncogene 2017, 36, 652–666. [Google Scholar] [CrossRef]
- Hamzeh, O.; Alkhateeb, A.; Zheng, J.Z.; Kandalam, S.; Leung, C.; Atikukke, G.; Cavallo-Medved, D.; Palanisamy, N.; Rueda, L. A Hierarchical Machine Learning Model to Discover Gleason Grade-Specific Biomarkers in Prostate Cancer. Diagnostics 2019, 9, 219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, J.S.; Hickey, T.E.; Tarulli, G.A.; Williams, M.; Tilley, W.D. Deciphering the Divergent Roles of Progestogens in Breast Cancer. Nat. Rev. Cancer. 2017, 17, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhou, S.; Gustafsson, J.A. Nuclear Receptors: Recent Drug Discovery for Cancer Therapies. Endocr. Rev. 2019, 40, 1207–1249. [Google Scholar] [CrossRef]
- Foidart, J.M.; Colin, C.; Denoo, X.; Desreux, J.; Beliard, A.; Fournier, S.; de Lignieres, B. Estradiol and Progesterone Regulate the Proliferation of Human Breast Epithelial Cells. Fertil. Steril. 1998, 69, 963–969. [Google Scholar] [CrossRef]
- Chang, K.J.; Lee, T.T.; Linares-Cruz, G.; Fournier, S.; de Lignieres, B. Influences of Percutaneous Administration of Estradiol and Progesterone on Human Breast Epithelial Cell Cycle in Vivo. Fertil. Steril. 1995, 63, 785–791. [Google Scholar] [CrossRef]
- Giulianelli, S.; Vaqué, J.P.; Soldati, R.; Wargon, V.; Vanzulli, S.I.; Martins, R.; Zeitlin, E.; Molinolo, A.A.; Helguero, L.A.; Lamb, C.A.; et al. Estrogen Receptor Alpha Mediates Progestin-Induced Mammary Tumor Growth by Interacting with Progesterone Receptors at the Cyclin D1/MYC Promoters. Cancer Res. 2012, 72, 2416–2427. [Google Scholar] [CrossRef] [Green Version]
- Oh, H.; Eliassen, A.H.; Wang, M.; Smith-Warner, S.A.; Beck, A.H.; Schnitt, S.J.; Collins, L.C.; Connolly, J.L.; Montaser-Kouhsari, L.; Polyak, K.; et al. Expression of Estrogen Receptor, Progesterone Receptor, and Ki67 in Normal Breast Tissue in Relation to Subsequent Risk of Breast Cancer. NPJ Breast Cancer 2016, 2. [Google Scholar] [CrossRef] [Green Version]
- Campbell, E.J.; Tesson, M.; Doogan, F.; Mohammed, Z.M.A.; Mallon, E.; Edwards, J. The Combined Endocrine Receptor in Breast Cancer, a Novel Approach to Traditional Hormone Receptor Interpretation and a Better Discriminator of Outcome than ER and PR Alone. Br. J. Cancer 2016, 115, 967–973. [Google Scholar] [CrossRef] [Green Version]
- Lim, E.; Palmieri, C.; Tilley, W.D. Renewed Interest in the Progesterone Receptor in Breast Cancer. Br. J. Cancer 2016, 115, 909–911. [Google Scholar] [CrossRef] [Green Version]
- Hickey, T.E.; Robinson, J.L.; Carroll, J.S.; Tilley, W.D. Minireview: The Androgen Receptor in Breast Tissues: Growth Inhibitor, Tumor Suppressor, Oncogene? Mol. Endocrinol. 2012, 26, 1252–1267. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, D.R.; Bernales, S.; Jacobsen, B.M.; Cittelly, D.M.; Howe, E.N.; D’Amato, N.C.; Spoelstra, N.S.; Edgerton, S.M.; Jean, A.; Guerrero, J.; et al. Role of the Androgen Receptor in Breast Cancer and Preclinical Analysis of Enzalutamide. Breast Cancer Res. 2014, 16, R7. [Google Scholar] [CrossRef] [Green Version]
