Predicting and Quantifying Antagonistic Effects of Natural Compounds Given with Chemotherapeutic Agents: Applications for High-Throughput Screening
Abstract
:Simple Summary
Abstract
1. Introduction
2. Drug Synergy and Antagonism
3. Natural Products that Inhibit Chemotherapeutics
3.1. Genistein
3.2. (-)–Epigallocatechin Gallate (EGCG)
3.3. Curcumin
3.4. Vitamin C
3.5. Tangeretin
3.6. Xanthorrhizol
3.7. Si-Wu-Tang (SWT)
3.8. Quercetin and Myricetin
3.9. Tannic Acid, Gallic Acid, Caffeic Acid
4. Tools for Evaluating or Predicting Drug Synergy and Antagonism and Their Applications
4.1. High Throughput Screening for Natural Product–Drug Antagonism
4.2. Deep Learning for Prediction of Natural Product Antagonism
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cancer.org. Available online: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2020.html (accessed on 5 June 2020).
- Who.int. World Health Organization: WHO. The Top 10 Causes of Death. Available online: https://www.who.int/en/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 22 May 2020).
- Zugazagoitia, J.; Guedes, C.; Ponce, S.; Ferrer, I.; Molina-Pinelo, S.; Paz-Ares, L. Current Challenges in Cancer Treatment. Clin. Ther. 2016, 38, 1551–1566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, S.E.; Chester, J.D. Personalised cancer medicine. Int. J. Cancer 2015, 137, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Beyer, A.M.; Bonini, M.G.; Moslehi, J. Cancer therapy-induced cardiovascular toxicity: Old/new problems and old drugs. Am. J. Physiol. Heart Circ. Physiol. 2019, 317, H164–H167. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L.; Vaishnav, P. Natural products for cancer chemotherapy. Microb. Biotechnol. 2011, 4, 687–699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutta, S.; Mahalanobish, S.; Saha, S.; Ghosh, S.; Sil, P.C. Natural products: An upcoming therapeutic approach to cancer. Food Chem. Toxicol. 2019, 128, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, M.K.; Warrier, S.; Kumar, A.P.; Sethi, G.; Arfuso, F. Potential Role of Natural Compounds as Anti-Angiogenic Agents in Cancer. Curr. Vasc. Pharmacol. 2017, 15, 503–519. [Google Scholar] [CrossRef] [PubMed]
- Karikas, G.A. Anticancer and chemopreventing natural products: Some biochemical and therapeutic aspects. J. Buon 2010, 15, 627–638. [Google Scholar]
- Yuan, R.; Hou, Y.; Sun, W.; Yu, J.; Liu, X.; Niu, Y.; Lu, J.J.; Chen, X. Natural products to prevent drug resistance in cancer chemotherapy: A review. Ann. N. Y. Acad. Sci. 2017, 1401, 19–27. [Google Scholar] [CrossRef]
- Vidoni, C.; Ferraresi, A.; Secomandi, E.; Vallino, L.; Dhanasekaran, D.N.; Isidoro, C. Epigenetic targeting of autophagy for cancer prevention and treatment by natural compounds. Semin. Cancer Biol. 2020, 66, 34–44. [Google Scholar] [CrossRef]
- Medina-Franco, J. New Approaches for the Discovery of Pharmacologically-Active Natural Compounds. Biomolecules 2019, 9, 115. [Google Scholar] [CrossRef] [Green Version]
- Institute of Medicine Committee on the Use of Complementary and Alternative Medicine by the American Public. 2005. Available online: https://www-ncbi-nlm-nih-gov.ezproxy.lib.utexas.edu/books/NBK83804/ (accessed on 6 June 2020).
- Rajesh, E.; Sankari, L.S.; Malathi, L.; Krupaa, J.R. Naturally occurring products in cancer therapy. J. Pharm. Bioallied Sci. 2015, 7, S181–S183. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, J.; Cui, R.; Lin, J.; Ding, X. Curcumin in Treating Breast Cancer: A Review. J. Lab. Autom. 2016, 21, 723–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Momtazi-Borojeni, A.A.; Ghasemi, F.; Hesari, A.; Majeed, M.; Caraglia, M.; Sahebkar, A. Anti-Cancer and Radio-Sensitizing Effects of Curcumin in Nasopharyngeal Carcinoma. Curr. Pharm. Des. 2018, 24, 2121–2128. [Google Scholar] [CrossRef] [PubMed]
- Pezzani, R.; Salehi, B.; Vitalini, S.; Iriti, M.; Zuniga, F.A.; Sharifi-Rad, J.; Martorell, M.; Martins, N. Synergistic Effects of Plant Derivatives and Conventional Chemotherapeutic Agents: An Update on the Cancer Perspective. Medicina 2019, 55, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lodi, A.; Saha, A.; Lu, X.; Wang, B.; Sentandreu, E.; Collins, M.; Kolonin, M.G.; Digiovanni, J.; Tiziani, S. Combinatorial treatment with natural compounds in prostate cancer inhibits prostate tumor growth and leads to key modulations of cancer cell metabolism. Npj Precis. Oncol. 2017, 1, 1–12. [Google Scholar] [CrossRef]
