The Urinary Transcriptome as a Source of Biomarkers for Prostate Cancer
Abstract
:1. Introduction
2. Results
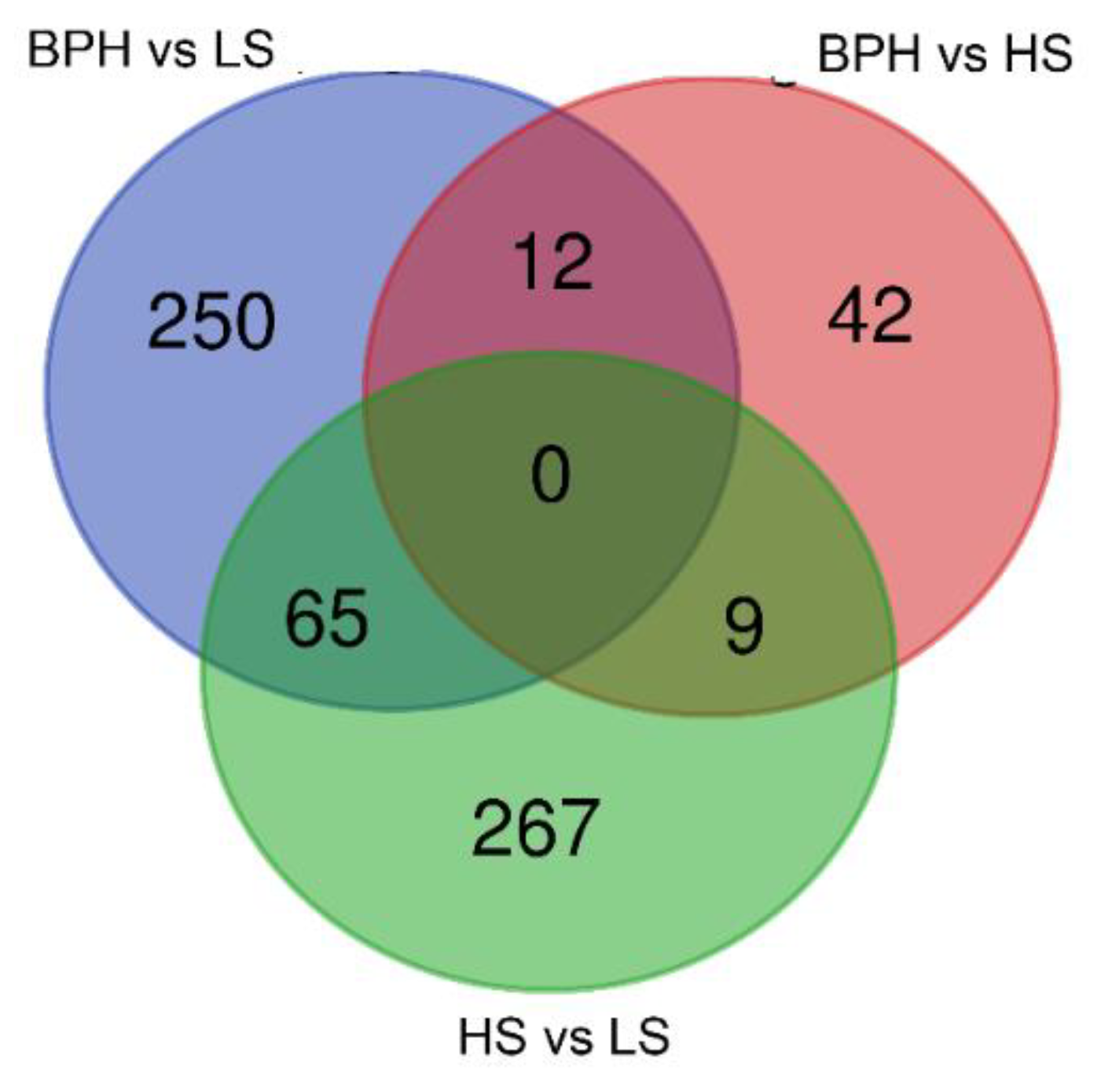
2.1. Sequencing the Circulating Transcriptome of Urine from Prostate Cancer Patients
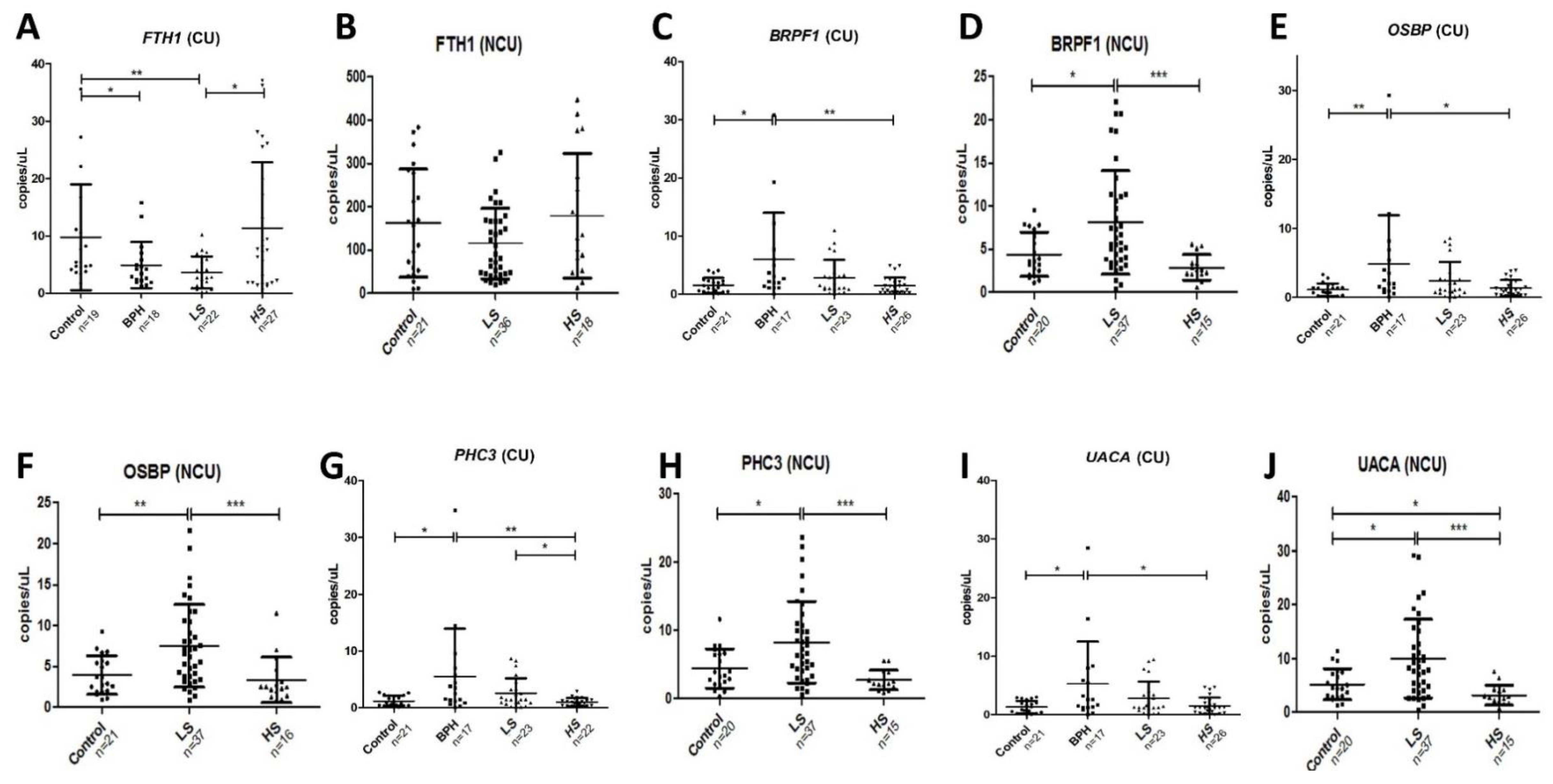
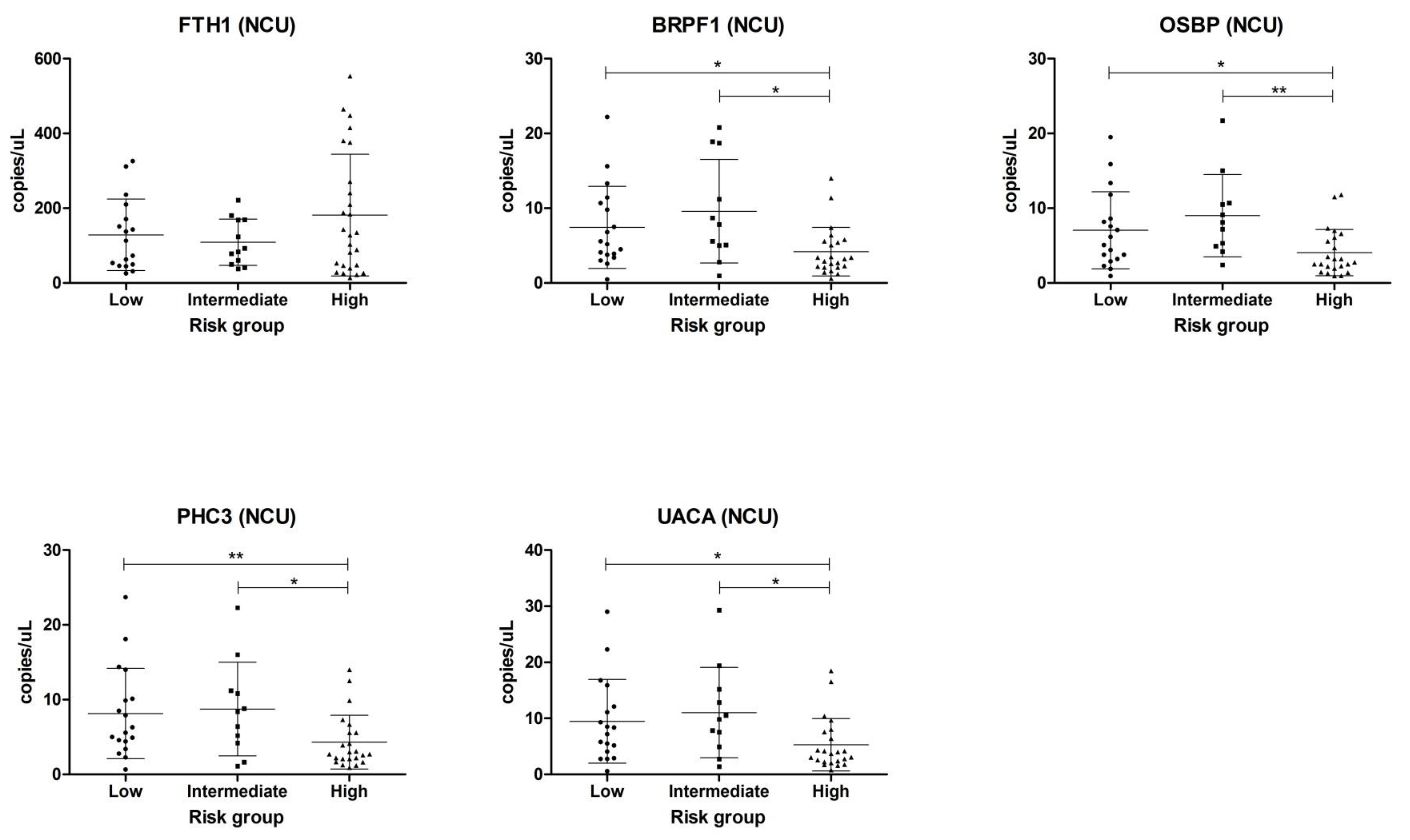
2.2. Validation in Centrifuged and Non-Centrifuged Urine Cohorts
3. Discussion
4. Materials and Methods
4.1. Patient Selection
4.2. RNA Purification and Library Construction
4.3. Bioinformatic Analysis
4.4. Droplet Digital PCR (ddPCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Auvinen, A.; Moss, S.M.; Tammela, T.L.; Taari, K.; Roobol, M.J.; Schroder, F.H.; Bangma, C.H.; Carlsson, S.; Aus, G.; Zappa, M.; et al. Absolute Effect of Prostate Cancer Screening: Balance of Benefits and Harms by Center within the European Randomized Study of Prostate Cancer Screening. Clin. Cancer Res. 2016, 22, 243–249. [Google Scholar] [CrossRef] [Green Version]
- Sokoll, L.J.; Sanda, M.G.; Feng, Z.; Kagan, J.; Mizrahi, I.A.; Broyles, D.L.; Partin, A.W.; Srivastava, S.; Thompson, I.M.; Wei, J.T.; et al. A prospective, multicenter, National Cancer Institute Early Detection Research Network study of [-2]proPSA: Improving prostate cancer detection and correlating with cancer aggressiveness. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1193–1200. [Google Scholar] [CrossRef] [Green Version]
- Heidegger, I.; Klocker, H.; Steiner, E.; Skradski, V.; Ladurner, M.; Pichler, R.; Schafer, G.; Horninger, W.; Bektic, J. [-2]proPSA is an early marker for prostate cancer aggressiveness. Prostate Cancer Prostatic Dis. 2014, 17, 70–74. [Google Scholar] [CrossRef]
- Thompson, I.M.; Pauler, D.K.; Goodman, P.J.; Tangen, C.M.; Lucia, M.S.; Parnes, H.L.; Minasian, L.M.; Ford, L.G.; Lippman, S.M.; Crawford, E.D.; et al. Prevalence of prostate cancer among men with a prostate-specific antigen level ≤ 4.0 ng per milliliter. N. Engl. J. Med. 2004, 350, 2239–2246. [Google Scholar] [CrossRef] [Green Version]