- Bardia, A.; Gucalp, A.; DaCosta, N.; Gabrail, N.; Danso, M.; Ali, H.; Blackwell, K.L.; Carey, L.A.; Eisner, J.R.; Baskin-Bey, E.S.; et al. Phase 1 Study of Seviteronel, a Selective CYP17 Lyase and Androgen Receptor Inhibitor, in Women with Estrogen Receptor-Positive Or Triple-Negative Breast Cancer. Breast Cancer Res. Treat. 2018, 171, 111–120. [Google Scholar] [CrossRef]
- Nadji, M.; Gomez-Fernandez, C.; Ganjei-Azar, P.; Morales, A.R. Immunohistochemistry of Estrogen and Progesterone Receptors Reconsidered: Experience with 5,993 Breast Cancers. Am. J. Clin. Pathol. 2005, 123, 21–27. [Google Scholar] [CrossRef]
- Hefti, M.M.; Hu, R.; Knoblauch, N.W.; Collins, L.C.; Haibe-Kains, B.; Tamimi, R.M.; Beck, A.H. Estrogen Receptor negative/progesterone Receptor Positive Breast Cancer is Not a Reproducible Subtype. Breast Cancer Res. 2013, 15, R68. [Google Scholar] [CrossRef] [Green Version]
- Zaha, D.C. Significance of Immunohistochemistry in Breast Cancer. World J. Clin. Oncol. 2014, 5, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Chaudhri, R.A.; Olivares-Navarrete, R.; Cuenca, N.; Hadadi, A.; Boyan, B.D.; Schwartz, Z. Membrane Estrogen Signaling Enhances Tumorigenesis and Metastatic Potential of Breast Cancer Cells Via Estrogen Receptor-α36 (ERα36). J. Biol. Chem. 2012, 287, 7169–7181. [Google Scholar] [CrossRef] [Green Version]
- Badve, S.; Vladislav, I.T.; Spaulding, B.; Strickland, A.; Hernandez, S.; Bird-Turner, L.; Dodson, C.; Elleby, B.; Phillips, T. EP1: A Novel Rabbit Monoclonal Antibody for Detection of Oestrogen Receptor Alpha. J. Clin. Pathol. 2013, 66, 1051–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giovannelli, P.; Di Donato, M.; Galasso, G.; Di Zazzo, E.; Medici, N.; Bilancio, A.; Migliaccio, A.; Castoria, G. Breast Cancer Stem Cells: The Role of Sex Steroid Receptors. World J. Stem Cells 2019, 11, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Giovannelli, P.; Di Donato, M.; Galasso, G.; Di Zazzo, E.; Bilancio, A.; Migliaccio, A. The Androgen Receptor in Breast Cancer. Front. Endocrinol. (Lausanne) 2018, 9, 492. [Google Scholar] [CrossRef] [Green Version]
- Ballare, C.; Uhrig, M.; Bechtold, T.; Sancho, E.; Di Domenico, M.; Migliaccio, A.; Auricchio, F.; Beato, M. Two Domains of the Progesterone Receptor Interact with the Estrogen Receptor and are Required for Progesterone Activation of the c-Src/Erk Pathway in Mammalian Cells. Mol. Cell. Biol. 2003, 23, 1994–2008. [Google Scholar] [CrossRef] [Green Version]
- Maselli, A.; Pierdominici, M.; Vitale, C.; Ortona, E. Membrane Lipid Rafts and Estrogenic Signalling: A Functional Role in the Modulation of Cell Homeostasis. Apoptosis 2015, 20, 671–678. [Google Scholar] [CrossRef]
- Marquez, D.C.; Chen, H.W.; Curran, E.M.; Welshons, W.V.; Pietras, R.J. Estrogen Receptors in Membrane Lipid Rafts and Signal Transduction in Breast Cancer. Mol. Cell. Endocrinol. 2006, 246, 91–100. [Google Scholar] [CrossRef]