- Tremmel, L.; Rho, O.; Slaga, T.J.; DiGiovanni, J. Inhibition of skin tumor promotion by TPA using a combination of topically applied ursolic acid and curcumin. Mol. Carcinog. 2019, 58, 185–195. [Google Scholar] [CrossRef]
- Werneke, U.; Earl, J.; Seydel, C.; Horn, O.; Crichton, P.; Fannon, D. Potential health risks of complementary alternative medicines in cancer patients. Br. J. Cancer 2004, 90, 408–413. [Google Scholar] [CrossRef]
- Keene, M.R.; Heslop, I.M.; Sabesan, S.S.; Glass, B.D. Complementary and alternative medicine use in cancer: A systematic review. Complement. Ther. Clin. Pract. 2019, 35, 33–47. [Google Scholar] [CrossRef]
- Gaston, T.E.; Mendrick, D.L.; Paine, M.F.; Roe, A.L.; Yeung, C.K. “Natural” is not synonymous with “Safe”: Toxicity of natural products alone and in combination with pharmaceutical agents. Regul. Toxicol. Pharmacol. 2020, 113, 104642. [Google Scholar] [CrossRef]
- Bailey, D.G.; Dresser, G.; Arnold, J.M. Grapefruit-medication interactions: Forbidden fruit or avoidable consequences? Cmaj 2013, 185, 309–316. [Google Scholar] [CrossRef] [Green Version]
- Fugh-Berman, A. Herb-drug interactions. Lancet 2000, 355, 134–138. [Google Scholar] [CrossRef]
- Atwood, K.C.t. Naturopathy: A critical appraisal. MedGenMed 2003, 5, 39. [Google Scholar] [PubMed]
- Buckner, C.A.; Lafrenie, R.M.; Dénommée, J.A.; Caswell, J.M.; Want, D.A. Complementary and alternative medicine use in patients before and after a cancer diagnosis. Curr. Oncol. 2018, 25, e275–e281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenberg, D.M.; Kessler, R.C.; Foster, C.; Norlock, F.E.; Calkins, D.R.; Delbanco, T.L. Unconventional medicine in the United States. Prevalence, costs, and patterns of use. N. Engl. J. Med. 1993, 328, 246–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanford, N.N.; Sher, D.J.; Ahn, C.; Aizer, A.A.; Mahal, B.A. Prevalence and Nondisclosure of Complementary and Alternative Medicine Use in Patients With Cancer and Cancer Survivors in the United States. JAMA Oncol. 2019, 5, 735. [Google Scholar] [CrossRef] [PubMed]
- White, A.; Boon, H.; Alraek, T.; Lewith, G.; Liu, J.-P.; Norheim, A.-J.; Steinsbekk, A.; Yamashita, H.; Fønnebø, V. Reducing the risk of complementary and alternative medicine (CAM): Challenges and priorities. Eur. J. Integr. Med. 2014, 6, 404–408. [Google Scholar] [CrossRef] [Green Version]
- Ju, Y.H.; Doerge, D.R.; Woodling, K.A.; Hartman, J.A.; Kwak, J.; Helferich, W.G. Dietary genistein negates the inhibitory effect of letrozole on the growth of aromatase-expressing estrogen-dependent human breast cancer cells (MCF-7Ca) in vivo. Carcinogenesis 2008, 29, 2162–2168. [Google Scholar] [CrossRef] [Green Version]
- Ju, Y.H.; Doerge, D.R.; Allred, K.F.; Allred, C.D.; Helferich, W.G. Dietary genistein negates the inhibitory effect of tamoxifen on growth of estrogen-dependent human breast cancer (MCF-7) cells implanted in athymic mice. Cancer Res. 2002, 62, 2474–2477. [Google Scholar]
- Warth, B.; Raffeiner, P.; Granados, A.; Huan, T.; Fang, M.; Forsberg, E.M.; Benton, H.P.; Goetz, L.; Johnson, C.H.; Siuzdak, G. Metabolomics Reveals that Dietary Xenoestrogens Alter Cellular Metabolism Induced by Palbociclib/Letrozole Combination Cancer Therapy. Cell Chem. Biol. 2018, 25, 291–300.e293. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Edgerton, S.; Yang, X.; Kim, A.; Ordonez-Ercan, D.; Mason, T.; Alvarez, K.; McKimmey, C.; Liu, N.; Thor, A. Low-dose dietary phytoestrogen abrogates tamoxifen-associated mammary tumor prevention. Cancer Res. 2005, 65, 879–886. [Google Scholar]
- Modernelli, A.; Naponelli, V.; Giovanna Troglio, M.; Bonacini, M.; Ramazzina, I.; Bettuzzi, S.; Rizzi, F. EGCG antagonizes Bortezomib cytotoxicity in prostate cancer cells by an autophagic mechanism. Sci. Rep. 2015, 5, 15270. [Google Scholar] [CrossRef] [PubMed]
