Abstract
Determining which patients with early-stage breast cancer should receive chemotherapy is an important clinical issue. Chemotherapy has several adverse side effects, impacting on quality of life, along with significant economic consequences. There are a number of multi-gene prognostic signatures for breast cancer recurrence but there is less evidence that these prognostic signatures are predictive of therapy benefit. Biomarkers that can predict patient response to chemotherapy can help avoid ineffective over-treatment. The aim of this work was to assess if the OncoMasTR prognostic signature can predict pathological complete response (pCR) to neoadjuvant chemotherapy, and to compare its predictive value with other prognostic signatures: EndoPredict, Oncotype DX and Tumor Infiltrating Leukocytes. Gene expression datasets from ER-positive, HER2-negative breast cancer patients that had pre-treatment biopsies, received neoadjuvant chemotherapy and an assessment of pCR were obtained from the Gene Expression Omnibus repository. A total of 813 patients with 66 pCR events were included in the analysis. OncoMasTR, EndoPredict, Oncotype DX and Tumor Infiltrating Leukocytes numeric risk scores were approximated by applying the gene coefficients to the corresponding mean probe expression values. OncoMasTR, EndoPredict and Oncotype DX prognostic scores were moderately well correlated according to the Pearson’s correlation coefficient. Association with pCR was estimated using logistic regression. The odds ratio for a 1 standard deviation increase in risk score, adjusted for cohort, were similar in magnitude for all four signatures. Additionally, the four signatures were significant predictors of pCR. OncoMasTR added significant predictive value to Tumor Infiltrating Leukocytes signatures as determined by bivariable and trivariable analysis. In this in silico analysis, OncoMasTR, EndoPredict, Oncotype DX, and Tumor Infiltrating Leukocytes were significantly predictive of pCR to neoadjuvant chemotherapy in ER-positive and HER2-negative breast cancer patients.
1. Introduction
Breast cancer is the most frequently diagnosed cancer in women, with more than million new diagnoses worldwide every year, and the second leading cause of cancer death. The American Cancer Society and the International Agency for Research on Cancer reported that mortality rates are on the decline in certain regions of the world as a result of earlier diagnosis and improved therapies [1,2]. Accurate tools to help with optimal treatment decisions for individual patients to improve their prognosis, survival and quality of life are needed, whilst also reducing associated healthcare costs [3]. Breast cancer patients who receive chemotherapy can experience several side effects and symptoms that have a negative effect on their quality of life during and after the completion of treatment [4]. Biomarkers that can predict patient response to chemotherapy can help identify which patients are likely to benefit from chemotherapy, thereby potentially reducing the adverse effects of over-treatment.
Multi-gene prognostic signatures may be used to estimate risk of recurrence following surgery and endocrine treatment to make decisions about the suitability of chemotherapy. A patient who is predicted to be at low risk of breast cancer recurrence can safely forego aggressive treatment plans such as chemotherapy. OncoMasTR (OM) is a new 6-gene assay (3 prognostic genes plus 3 reference genes) discovered using a novel transcriptional network analysis approach that identified genes—Master Transcriptional Regulators (MTRs)—that putatively regulate previously known prognostic signatures [5,6]. OM has been analytically validated [7] in terms of assay robustness and clinically validated [8,9] in terms of accurate risk stratification, providing significant additional prognostic value to clinical information. The Oncotype DX Recurrence Score (RS) is a 21-gene prognostic assay that is widely used to predict risk of recurrence [10]. The EndoPredict assay (EP) is a 12-gene prognostic assay which incorporates tumor size and nodal status [11]. The Tumor Infiltrating Leukocytes (TILs) signature [12] determines risk of recurrence using expression correlates of 60 genes representing 15 immune cell sub-populations. Elevated TILs levels have been associated with better survival in patients with either ER-positive, HER2-positive disease or Triple-Negative Breast Cancer (TNBC) [13,14,15,16,17]. Furthermore, high TILs levels are associated with increased sensitivity to chemotherapy, reflected by higher pCR rates to neoadjuvant chemotherapy [18,19]. Indeed, a critical question remains as to whether one can predict a particular benefit from chemotherapy, as opposed to solely prognostic information, in breast cancer patients using multi-gene signatures.
The focus of this work is on ER-positive, HER2-negative breast cancer patients as the pCR to neoadjuvant chemotherapy is normally quite low in these patients compared to other breast cancer subtypes. We present an approach to expand on previous in silico analyses of other multi-gene prognostic signatures [20]. We assessed if OM could predict pCR of early-stage breast cancer patients to neoadjuvant chemotherapy and subsequently compared its predictive performance with EP, RS and TILs. Additionally, we assessed if TILs added significant predictive value to RS, EP and OM using univariable, bivariable and trivariable logistic regression analysis.