- Andriole, G.L.; Levin, D.L.; Crawford, E.D.; Gelmann, E.P.; Pinsky, P.F.; Chia, D.; Kramer, B.S.; Reding, D.; Church, T.R.; Grubb, R.L.; et al. Prostate Cancer Screening in the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial: Findings from the initial screening round of a randomized trial. J. Natl. Cancer Inst. 2005, 97, 433–438. [Google Scholar] [CrossRef]
- Heidenreich, A.; Pfister, D.; Merseburger, A.; Bartsch, G.; German Working Group on Castration-Resistant Prostate Cancer. Castration-resistant prostate cancer: Where we stand in 2013 and what urologists should know. Eur. Urol. 2013, 64, 260–265. [Google Scholar] [CrossRef]
- Miranda, K.C.; Bond, D.T.; Levin, J.Z.; Adiconis, X.; Sivachenko, A.; Russ, C.; Brown, D.; Nusbaum, C.; Russo, L.M. Massively parallel sequencing of human urinary exosome/microvesicle RNA reveals a predominance of non-coding RNA. PLoS ONE 2014, 9, e96094. [Google Scholar] [CrossRef] [Green Version]
- Bryant, R.J.; Pawlowski, T.; Catto, J.W.; Marsden, G.; Vessella, R.L.; Rhees, B.; Kuslich, C.; Visakorpi, T.; Hamdy, F.C. Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer 2012, 106, 768–774. [Google Scholar] [CrossRef] [Green Version]
- Sokoll, L.J.; Ellis, W.; Lange, P.; Noteboom, J.; Elliott, D.J.; Deras, I.L.; Blase, A.; Koo, S.; Sarno, M.; Rittenhouse, H.; et al. A multicenter evaluation of the PCA3 molecular urine test: Pre-analytical effects, analytical performance, and diagnostic accuracy. Clin. Chim. Acta 2008, 389, 1–6. [Google Scholar] [CrossRef]
- Wei, W.; Leng, J.; Shao, H.; Wang, W. High PCA3 scores in urine correlate with poor-prognosis factors in prostate cancer patients. Int. J. Clin. Exp. Med. 2015, 8, 16606–16612. [Google Scholar] [PubMed]
- Govers, T.M.; Hessels, D.; Vlaeminck-Guillem, V.; Schmitz-Drager, B.J.; Stief, C.G.; Martinez-Ballesteros, C.; Ferro, M.; Borque-Fernando, A.; Rubio-Briones, J.; Sedelaar, J.P.M.; et al. Cost-effectiveness of SelectMDx for prostate cancer in four European countries: A comparative modeling study. Prostate Cancer Prostatic Dis. 2019, 22, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Guelfi, G.; Cochetti, G.; Stefanetti, V.; Zampini, D.; Diverio, S.; Boni, A.; Mearini, E. Next Generation Sequencing of urine exfoliated cells: An approach of prostate cancer microRNAs research. Sci. Rep. 2018, 8, 7111. [Google Scholar] [CrossRef] [PubMed]