- Golden, E.B.; Lam, P.Y.; Kardosh, A.; Gaffney, K.J.; Cadenas, E.; Louie, S.G.; Petasis, N.A.; Chen, T.C.; Schönthal, A.H. Green tea polyphenols block the anticancer effects of bortezomib and other boronic acid-based proteasome inhibitors. Blood 2009, 113, 5927–5937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.Y.; Park, J.; Oh, B.; Min, H.J.; Jeong, T.-S.; Lee, J.H.; Suh, C.; Cheong, J.-W.; Kim, H.J.; Yoon, S.-S.; et al. Natural polyphenols antagonize the antimyeloma activity of proteasome inhibitor bortezomib by direct chemical interaction. Br. J. Haematol. 2009, 146, 270–281. [Google Scholar] [CrossRef] [PubMed]
- Saleh, E.M.; El-Awady, R.A.; Eissa, N.A.; Abdel-Rahman, W.M. Antagonism between curcumin and the topoisomerase II inhibitor etoposide. Cancer Biol. Ther. 2012, 13, 1058–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, J.-L.; Chiang, P.-C.; Guh, J.-H. Tunicamycin induces resistance to camptothecin and etoposide in human hepatocellular carcinoma cells: Role of cell-cycle arrest and GRP78. Naunyn Schmiedebergs Arch. Pharmacol. 2009, 380, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Somasundaram, S.; Edmund, N.A.; Moore, D.T.; Small, G.W.; Shi, Y.Y.; Orlowski, R.Z. Dietary curcumin inhibits chemotherapy-induced apoptosis in models of human breast cancer. Cancer Res. 2002, 62, 3868–3875. [Google Scholar] [PubMed]
- Heaney, M.L.; Gardner, J.R.; Karasavvas, N.; Golde, D.W.; Scheinberg, D.A.; Smith, E.A.; O’Connor, O.A. Vitamin C Antagonizes the Cytotoxic Effects of Antineoplastic Drugs. Cancer Res. 2008, 68, 8031–8038. [Google Scholar] [CrossRef] [Green Version]
- Zou, W. Vitamin C Inactivates the Proteasome Inhibitor PS-341 in Human Cancer Cells. Clin. Cancer Res. 2006, 12, 273–280. [Google Scholar] [CrossRef] [Green Version]
- Perrone, G.; Hideshima, T.; Ikeda, H.; Okawa, Y.; Calabrese, E.; Gorgun, G.; Santo, L.; Cirstea, D.; Raje, N.; Chauhan, D.; et al. Ascorbic acid inhibits antitumor activity of bortezomib in vivo. Leukemia 2009, 23, 1679–1686. [Google Scholar] [CrossRef]
- Llobet, D.; Eritja, N.; Encinas, M.; Sorolla, A.; Yeramian, A.; Schoenenberger, J.A.; Llombart-Cussac, A.; Marti, R.M.; Matias-Guiu, X.; Dolcet, X. Antioxidants block proteasome inhibitor function in endometrial carcinoma cells. Anticancer Drugs 2008, 19, 115–124. [Google Scholar] [CrossRef]
- Bracke, M.E.; Depypere, H.T.; Boterberg, T.; Van Marck, V.L.; Vennekens, K.L.M.; Vanluchene, E.; Nuytinck, M.; Serreyn, R.; Mareel, M.M. Influence of Tangeretin on Tamoxifen’s Therapeutic Benefit in Mammary Cancer. JNCI J. Natl. Cancer Inst. 1999, 91, 354–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Depypere, H.T.; Bracke, M.E.; Boterberg, T.; Mareel, M.M.; Nuytinck, M.; Vennekens, K.; Serreyn, R. Inhibition of tamoxifen’s therapeutic benefit by tangeretin in mammary cancer. Eur. J. Cancer 2000, 36 (Suppl. S4), S73. [Google Scholar] [CrossRef]
- Noomhorm, N.; Chang, C.-J.; Wen, C.-S.; Wang, J.-Y.; Chen, J.-L.; Tseng, L.-M.; Chen, W.-S.; Chiu, J.-H.; Shyr, Y.-M. In Vitro and In Vivo Effects of Xanthorrhizol on Human Breast Cancer MCF-7 Cells Treated With Tamoxifen. J. Pharmacol. Sci. 2014, 125, 375–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.-T.; Agrawal, S.G.; Movasaghi, Z.; Wyatt, P.B.; Rehman, I.U.; Gribben, J.G.; Newland, A.C.; Jia, L. Dietary flavonoids inhibit the anticancer effects of the proteasome inhibitor bortezomib. Blood 2008, 112, 3835–3846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.L.; Wang, J.Y.; Tsai, Y.F.; Lin, Y.H.; Tseng, L.M.; Chang, W.C.; King, K.L.; Chen, W.S.; Chiu, J.H.; Shyr, Y.M. In vivo and in vitro demonstration of herb-drug interference in human breast cancer cells treated with tamoxifen and trastuzumab. Menopause 2013, 20, 646–654. [Google Scholar] [CrossRef] [PubMed]
- Foucquier, J.; Guedj, M. Analysis of drug combinations: Current methodological landscape. Pharmacol. Res. Perspect. 2015, 3, e00149. [Google Scholar] [CrossRef] [PubMed]
- Tallarida, R.J.; Raffa, R.B. Testing for synergism over a range of fixed ratio drug combinations: Replacing the isobologram. Life Sci. 1995, 58, PL23–PL28. [Google Scholar] [CrossRef]
- Chou, T.C. Drug Combination Studies and Their Synergy Quantification Using the Chou-Talalay Method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef] [Green Version]
- Tallarida, R.J. Quantitative Methods for Assessing Drug Synergism. Genes Cancer 2011, 2, 1003–1008. [Google Scholar] [CrossRef] [Green Version]