2. Method
First, a systematic search was carried out to select the datasets for this study. Following this, the relationship between OM, RS, EP, and TILs standardised risk scores was assessed by the Pearson’s correlation coefficient [21,22]. Finally, logistic regression [23] was used to estimate the associations between the signatures and pCR.
2.1. Systematic Search
The following sources were used to identify relevant breast cancer datasets: (i) dataset engines such as the Gene Expression Omnibus (GEO) repository [24], ArrayExpress [25], GDC data portal [26] and EGAS [27]; (ii) related papers and similar studies, as well as peer reviewed papers that performed a systematic search for gene expression datasets in breast cancer.
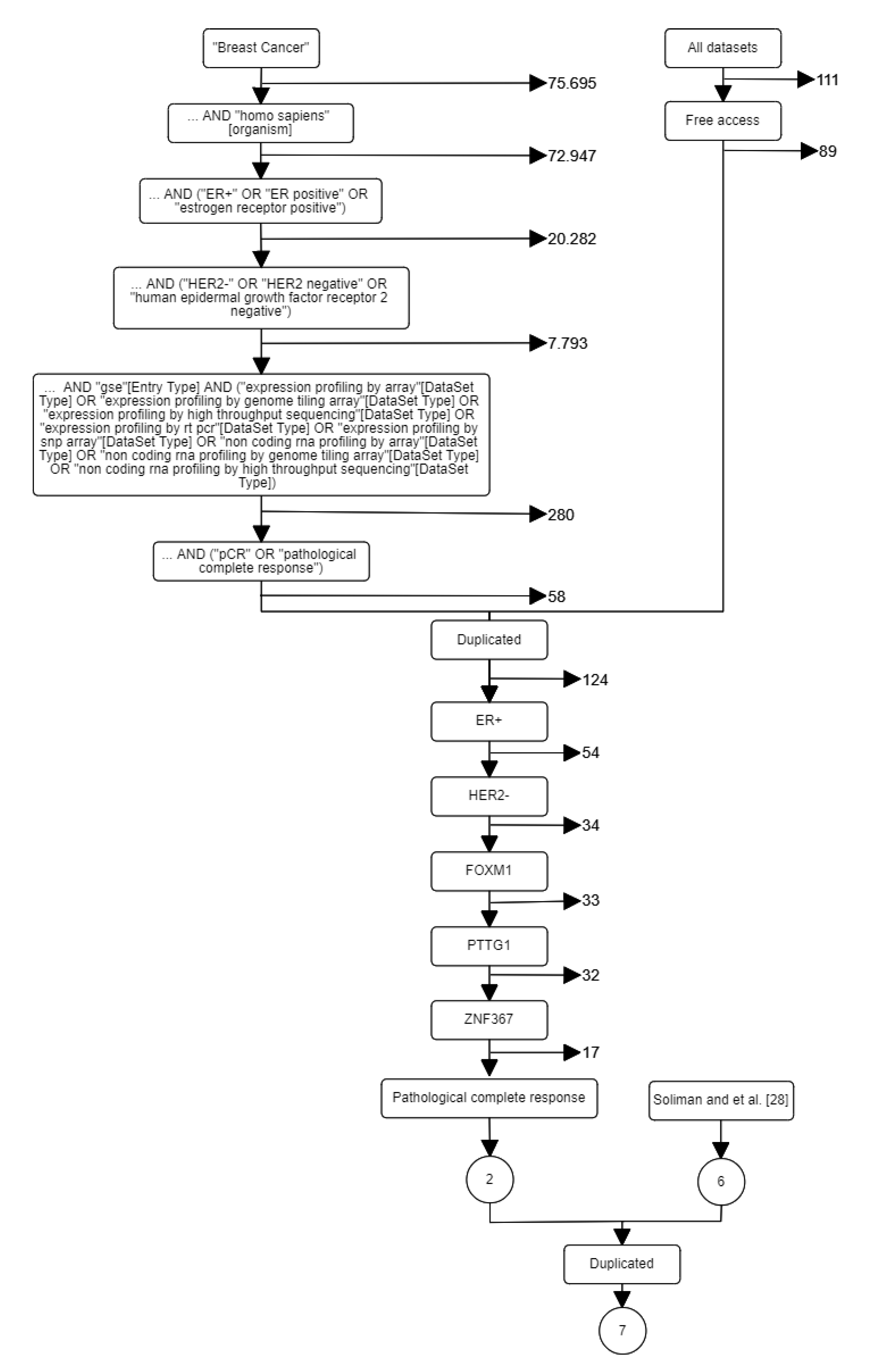
After the resources were specified, the following key words were selected: breast cancer, early, Homo sapiens, ER+, and HER2-. Particularly in GEO, there are many keywords and hierarchical orders that may be used in a systematic search resulting in different results. Furthermore, including the synonyms of the keywords or GEO’s query fields may alter the results obtained. Additional constraints were applied according to our project’s need: (i) including FOXM1, PTTG1, and ZNF367 genes; (ii) endpoint or outcome (e.g., pCR, distant recurrence, death, response to therapy); (iii) number of endpoint events; (iv) lymph node status; (v) therapy/treatment (e.g., none, endocrine, chemotherapy) (vi) platform; and (vii) public access. Figure 1 shows the flow diagram according to the project’s objectives after different search criteria. Finally, joining the results from the systematic search and the reference-work presented in [28], seven datasets (GSE16716, GSE20271, GSE25066, GSE32646, GSE34138, GSE41998, and GSE22226) were identified (see Table 1).