- Giovannelli, P.; Di Donato, M.; Auricchio, F.; Castoria, G.; Migliaccio, A. Androgens Induce Invasiveness of Triple Negative Breast Cancer Cells through AR/Src/PI3-K Complex Assembly. Sci. Rep. 2019, 9, 4490. [Google Scholar] [CrossRef] [Green Version]
- Leav, I.; Lau, K.M.; Adams, J.Y.; McNeal, J.E.; Taplin, M.E.; Wang, J.; Singh, H.; Ho, S.M. Comparative Studies of the Estrogen Receptors Beta and Alpha and the Androgen Receptor in Normal Human Prostate Glands, Dysplasia, and in Primary and Metastatic Carcinoma. Am. J. Pathol. 2001, 159, 79–92. [Google Scholar] [CrossRef]
- Sehgal, P.D.; Bauman, T.M.; Nicholson, T.M.; Vellky, J.E.; Ricke, E.A.; Tang, W.; Xu, W.; Huang, W.; Ricke, W.A. Tissue-Specific Quantification and Localization of Androgen and Estrogen Receptors in Prostate Cancer. Hum. Pathol. 2019, 89, 99–108. [Google Scholar] [CrossRef]
- Di Zazzo, E.; Galasso, G.; Giovannelli, P.; Di Donato, M.; Castoria, G. Estrogens and their Receptors in Prostate Cancer: Therapeutic Implications. Front. Oncol. 2018, 8, 2. [Google Scholar] [CrossRef] [Green Version]
- Risbridger, G.P.; Wang, H.; Frydenberg, M.; Cunha, G. The Metaplastic Effects of Estrogen on Mouse Prostate Epithelium: Proliferation of Cells with Basal Cell Phenotype. Endocrinology 2001, 142, 2443–2450. [Google Scholar] [CrossRef]
- Majumdar, S.; Rinaldi, J.C.; Malhotra, N.R.; Xie, L.; Hu, D.P.; Gauntner, T.D.; Grewal, H.S.; Hu, W.Y.; Kim, S.H.; Katzenellenbogen, J.A.; et al. Differential Actions of Estrogen Receptor Alpha and Beta Via Nongenomic Signaling in Human Prostate Stem and Progenitor Cells. Endocrinology 2019, 160, 2692–2708. [Google Scholar] [CrossRef]
- Morais-Santos, M.; Werneck-Gomes, H.; Campolina-Silva, G.; Santos, L.C.; Mahecha, G.A.B.; Hess, R.A.; Oliveira, C.A. Basal Cells show Increased Expression of Aromatase and Estrogen Receptor α in Prostate Epithelial Lesions of Male Aging Rats. Endocrinology 2018, 159, 723–732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grindstad, T.; Skjefstad, K.; Andersen, S.; Ness, N.; Nordby, Y.; Al-Saad, S.; Fismen, S.; Donnem, T.; Khanehkenari, M.R.; Busund, L.; et al. Estrogen Receptors α and β and Aromatase as Independent Predictors for Prostate Cancer Outcome. Sci. Rep. 2016, 6, 33114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Megas, G.; Chrisofos, M.; Anastasiou, I.; Tsitlidou, A.; Choreftaki, T.; Deliveliotis, C. Estrogen Receptor (α and β) but Not Androgen Receptor Expression is Correlated with Recurrence, Progression and Survival in Post Prostatectomy T3N0M0 Locally Advanced Prostate Cancer in an Urban Greek Population. Asian J. Androl. 2015, 17, 98–105. [Google Scholar] [CrossRef]
- Di Zazzo, E.; Galasso, G.; Giovannelli, P.; Di Donato, M.; Bilancio, A.; Perillo, B.; Sinisi, A.A.; Migliaccio, A.; Castoria, G. Estrogen Receptors in Epithelial-Mesenchymal Transition of Prostate Cancer. Cancers 2019, 11, 1418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, S.; Prasad, M.L. Comparative Evaluation of High-Throughput Small-Core (0.6-mm) and Large-Core (2-mm) Thyroid Tissue Microarray: Is Larger Better? Arch. Pathol. Lab. Med. 2012, 136, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Allred, D.C.; Harvey, J.M.; Berardo, M.; Clark, G.M. Prognostic and Predictive Factors in Breast Cancer by Immunohistochemical Analysis. Mod. Pathol. 1998, 11, 155–168. [Google Scholar] [PubMed]