- Meyer, C.T.; Wooten, D.J.; Paudel, B.B.; Bauer, J.; Hardeman, K.N.; Westover, D.; Lovly, C.M.; Harris, L.A.; Tyson, D.R.; Quaranta, V. Quantifying Drug Combination Synergy along Potency and Efficacy Axes. Cell Syst. 2019, 8, 97–108.e116. [Google Scholar] [CrossRef] [Green Version]
- Wagner, H.; Ulrich-Merzenich, G. Synergy research: Approaching a new generation of phytopharmaceuticals. Phytomedicine 2009, 16, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, Z.; Li, S.; Ye, X.; Li, X.; He, K. Synergy effects of herb extracts: Pharmacokinetics and pharmacodynamic basis. Fitoterapia 2014, 92, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.H.; Zheng, C.J.; Han, L.Y.; Xie, B.; Jia, J.; Cao, Z.W.; Li, Y.X.; Chen, Y.Z. Synergistic therapeutic actions of herbal ingredients and their mechanisms from molecular interaction and network perspectives. Drug Discov. Today 2009, 14, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Hemaiswarya, S.; Kruthiventi, A.K.; Doble, M. Synergism between natural products and antibiotics against infectious diseases. Phytomedicine 2008, 15, 639–652. [Google Scholar] [CrossRef] [PubMed]
- Van Vuuren, S.; Viljoen, A. Plant-Based Antimicrobial Studies—Methods and Approaches to Study the Interaction between Natural Products. Planta Med. 2011, 77, 1168–1182. [Google Scholar] [CrossRef] [Green Version]
- Jia, J.; Zhu, F.; Ma, X.; Cao, Z.W.; Li, Y.X.; Chen, Y.Z. Mechanisms of drug combinations: Interaction and network perspectives. Nat. Rev. Drug Discov. 2009, 8, 111–128. [Google Scholar] [CrossRef]
- Caesar, L.K.; Cech, N.B. Synergy and antagonism in natural product extracts: When 1 + 1 does not equal 2. Nat. Prod. Rep. 2019, 36, 869–888. [Google Scholar] [CrossRef] [Green Version]
- Péter, S.; Navis, G.; De Borst, M.H.; Von Schacky, C.; Van Orten-Luiten, A.C.B.; Zhernakova, A.; Witkamp, R.F.; Janse, A.; Weber, P.; Bakker, S.J.L.; et al. Public health relevance of drug–nutrition interactions. Eur. J. Nutr. 2017, 56, 23–36. [Google Scholar] [CrossRef] [Green Version]
- Siu, D. Natural products and their role in cancer therapy. Med. Oncol. 2011, 28, 888–900. [Google Scholar] [CrossRef]
- Meyer, C.T.; Wooten, D.J.; Lopez, C.F.; Quaranta, V. Charting the Fragmented Landscape of Drug Synergy. Trends Pharmacol. Sci. 2020, 41, 266–280. [Google Scholar] [CrossRef]
- Odds, F.C. Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 2003, 52, 1. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Wennerberg, K.; Aittokallio, T. What is synergy? The Saariselkä agreement revisited. Front. Pharmacol. 2015, 6, 181. [Google Scholar] [CrossRef] [PubMed]
- Di Veroli, G.Y.; Fornari, C.; Wang, D.; Mollard, S.; Bramhall, J.L.; Richards, F.M.; Jodrell, D.I. Combenefit: An interactive platform for the analysis and visualization of drug combinations. Bioinformatics 2016, 32, 2866–2868. [Google Scholar] [CrossRef] [PubMed]
- Ianevski, A.; He, L.; Aittokallio, T.; Tang, J. SynergyFinder: A web application for analyzing drug combination dose–response matrix data. Bioinformatics 2017, 33, 2413–2415. [Google Scholar] [CrossRef]
- Flobak, Å.; Vazquez, M.; Lægreid, A.; Valencia, A. CImbinator: A web-based tool for drug synergy analysis in small- and large-scale datasets. Bioinformatics 2017, 33, 2410–2412. [Google Scholar] [CrossRef] [Green Version]
- Chou, T.C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef]
- Thangavel, P.; Puga-Olguín, A.; Rodríguez-Landa, J.F.; Zepeda, R.C. Genistein as Potential Therapeutic Candidate for Menopausal Symptoms and Other Related Diseases. Molecules 2019, 24, 3892. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Zhang, Y.; Ali, S.; Bhuiyan, M.; Wang, Z.; Chiao, P.J.; Philip, P.A.; Abbruzzese, J.; Sarkar, F.H. Molecular Evidence for Increased Antitumor Activity of Gemcitabine by Genistein In vitro and In vivo Using an Orthotopic Model of Pancreatic Cancer. Cancer Res. 2005, 65, 9064–9072. [Google Scholar] [CrossRef] [Green Version]
- HemaIswarya, S.; Doble, M. Potential synergism of natural products in the treatment of cancer. Phytother. Res. PTR 2006, 20, 239–249. [Google Scholar] [CrossRef]