- Cochetti, G.; Poli, G.; Guelfi, G.; Boni, A.; Egidi, M.G.; Mearini, E. Different levels of serum microRNAs in prostate cancer and benign prostatic hyperplasia: Evaluation of potential diagnostic and prognostic role. Onco Targets Ther. 2016, 9, 7545–7553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, M.; Bajo-Santos, C.; Hessvik, N.P.; Lorenz, S.; Fromm, B.; Berge, V.; Sandvig, K.; Line, A.; Llorente, A. Identification of non-invasive miRNAs biomarkers for prostate cancer by deep sequencing analysis of urinary exosomes. Mol. Cancer 2017, 16, 156. [Google Scholar] [CrossRef] [PubMed]
- Robin, X. PanelomiX for the Combination of Biomarkers. Methods Mol. Biol. 2019, 1959, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Sole, C.; Arnaiz, E.; Manterola, L.; Otaegui, D.; Lawrie, C.H. The circulating transcriptome as a source of cancer liquid biopsy biomarkers. Semin. Cancer Biol. 2019, 58, 100–108. [Google Scholar] [CrossRef]
- Royo, F.; Zuniga-Garcia, P.; Torrano, V.; Loizaga, A.; Sanchez-Mosquera, P.; Ugalde-Olano, A.; Gonzalez, E.; Cortazar, A.R.; Palomo, L.; Fernandez-Ruiz, S.; et al. Transcriptomic profiling of urine extracellular vesicles reveals alterations of CDH3 in prostate cancer. Oncotarget 2016, 7, 6835–6846. [Google Scholar] [CrossRef] [Green Version]
- Overbye, A.; Skotland, T.; Koehler, C.J.; Thiede, B.; Seierstad, T.; Berge, V.; Sandvig, K.; Llorente, A. Identification of prostate cancer biomarkers in urinary exosomes. Oncotarget 2015, 6, 30357–30376. [Google Scholar] [CrossRef] [Green Version]
- Lekchnov, E.A.; Amelina, E.V.; Bryzgunova, O.E.; Zaporozhchenko, I.A.; Konoshenko, M.Y.; Yarmoschuk, S.V.; Murashov, I.S.; Pashkovskaya, O.A.; Gorizkii, A.M.; Zheravin, A.A.; et al. Searching for the Novel Specific Predictors of Prostate Cancer in Urine: The Analysis of 84 miRNA Expression. Int. J. Mol. Sci. 2018, 19, 4088. [Google Scholar] [CrossRef] [Green Version]