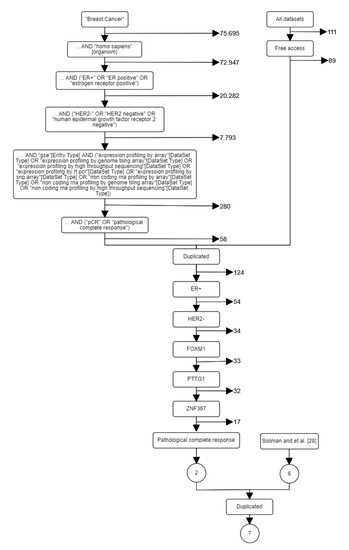
Figure 1.
A systematic search for breast cancer datasets. Left path corresponds to GEO search. Right path corresponds to related papers search.

Table 1.
GEO datasets included in the analysis *.
2.2. Dataset
Seven GEO dataset were used—GSE16716, GSE20271, GSE25066, GSE32646, GSE34138, GSE41998, and GSE22226. OM, EP and RS numeric risk scores were approximated by applying the signatures’ gene coefficients to the mean of the corresponding probe expression values. TILs scores is computed as the simple average expression of their marker genes. Risk scores were standardised within each dataset to have a mean of 0 and a standard deviation of 1. This allowed us to compare different risk scores from different datasets on a similar scale. A total of 813 patients with 66 pCR events were analysed. All the datasets except GSE32646 were missing at least one gene from at least one signature (see Table 1).
2.3. Predicting Probability of pCR Using Logistic Regression Analysis
Logistic regression is a statistical model that uses a logistic function to model a binary dependent variable. Logistic regression is used for different scenarios; in this case to estimate the probability that an event will occur, the outcome, using information thought to influence or be related to such events called predictors [23]. The outcome is a binary variable (1/0, Yes/No, True/False). The odds are defined as the probability that the event will occur divided by the probability that the event will not occur. Odds > 1 shows an association or correlation between risk scores and pCR, in our case. The Confidence Interval (CI) is used to estimate the precision of the odds. Additionally, a p-value is used as probability measure. The smaller the p-value (p) is the more significant the result is considered to be. is chosen as an arbitrary threshold where p is considered not to be significant. Univariable analysis was used to examine the relationship between pCR and each risk score while multivariable analysis was used to examine the relationship between pCR and each risk score, adjusted for cohort/GSE dataset or signature.