- Phillips, T.; Murray, G.; Wakamiya, K.; Askaa, J.; Huang, D.; Welcher, R.; Pii, K.; Allred, D.C. Development of Standard Estrogen and Progesterone Receptor Immunohistochemical Assays for Selection of Patients for Antihormonal Therapy. Appl. Immunohistochem. Mol. Morphol. 2007, 15, 325–331. [Google Scholar]
- Choudhury, K.R.; Yagle, K.J.; Swanson, P.E.; Krohn, K.A.; Rajendran, J.G. A Robust Automated Measure of Average Antibody Staining in Immunohistochemistry Images. J. Histochem. Cytochem. 2010, 58, 95–107. [Google Scholar] [CrossRef] [Green Version]
- Speel, E.J.; Hopman, A.H.; Komminoth, P. Amplification Methods to Increase the Sensitivity of in Situ Hybridization: Play Card(s). J. Histochem. Cytochem. 1999, 47, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Von Wasielewski, R.; Mengel, M.; Gignac, S.; Wilkens, L.; Werner, M.; Georgii, A. Tyramine Amplification Technique in Routine Immunohistochemistry. J. Histochem. Cytochem. 1997, 45, 1455–1459. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamentsky, L.; Jones, T.R.; Fraser, A.; Bray, M.; Logan, D.J.; Madden, K.L.; Ljosa, V.; Rueden, C.; Eliceiri, K.W.; Carpenter, A.E. Improved Structure, Function and Compatibility for CellProfiler: Modular High-Throughput Image Analysis Software. Bioinformatics 2011, 27, 1179–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chenlo, M.; Aliyev, E.; Rodrigues, J.S.; Vieiro-Balo, P.; Blanco Freire, M.N.; Cameselle-Teijeiro, J.M.; Alvarez, C.V. Sequential Colocalization of ERa, PR, and AR Hormone Receptors Using Confocal Microscopy Enables New Insights into Normal Breast and Prostate Tissue and Cancers. Cancers 2020, 12, 3591. https://doi.org/10.3390/cancers12123591
Chenlo M, Aliyev E, Rodrigues JS, Vieiro-Balo P, Blanco Freire MN, Cameselle-Teijeiro JM, Alvarez CV. Sequential Colocalization of ERa, PR, and AR Hormone Receptors Using Confocal Microscopy Enables New Insights into Normal Breast and Prostate Tissue and Cancers. Cancers. 2020; 12(12):3591. https://doi.org/10.3390/cancers12123591
Chicago/Turabian StyleChenlo, Miguel, Elvin Aliyev, Joana S. Rodrigues, Paula Vieiro-Balo, Manuel N. Blanco Freire, José Manuel Cameselle-Teijeiro, and Clara V. Alvarez. 2020. "Sequential Colocalization of ERa, PR, and AR Hormone Receptors Using Confocal Microscopy Enables New Insights into Normal Breast and Prostate Tissue and Cancers" Cancers 12, no. 12: 3591. https://doi.org/10.3390/cancers12123591
APA StyleChenlo, M., Aliyev, E., Rodrigues, J. S., Vieiro-Balo, P., Blanco Freire, M. N., Cameselle-Teijeiro, J. M., & Alvarez, C. V. (2020). Sequential Colocalization of ERa, PR, and AR Hormone Receptors Using Confocal Microscopy Enables New Insights into Normal Breast and Prostate Tissue and Cancers. Cancers, 12(12), 3591. https://doi.org/10.3390/cancers12123591