- Kutwin, P.; Konecki, T.; Borkowska, E.M.; Traczyk-Borszynska, M.; Jablonowski, Z. Urine miRNA as a potential biomarker for bladder cancer detection—A meta-analysis. Cent. Eur. J. Urol. 2018, 71, 177–185. [Google Scholar] [CrossRef]
- Mall, C.; Rocke, D.M.; Durbin-Johnson, B.; Weiss, R.H. Stability of miRNA in human urine supports its biomarker potential. Biomark. Med. 2013, 7, 623–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, K.; Spillman, M.A.; Behbakht, K.; Komatsu, J.M.; Abrahante, J.E.; Hicks, D.; Schotl, B.; Odean, E.; Jones, K.L.; Graner, M.W.; et al. A method for extracting and characterizing RNA from urine: For downstream PCR and RNAseq analysis. Anal. Biochem. 2017, 536, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Rivas, A.; Burzio, V.; Landerer, E.; Borgna, V.; Gatica, S.; Avila, R.; Lopez, C.; Villota, C.; de la Fuente, R.; Echenique, J.; et al. Determination of the differential expression of mitochondrial long non-coding RNAs as a noninvasive diagnosis of bladder cancer. BMC Urol. 2012, 12, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urbanucci, A.; Waltering, K.K.; Suikki, H.E.; Helenius, M.A.; Visakorpi, T. Androgen regulation of the androgen receptor coregulators. BMC Cancer 2008, 8, 219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, H.C.; Chen, S.C.; Chen, J.; Hsieh, J.T. In vitro gene expression changes of androgen receptor coactivators after hormone deprivation in an androgen-dependent prostate cancer cell line. J. Formos. Med. Assoc. 2005, 104, 652–658. [Google Scholar]
- Chen, Z.; Lu, W. Roles of ubiquitination and SUMOylation on prostate cancer: Mechanisms and clinical implications. Int. J. Mol. Sci. 2015, 16, 4560–4580. [Google Scholar] [CrossRef] [Green Version]
- Papandreou, C.N.; Logothetis, C.J. Bortezomib as a potential treatment for prostate cancer. Cancer Res. 2004, 64, 5036–5043. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.J. MOZ and MORF acetyltransferases: Molecular interaction, animal development and human disease. Biochim. Biophys. Acta 2015, 1853, 1818–1826. [Google Scholar] [CrossRef] [Green Version]