3. Results
3.1. Correlation Analysis
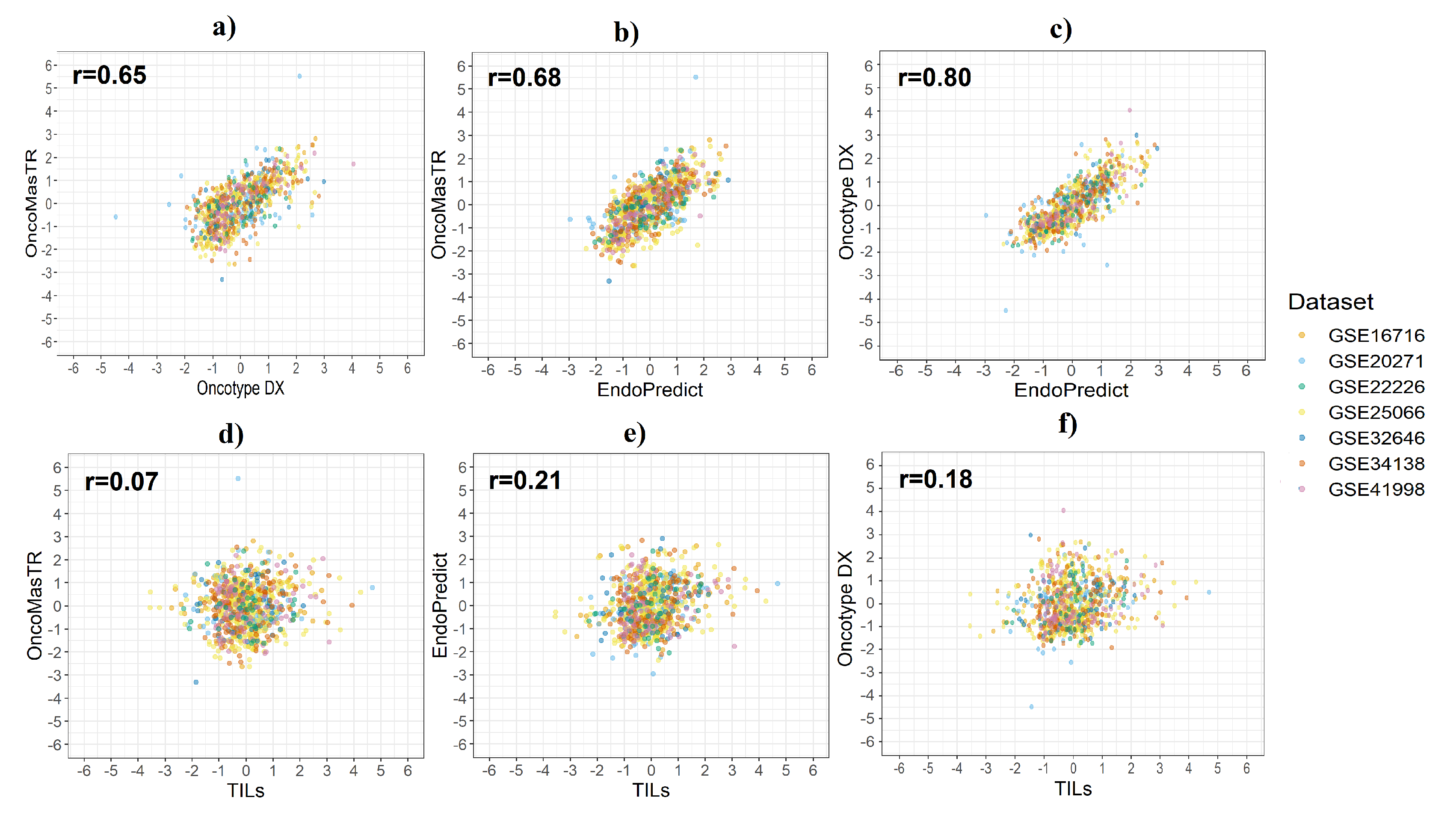
A Pearson’s correlation coefficient was used to assess the linear relationship between the standardised scores from the four signatures. We evaluated the correlation between six pairs of the four signatures: OM vs. RS, OM vs. EP, RS vs. EP, TILs vs. OM, TILs vs. EP, and TILs vs. RS for the seven datasets combined (Table 2).

Table 2.
Correlation coefficient r among the four signatures * **.
OM, RS and EP were moderately well correlated. However, there was very low correlation between TILs and the other three signatures. The GSE20271 dataset provided the lowest correlation in most cases. Figure 2 shows the results of the correlation analysis between risk score in the seven datasets combined.
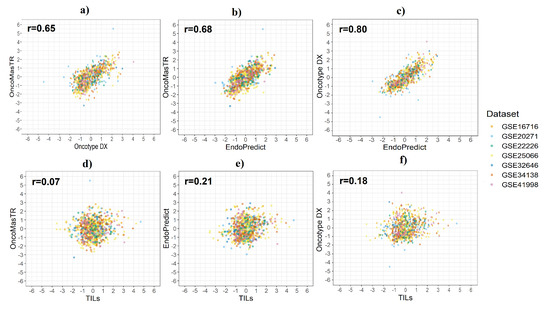
Figure 2.
Correlation between risk scores by dataset. (a) OM vs. RS; (b) OM vs. EP; (c) RS vs. EP; (d) OM vs. TILs; (e) EP vs. TILs; (f) RS vs. TILs.