- Papazisis, K.T.; Kalemi, T.G.; Zambouli, D.; Geromichalos, G.D.; Lambropoulos, A.F.; Kotsis, A.; Boutis, L.L.; Kortsaris, A.H. Synergistic effects of protein tyrosine kinase inhibitor genistein with camptothecins against three cell lines in vitro. Cancer Lett. 2006, 233, 255–264. [Google Scholar] [CrossRef]
- Mai, Z.; Blackburn, G.L.; Zhou, J.-R. Genistein sensitizes inhibitory effect of tamoxifen on the growth of estrogen receptor-positive and HER2-overexpressing human breast cancer cells. Mol. Carcinog. 2007, 46, 534–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Chen, J.; Sun, X.; Shi, X.; Wang, L.; Huang, L.; Zhou, W. Evaluation of the neuroprotective effect of EGCG: A potential mechanism of mitochondrial dysfunction and mitochondrial dynamics after subarachnoid hemorrhage. Food Funct. 2018, 9, 6349–6359. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Han, Z.; Li, X.; Xie, H.H.; Zhu, S.S. Mechanism of EGCG promoting apoptosis of MCF-7 cell line in human breast cancer. Oncol. Lett. 2017, 14, 3623–3627. [Google Scholar] [CrossRef] [PubMed]
- Le, C.T.; Leenders, W.P.J.; Molenaar, R.J.; van Noorden, C.J.F. Effects of the Green Tea Polyphenol Epigallocatechin-3-Gallate on Glioma: A Critical Evaluation of the Literature. Nutr. Cancer 2018, 70, 317–333. [Google Scholar] [CrossRef] [PubMed]
- Luo, K.W.; Lung, W.Y.; Chun, X.; Luo, X.L.; Huang, W.R. EGCG inhibited bladder cancer T24 and 5637 cell proliferation and migration via PI3K/AKT pathway. Oncotarget 2018, 9, 12261–12272. [Google Scholar] [CrossRef] [Green Version]
- Stuart, E.C.; Scandlyn, M.J.; Rosengren, R.J. Role of epigallocatechin gallate (EGCG) in the treatment of breast and prostate cancer. Life Sci. 2006, 79, 2329–2336. [Google Scholar] [CrossRef]
- Wei, R.; Hackman, R.M.; Wang, Y.; Mackenzie, G.G. Targeting Glycolysis with Epigallocatechin-3-Gallate Enhances the Efficacy of Chemotherapeutics in Pancreatic Cancer Cells and Xenografts. Cancers 2019, 11, 1496. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Xie, J.; Gan, R.; Wu, Z.; Luo, H.; Chen, X.; Lu, Y.; Wu, L.; Zheng, D. Synergistic inhibition of lung cancer cells by EGCG and NF-κB inhibitor BAY11-7082. J. Cancer 2019, 10, 6543–6556. [Google Scholar] [CrossRef]
- Bannerman, B.; Xu, L.; Jones, M.; Tsu, C.; Yu, J.; Hales, P.; Monbaliu, J.; Fleming, P.; Dick, L.; Manfredi, M.; et al. Preclinical evaluation of the antitumor activity of bortezomib in combination with vitamin C or with epigallocatechin gallate, a component of green tea. Cancer Chemother. Pharmacol. 2011, 68, 1145–1154. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.C.; Patchva, S.; Aggarwal, B.B. Therapeutic roles of curcumin: Lessons learned from clinical trials. Aaps J. 2013, 15, 195–218. [Google Scholar] [CrossRef] [Green Version]
- Giordano, A.; Tommonaro, G. Curcumin and Cancer. Nutrients 2019, 11, 2376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassanalilou, T.; Ghavamzadeh, S.; Khalili, L. Curcumin and Gastric Cancer: A Review on Mechanisms of Action. J. Gastrointest. Cancer 2019, 50, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Hesari, A.; Azizian, M.; Sheikhi, A.; Nesaei, A.; Sanaei, S.; Mahinparvar, N.; Derakhshani, M.; Hedayt, P.; Ghasemi, F.; Mirzaei, H. Chemopreventive and therapeutic potential of curcumin in esophageal cancer: Current and future status. Int. J. Cancer 2019, 144, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Selvam, C.; Prabu, S.L.; Jordan, B.C.; Purushothaman, Y.; Umamaheswari, A.; Hosseini Zare, M.S.; Thilagavathi, R. Molecular mechanisms of curcumin and its analogs in colon cancer prevention and treatment. Life Sci. 2019, 239, 117032. [Google Scholar] [CrossRef] [PubMed]
- Wan Mohd Tajuddin, W.N.B.; Lajis, N.H.; Abas, F.; Othman, I.; Naidu, R. Mechanistic Understanding of Curcumin’s Therapeutic Effects in Lung Cancer. Nutrients 2019, 11, 2989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, W.-H.; Wu, C.-C.; Yu, J.-S. Curcumin inhibits UV irradiation-induced oxidative stress and apoptotic biochemical changes in human epidermoid carcinoma A431 cells. J. Cell. Biochem. 2003, 90, 327–338. [Google Scholar] [CrossRef]
- Ma, W.; Guo, Q.; Li, Y.; Wang, X.; Wang, J.; Tu, P. Co-assembly of doxorubicin and curcumin targeted micelles for synergistic delivery and improving anti-tumor efficacy. J. Pharm. Biopharm. 2017, 112, 209–223. [Google Scholar] [CrossRef]