- Shima, H.; Yamagata, K.; Aikawa, Y.; Shino, M.; Koseki, H.; Shimada, H.; Kitabayashi, I. Bromodomain-PHD finger protein 1 is critical for leukemogenesis associated with MOZ-TIF2 fusion. Int. J. Hematol. 2014, 99, 21–31. [Google Scholar] [CrossRef]
- Grasso, C.S.; Wu, Y.M.; Robinson, D.R.; Cao, X.; Dhanasekaran, S.M.; Khan, A.P.; Quist, M.J.; Jing, X.; Lonigro, R.J.; Brenner, J.C.; et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012, 487, 239–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietrangelo, A.; Ridgway, N.D. Bridging the molecular and biological functions of the oxysterol-binding protein family. Cell. Mol. Life Sci. 2018, 75, 3079–3098. [Google Scholar] [CrossRef] [PubMed]
- Koga, Y.; Ishikawa, S.; Nakamura, T.; Masuda, T.; Nagai, Y.; Takamori, H.; Hirota, M.; Kanemitsu, K.; Baba, Y.; Baba, H. Oxysterol binding protein-related protein-5 is related to invasion and poor prognosis in pancreatic cancer. Cancer Sci. 2008, 99, 2387–2394. [Google Scholar] [CrossRef] [PubMed]
- Nagano, K.; Imai, S.; Zhao, X.; Yamashita, T.; Yoshioka, Y.; Abe, Y.; Mukai, Y.; Kamada, H.; Nakagawa, S.; Tsutsumi, Y.; et al. Identification and evaluation of metastasis-related proteins, oxysterol binding protein-like 5 and calumenin, in lung tumors. Int. J. Oncol. 2015, 47, 195–203. [Google Scholar] [CrossRef] [Green Version]
- Silva, J.; Beckedorf, A.; Bieberich, E. Osteoblast-derived oxysterol is a migration-inducing factor for human breast cancer cells. J. Biol. Chem. 2003, 278, 25376–25385. [Google Scholar] [CrossRef] [Green Version]
- Loilome, W.; Wechagama, P.; Namwat, N.; Jusakul, A.; Sripa, B.; Miwa, M.; Kuver, R.; Yongvanit, P. Expression of oxysterol binding protein isoforms in opisthorchiasis-associated cholangiocarcinoma: A potential molecular marker for tumor metastasis. Parasitol. Int. 2012, 61, 136–139. [Google Scholar] [CrossRef]
- Josson, S.; Chung, L.W.; Gururajan, M. microRNAs and Prostate Cancer. Adv. Exp. Med. Biol. 2015, 889, 105–118. [Google Scholar] [CrossRef] [Green Version]