3.2. Predicted Probability of pCR
Univariable, bivariable and trivariable logistic regression was used to estimate the relationship of OM, RS, EP and TILs with pCR. Table 3 contains a summary of the odds ratio (for a 1 standard deviation in risk score), confidence intervals, and p-values for the relationship between risk scores and pCR.

Table 3.
Odds ratio ( confidence intervals) for pCR by risk score * **.
Table 3 shows that the odds ratios of the signatures were similar in magnitude. OM, RS, EP and TILs were significant predictors of pCR, with TILs being less predictive. OM had an odds ratio of with a confidence interval of to . EP had an odds ratio of with a confidence interval of to . RS had an odds ratio of with confidence interval of to . TILs had an odds ratio of with a confidence interval of to .
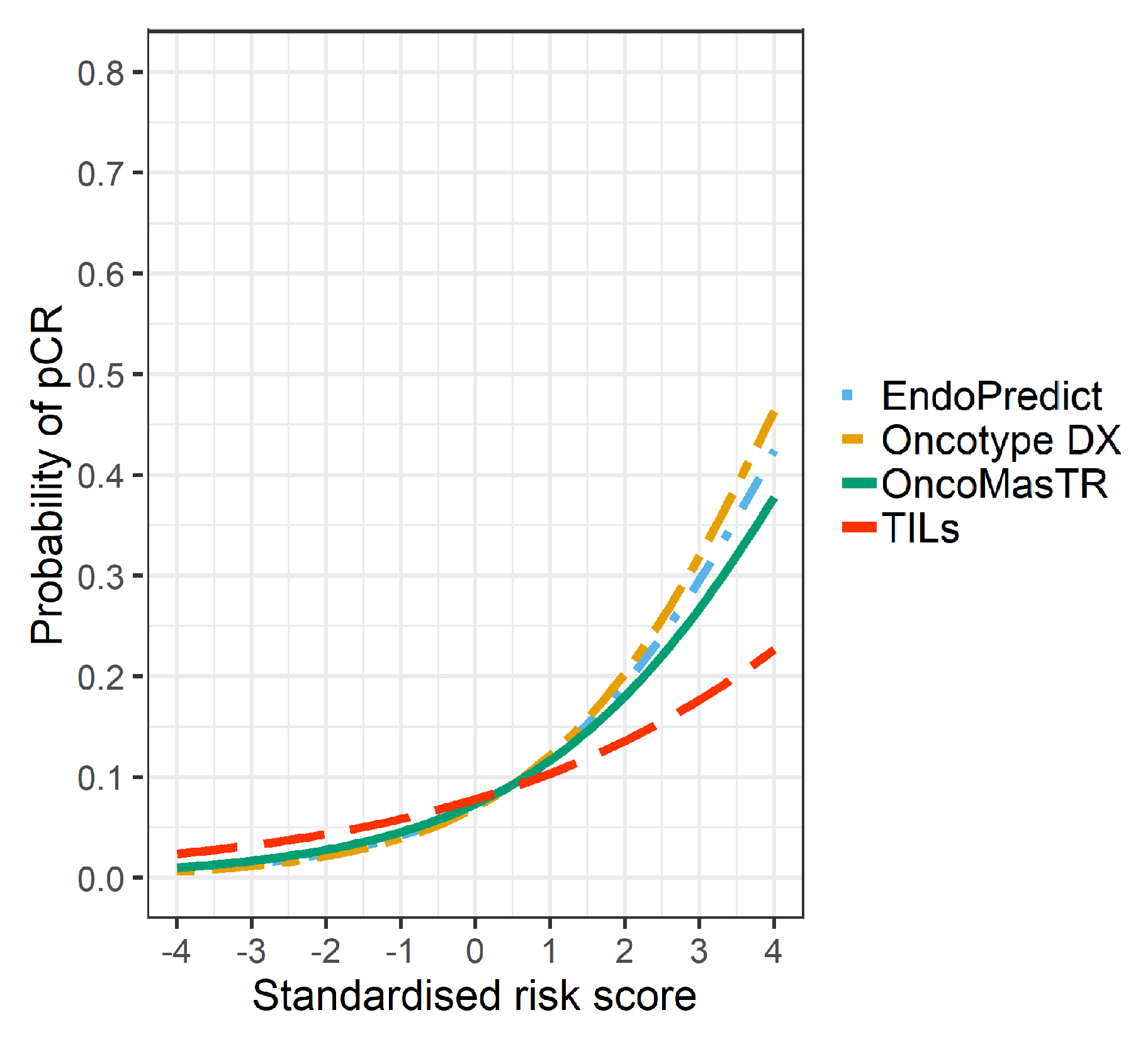
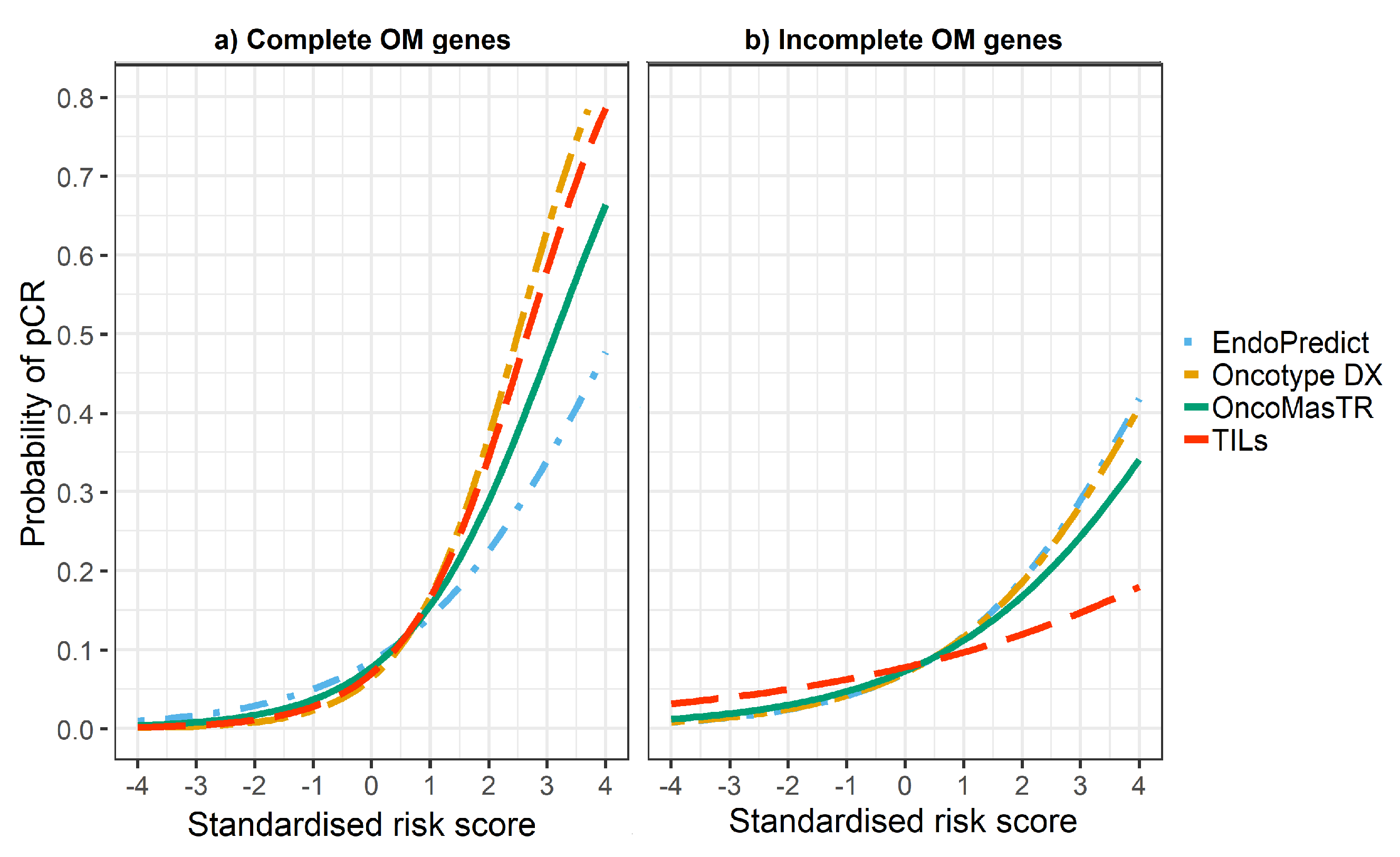
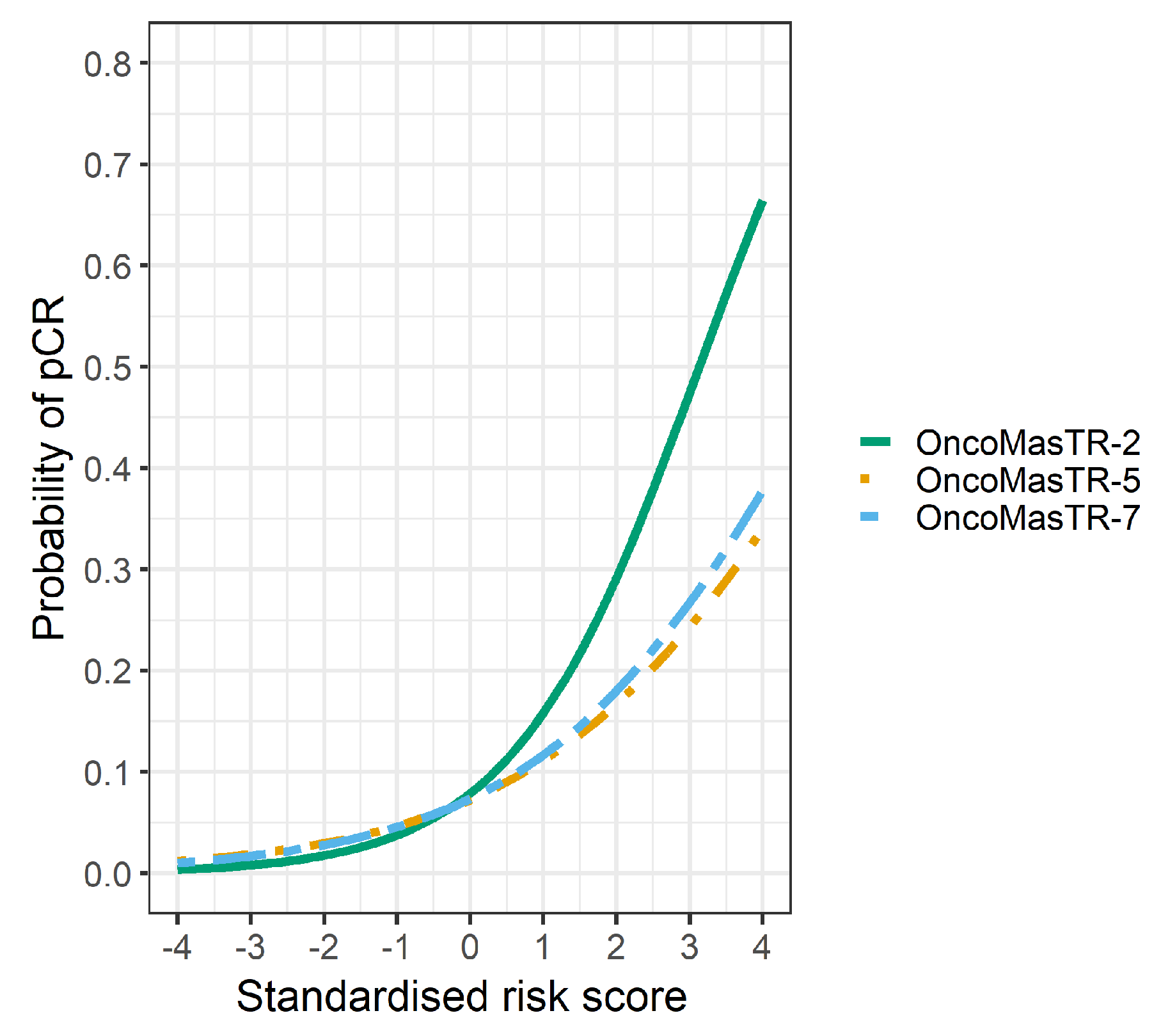
The univariable odds ratios for OM, RS, EP and TILs were similar when controlled for dataset and/or other signatures (bivariable and trivariable models). Accordingly, we based our predicted probabilities on