- Dickinson, A.; Blatman, J.; El-Dash, N.; Franco, J.C. Consumer usage and reasons for using dietary supplements: Report of a series of surveys. J. Am. Coll. Nutr. 2014, 33, 176–182. [Google Scholar] [CrossRef]
- Shenoy, N.; Creagan, E.; Witzig, T.; Levine, M. Ascorbic Acid in Cancer Treatment: Let the Phoenix Fly. Cancer Cell 2018, 34, 700–706. [Google Scholar] [CrossRef] [Green Version]
- Mahmoud, A.M.; Hernández Bautista, R.J.; Sandhu, M.A.; Hussein, O.E. Beneficial Effects of Citrus Flavonoids on Cardiovascular and Metabolic Health. Oxidative Med. Cell. Longev. 2019, 2019, 5484138. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.-J.; Park, K.-K.; Chung, W.-Y.; Hwang, J.-K.; Lee, S.K. Xanthorrhizol, a Natural Sesquiterpenoid, Induces Apoptosis and Growth Arrest in HCT116 Human Colon Cancer Cells. J. Pharmacol. Sci. 2009, 111, 276–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, M.A.; Kim, S.H.; Chung, W.Y.; Hwang, J.K.; Park, K.K. Xanthorrhizol, a natural sesquiterpenoid from Curcuma xanthorrhiza, has an anti-metastatic potential in experimental mouse lung metastasis model. Biochem. Biophys. Res. Commun. 2005, 326, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Chung, W.Y.; Park, J.H.; Kim, M.J.; Kim, H.O.; Hwang, J.K.; Lee, S.K.; Park, K.K. Xanthorrhizol inhibits 12-O-tetradecanoylphorbol-13-acetate-induced acute inflammation and two-stage mouse skin carcinogenesis by blocking the expression of ornithine decarboxylase, cyclooxygenase-2 and inducible nitric oxide synthase through mitogen-activated protein kinases and/or the nuclear factor-kappa B. Carcinogenesis 2007, 28, 1224–1231. [Google Scholar] [PubMed] [Green Version]
- Oon, S.F.; Nallappan, M.; Tee, T.T.; Shohaimi, S.; Kassim, N.K.; Sa’Ariwijaya, M.S.F.; Cheah, Y.H. Xanthorrhizol: A review of its pharmacological activities and anticancer properties. Cancer Cell Int. 2015, 15, 100. [Google Scholar] [CrossRef] [Green Version]
- Yeh, L.L.L.; Liu, J.-Y.; Lin, K.-S.; Liu, Y.-S.; Chiou, J.-M.; Liang, K.-Y.; Tsai, T.-F.; Wang, L.-H.; Chen, C.-T.; Huang, C.-Y. A Randomised Placebo-Controlled Trial of a Traditional Chinese Herbal Formula in the Treatment of Primary Dysmenorrhoea. PLoS ONE 2007, 2, e719. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.-J.; Chiu, J.-H.; Tseng, L.-M.; Chang, C.-H.; Chien, T.-M.; Chen, C.-C.; Wu, C.-W.; Lui, W.-Y. Si-Wu-Tang and its constituents promote mammary duct cell proliferation by up-regulation of HER-2 signaling. Menopause 2006, 13, 967–976. [Google Scholar] [CrossRef]
- Ko, C.H.; Shen, S.C.; Hsu, C.S.; Chen, Y.C. Mitochondrial-dependent, reactive oxygen species-independent apoptosis by myricetin: Roles of protein kinase C, cytochrome c, and caspase cascade. Biochem. Pharmacol. 2005, 69, 913–927. [Google Scholar] [CrossRef]
- Vijayababu, M.R.; Kanagaraj, P.; Arunkumar, A.; Ilangovan, R.; Aruldhas, M.M.; Arunakaran, J. Quercetin-induced growth inhibition and cell death in prostatic carcinoma cells (PC-3) are associated with increase in p21 and hypophosphorylated retinoblastoma proteins expression. J. Cancer Res. Clin. Oncol. 2005, 131, 765–771. [Google Scholar] [CrossRef]
- Lee, T.J.; Kim, O.H.; Kim, Y.H.; Lim, J.H.; Kim, S.; Park, J.W.; Kwon, T.K. Quercetin arrests G2/M phase and induces caspase-dependent cell death in U937 cells. Cancer Lett. 2006, 240, 234–242. [Google Scholar] [CrossRef]
- Chen, D.; Daniel, K.G.; Chen, M.S.; Kuhn, D.J.; Landis-Piwowar, K.R.; Dou, Q.P. Dietary flavonoids as proteasome inhibitors and apoptosis inducers in human leukemia cells. Biochem. Pharmacol. 2005, 69, 1421–1432. [Google Scholar] [CrossRef]
- He, L.; Kulesskiy, E.; Saarela, J.; Turunen, L.; Wennerberg, K.; Aittokallio, T.; Tang, J. Methods for High-throughput Drug Combination Screening and Synergy Scoring. Methods Mol. Biol. 2018, 1711, 351–398. [Google Scholar] [PubMed]
- Attene-Ramos, M.S.; Austin, C.P.; Xia, M. High Throughput Screening; Elsevier: Amsterdam, The Netherlands, 2014; pp. 916–917. [Google Scholar] [CrossRef]