- Burikhanov, R.; Shrestha-Bhattarai, T.; Hebbar, N.; Qiu, S.; Zhao, Y.; Zambetti, G.P.; Rangnekar, V.M. Paracrine apoptotic effect of p53 mediated by tumor suppressor Par-4. Cell Rep. 2014, 6, 271–277. [Google Scholar] [CrossRef] [Green Version]
- Burikhanov, R.; Shrestha-Bhattarai, T.; Qiu, S.; Shukla, N.; Hebbar, N.; Lele, S.M.; Horbinski, C.; Rangnekar, V.M. Novel mechanism of apoptosis resistance in cancer mediated by extracellular PAR-4. Cancer Res. 2013, 73, 1011–1019. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.J.; Kwok, Z.H.; Chew, X.H.; Zhang, B.; Liu, C.; Soong, T.W.; Yang, H.; Tay, Y. A FTH1 gene:pseudogene:microRNA network regulates tumorigenesis in prostate cancer. Nucleic Acids Res. 2018, 46, 1998–2011. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moret, I.; Sanchez-Izquierdo, D.; Iborra, M.; Tortosa, L.; Navarro-Puche, A.; Nos, P.; Cervera, J.; Beltran, B. Assessing an improved protocol for plasma microRNA extraction. PLoS ONE 2013, 8, e82753. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Mercado, M.; Manterola, L.; Larrea, E.; Goicoechea, I.; Arestin, M.; Armesto, M.; Otaegui, D.; Lawrie, C.H. The circulating transcriptome as a source of non-invasive cancer biomarkers: Concepts and controversies of non-coding and coding RNA in body fluids. J. Cell. Mol. Med. 2015, 19, 2307–2323. [Google Scholar] [CrossRef] [PubMed]
| Sample Type | Stage | N | Age (Median) | Average PSA |
|---|---|---|---|---|
| NGS cohort | BPH | 5 | 68 | 4.4 |
| Low stage (LS) | 5 | 70 | 6.7 | |
| High stage (HS) | 5 | 68 | 16.1 | |
| Centrifuged urine (CU) | Control | 21 | 69 | - |
| BPH | 21 | 72 | - | |
| LS | 25 | 64 | NK | |
| HS | 27 | 69 | NK | |
| Non-centrifuged urine (NCU) | Control | 24 | 67 | - |
| LS | 41 | 69 | 8.49 | |
| HS | 19 | 70 | 21.31 | |
| Total | - | 193 | - | - |
| Probe | Cont.* | (CU) | BPH * | (CU) | Cont + BPH * | (CU) | Cont.* | (NCU) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BPH | LS | HS | LS + HS | LS | HS | LS + HS | LS | HS | LS + HS | LS | HS | LS + HS | |
| FTH1 | 0.715 | 0.773 | 0.506 | 0.626 | 0.773 | 0.618 | 0.523 | 0.654 | 0.510 | 0.579 | 0.557 | 0.515 | 0.534 |