univariable odds ratio for clarity of presentation. Plots of the predicted probability (Figure 3, Figure 4 and Figure 5) are derived from the univariable odds ratios in Table 3 and show that the probabilities of pCR increase as the risk scores increase, indicating that the risk scores predict response to chemotherapy. Figure 4 shows that signatures with fewer genes are more likely to be impacted by missing genes. As hypothesised, the predictive performance of OM is better in datasets with complete OM genes (Figure 5).
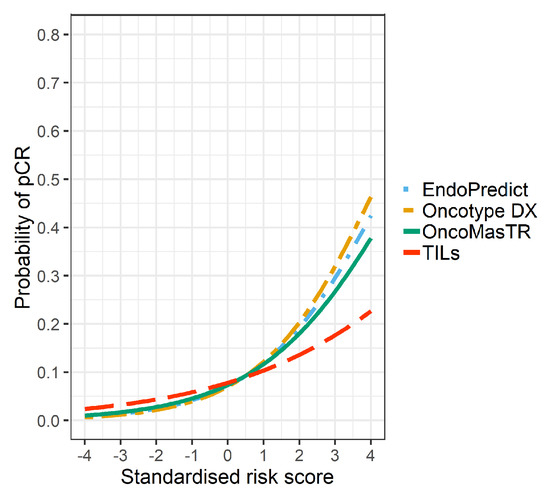
Figure 3.