- Pemovska, T.; Bigenzahn, J.W.; Superti-Furga, G. Recent advances in combinatorial drug screening and synergy scoring. Curr. Opin. Pharmacol. 2018, 42, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Vande Voorde, J.; Ackermann, T.; Pfetzer, N.; Sumpton, D.; Mackay, G.; Kalna, G.; Nixon, C.; Blyth, K.; Gottlieb, E.; Tardito, S. Improving the metabolic fidelity of cancer models with a physiological cell culture medium. Sci. Adv. 2019, 5, eaau7314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, G.R.; Lehár, J.; Keith, C.T. Multi-target therapeutics: When the whole is greater than the sum of the parts. Drug Discov. Today 2007, 12, 34–42. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Hackman, G.L.; Lodi, A.; Tiziani, S. Stable Isotope Tracing Metabolomics to Investigate the Metabolic Activity of Bioactive Compounds for Cancer Prevention and Treatment. Cancers 2020, 12, 2147. [Google Scholar] [CrossRef]
- Adam, G.; Rampášek, L.; Safikhani, Z.; Smirnov, P.; Haibe-Kains, B.; Goldenberg, A. Machine learning approaches to drug response prediction: Challenges and recent progress. Npj Precis. Oncol. 2020, 4, 1–10. [Google Scholar]
- Chen, H.; Engkvist, O.; Wang, Y.; Olivecrona, M.; Blaschke, T. The rise of deep learning in drug discovery. Drug Discov. Today 2018, 23, 1241–1250. [Google Scholar] [CrossRef]
- Xia, F.; Shukla, M.; Brettin, T.; Garcia-Cardona, C.; Cohn, J.; Allen, J.E.; Maslov, S.; Holbeck, S.L.; Doroshow, J.H.; Evrard, Y.A.; et al. Predicting tumor cell line response to drug pairs with deep learning. BMC Bioinform. 2018, 19, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Holbeck, S.L.; Camalier, R.; Crowell, J.A.; Govindharajulu, J.P.; Hollingshead, M.; Anderson, L.W.; Polley, E.; Rubinstein, L.; Srivastava, A.; Wilsker, D.; et al. The National Cancer Institute ALMANAC: A Comprehensive Screening Resource for the Detection of Anticancer Drug Pairs with Enhanced Therapeutic Activity. Cancer Res. 2017, 77, 3564–3576. [Google Scholar] [CrossRef] [Green Version]
- Preuer, K.; Lewis, R.P.I.; Hochreiter, S.; Bender, A.; Bulusu, K.C.; Klambauer, G. DeepSynergy: Predicting anti-cancer drug synergy with Deep Learning. Bioinformatics 2018, 34, 1538–1546. [Google Scholar] [CrossRef] [Green Version]

| Natural Product | Common Sources | Chemotherapy Drug | Cancer Type | Antagonism Mechanism | Antagonism Values (Cell Line–Value) | Ref | |
|---|---|---|---|---|---|---|---|
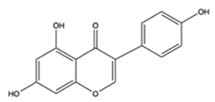 Genistein |  | Soybeans, fava beans, kudzu | Tamoxifen, letrozole, palbociclib + letrozole | Breast cancer | Reversed the anti-cancer effects of tamoxifen by inducing increased expression of estrogen responsive and cell cycle proteins pS2, PR, and cyclin D1 [30,31]; and activating mTOR by preventing amino acid depletion induced by palbociclib + letrozole [32] | Undetermined | [30,31,32,33] |
 EGCG |  | Green tea, berries, pears, apples, avocadoes | Bortezomib | Multiple myeloma, glioblastoma, prostate cancer | Protected against cancer cell death induced by bortezomib by preventing proteosome inhibition and ER stress induction and exacerbating autophagy activation to prevent apoptosis [34]; and by direct interaction with the drug’s boronic acid moiety that prevented proteosome inhibition [35,36] | U266-3.02 RPMI/8226-3.99 MC/CAR-4.01 | [34,35,36] |
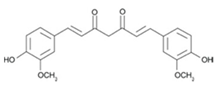 Curcumin |  | Turmeric | Etoposide, doxorubicin, mechlorethamine, camptothecin | Breast cancer | Prevented cancer cell death induced by etoposide and camptothecin by causing cell cycle arrest in the G1, S, or G2/M phases and allowing time for DNA repair prior to cell division [37,38]; and by inhibiting ROS generation and JNK activation induced by mechlorethamine and camptothecin [39] | MCF-7-2.5 HepG2-3.3 HCT116-67 HeLa-19 | [37,38,39] |
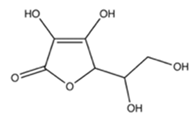 Vitamin C |  | Citrus fruits, potatoes, red and green peppers, broccoli, cauliflower, tomatoes | Bortezomib, doxorubicin, vinicristine, methotrexate, cisplatin, imatinib mesylate | Multiple myeloma, chronic myelogenous leukemia, B-cell lymphoma, breast, prostate, lung, oral, endometrial, and cervical cancer | Inhibited the anti-cancer effects of vinicristine, doxorubicin, methotrexate, imatinib mesylate, and cisplatin by preserving mitochondrial membrane potential and preventing apoptosis [40]; and by forming a chemical complex with bortezomib and blocking its activity [41,42] | Undetermined | [40,41,42,43] |