| BRPF1 | 0.741 | 0.596 | 0.551 | 0.518 | 0.642 | 0.768 | 0.709 | 0.521 | 0.648 | 0.568 | 0.641 | 0.577 | 0.572 |
| OSBP | 0.741 | 0.597 | 0.526 | 0.559 | 0.619 | 0.727 | 0.676 | 0.503 | 0.575 | 0.553 | 0.673 | 0.559 | 0.599 |
| PHC3 | 0.720 | 0.637 | 0.554 | 0.543 | 0.601 | 0.761 | 0.679 | 0.513 | 0.567 | 0.543 | 0.649 | 0.580 | 0.576 |
| UACA | 0.713 | 0.651 | 0.509 | 0.576 | 0.611 | 0.727 | 0.668 | 0.553 | 0.597 | 0.552 | 0.648 | 0.616 | 0.565 |
| Panel † | 0.782 (PHC3/FTH1) | 0.661 (OSBP/FTH1) | 0.832 (OSBP/FTH1/BRPF1) | 0.661 (OSBP/FTH1) | 0.773 (FTH1) | 0.771 (PHC3/FTH1) | 0.711 (PHC3/FTH1) | 0.654 (FTH1) | 0.618 (OSBP/FTH1) | 0.612 (BRPF1/FTH1/UACA) | 0.605 (OSBP/FTH1) | 0.621 (UACA/FTH1) | 0.638 (OSBP/FTH1) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solé, C.; Goicoechea, I.; Goñi, A.; Schramm, M.; Armesto, M.; Arestin, M.; Manterola, L.; Tellaetxe, M.; Alberdi, A.; Nogueira, L.; et al. The Urinary Transcriptome as a Source of Biomarkers for Prostate Cancer. Cancers 2020, 12, 513. https://doi.org/10.3390/cancers12020513
Solé C, Goicoechea I, Goñi A, Schramm M, Armesto M, Arestin M, Manterola L, Tellaetxe M, Alberdi A, Nogueira L, et al. The Urinary Transcriptome as a Source of Biomarkers for Prostate Cancer. Cancers. 2020; 12(2):513. https://doi.org/10.3390/cancers12020513
Chicago/Turabian StyleSolé, Carla, Ibai Goicoechea, Alai Goñi, Maike Schramm, María Armesto, María Arestin, Lorea Manterola, Maitena Tellaetxe, Aitor Alberdi, Leonor Nogueira, and et al. 2020. "The Urinary Transcriptome as a Source of Biomarkers for Prostate Cancer" Cancers 12, no. 2: 513. https://doi.org/10.3390/cancers12020513
APA StyleSolé, C., Goicoechea, I., Goñi, A., Schramm, M., Armesto, M., Arestin, M., Manterola, L., Tellaetxe, M., Alberdi, A., Nogueira, L., Roumiguie, M., López, J. I., Sanz Jaka, J. P., Urruticoechea, A., Vergara, I., Loizaga-Iriarte, A., Unda, M., Carracedo, A., Malavaud, B., & Lawrie, C. H. (2020). The Urinary Transcriptome as a Source of Biomarkers for Prostate Cancer. Cancers, 12(2), 513. https://doi.org/10.3390/cancers12020513