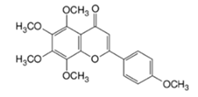 Tangeretin |  | Tangerines, mandarins, oranges, grapefruits | Tamoxifen | Breast cancer | Inhibited the anticancer effects of tamoxifen by downregulating NK cells and preventing tumor cell elimination in vivo | Undetermined | [44,45] |
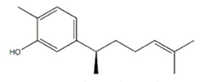 Xanthorrhizol |  | Curcuma xanthorriza (Javanese turmeric) | Tamoxifen | Breast cancer | May have reversed the anticancer effects of tamoxifen by activating the P38/MAPK pathway | Undetermined | [46] |
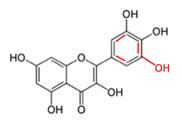 Quercetin and Myricetin |  | Onions, apples, grapes, berries, herbs | Bortezomib | B-cell lymphoma, chronic lymphocytic leukemia, multiple myeloma | Prevented the anticancer activity of bortezomib by directly interacting with the drug’s boronic moiety and inhibiting its activity [36,47] | U266-3.70 RPMI/8226-5.27 MC/CAR-5.04 (Quercetin) | [36,47] |
| Tannic acid, gallic acid, and caffeic acid |  | Coffee, tea, wine, grains, fruits, vegetables, berries, herbs | Bortezomib | Multiple myeloma | Blocked the anticancer activity of bortezomib by direct chemical interaction with its boronic acid moiety | U266- 6.70/3.08/4.22 RPMI/8226-11.39/3.34/8.54 MC/CAR-27.24/6.94/4.97 (TA/GA/CA) | [36] |
| Si-Wu-Tang |  | Combination: Radix Paeoniae Alba (bai shao yao), Rhizoma Ligusticum Chuanxiong (chuan xiong), Radix Angelica Sinensis (dang gui), and Radix Rehmanniae Preparata (shu di huang) | Tamoxifen, trastuzumab | Breast cancer | Reversed the cytotoxicity induced by tamoxifen by inactivating P27, and reversed the cytotoxicity of trastuzumab by activating AKT signaling and suppressing p27 by activating the P38/MAPK pathway | Undetermined | [48] |
| Synergy/Antagonism Model | Features | Strategy | Limitations | Equation | Software(s) |
|---|---|---|---|---|---|
| Loewe Additivity | Constant potency ratio Equal individual maximum effects Sham compliant: a drug cannot exhibit synergy with itself | Dose-effect based | Requires dose-effect curves for each compound Constant potency ratio and equal maximum effects is unlikely | Combenefit SynergyFinder CImbinator | |
| Bliss Independence | Drugs work by separate, non-overlapping mechanisms Assumes exponential dose-effect curves Expressed as a probability (0 1) | Effect-based | Most compounds do not have single, isolated mechanisms of action Exponential dose-effect curves unlikely | Combenefit SynergyFinder | |
| Highest Single Agent | A combined effect greater than that of the most effective single agent is synergistic Quantified by p-value of combined effect vs. highest individual agent | Effect-based | Fails to reflect true synergistic interactions (does not account for additivity) | Combenefit SynergyFinder | |
| Chou-Talalay | Based on the median effect equation derived from the mass action law Takes into account the Hill equation, Michaelis-Menten equation, Henderson-Hasselbalch equation, and Scatchard equations | Dose-effect based | Requires a dose-effect curve for each individual compound | CompuSyn |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hackman, G.L.; Collins, M.; Lu, X.; Lodi, A.; DiGiovanni, J.; Tiziani, S. Predicting and Quantifying Antagonistic Effects of Natural Compounds Given with Chemotherapeutic Agents: Applications for High-Throughput Screening. Cancers 2020, 12, 3714. https://doi.org/10.3390/cancers12123714
Hackman GL, Collins M, Lu X, Lodi A, DiGiovanni J, Tiziani S. Predicting and Quantifying Antagonistic Effects of Natural Compounds Given with Chemotherapeutic Agents: Applications for High-Throughput Screening. Cancers. 2020; 12(12):3714. https://doi.org/10.3390/cancers12123714
Chicago/Turabian StyleHackman, G. Lavender, Meghan Collins, Xiyuan Lu, Alessia Lodi, John DiGiovanni, and Stefano Tiziani. 2020. "Predicting and Quantifying Antagonistic Effects of Natural Compounds Given with Chemotherapeutic Agents: Applications for High-Throughput Screening" Cancers 12, no. 12: 3714. https://doi.org/10.3390/cancers12123714
APA StyleHackman, G. L., Collins, M., Lu, X., Lodi, A., DiGiovanni, J., & Tiziani, S. (2020). Predicting and Quantifying Antagonistic Effects of Natural Compounds Given with Chemotherapeutic Agents: Applications for High-Throughput Screening. Cancers, 12(12), 3714. https://doi.org/10.3390/cancers12123714