Predicted probability of pCR by risk score in 7 datasets (5 datasets missing ZNF367).
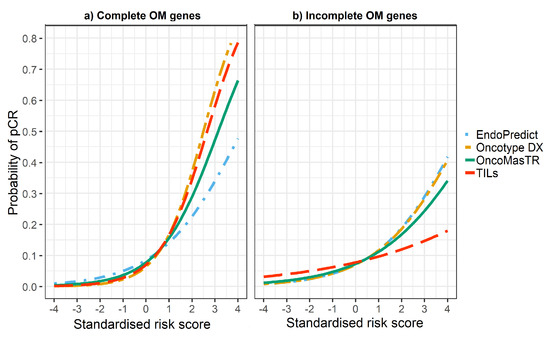
Figure 4.
Predicted probability of pCR by risk score in (a) datasets with complete OM genes (2 datasets); (b) datasets with incomplete OM genes (5 datasets missing ZNF367).
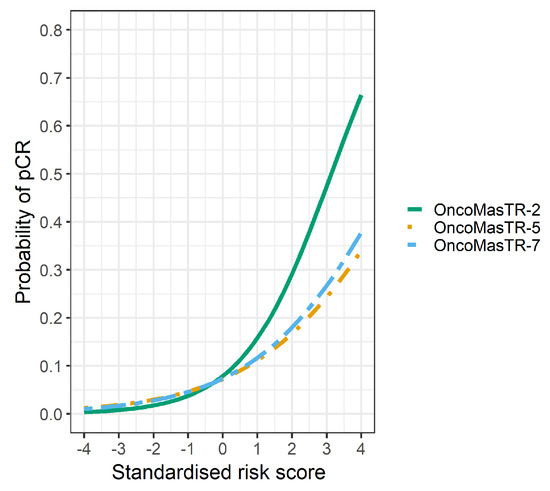
Figure 5.
Predicted probability of pCR by OM risk score using different dataset groups. Green line complete OM genes (2 datasets); Orange line incomplete OM genes (5 datasets missing ZNF367); Blue line combined datasets (7 datasets).
Residual deviance was used to assess how much TILs added to the other three risk scores. Residual deviance is a measure of goodness of fit of a model, a lower deviance means a better fit model and a 0 deviance is a perfect model. TILs added significant predictive value if the residual deviance decreased by >3.84 when TILs was added to a model (p(chi-sq > 3.84, = 1) < 0.05) (see Table 4).

Table 4.
Deviance statistic by model *.
TILs added significant predictive value to OM and vice versa and the significant predictive values are increased when the model is adjusted by dataset. TILs does not add significant predictive value to RS and EP (p = 0.0630 and 0.0886 respectively). However, RS and EP added significant predictive value to TILs. Controlling for the differences between the datasets further decrease the residual deviance of OM + TILs, RS + TILs and EP + TILs. The results show that the models adjusted for dataset are a better fit than unadjusted models. Finally, the best fit model was RS + TILs + Dataset with a residual deviance of .
4. Discussion
Multi-gene expression prognostic assays are commonly used to aid clinical decision-making in early-stage ER-positive, HER2-negative breast cancer as they provide complementary prognostic information to clinico-pathologic features [3]. There are many assays available, including RS, EP, TILs, MammaPrint [29], PAM50 [30], Breast Cancer Index (BCI) [31], along with the IHC4 test that is immunohistochemically based [32]. The amount of prognostic information provided in relation to risk of recurrence at an early (0–5 years) and late (beyond five years) time-point varies across these tests [33]. While the majority of data regarding these signatures focuses on prognostic value in the context of a background of adjuvant hormone therapy, there is comparatively little data available relating to their use in predicting benefit to neoadjuvant chemotherapy.
In this paper, we evaluated a new multi-gene prognostic signature, OM. We compared three other multi-gene expression prognostic signatures (RS, EP and TILs) with OM and evaluated the correlation between the four signatures (OM, RS, EP and TILs). Furthermore, we assessed if OM can predict pCR to neoadjuvant chemotherapy, and we compared the OM predictive performance with RS, EP and TILs. Finally, we evaluated if TILs added significant predictive value to OM, RS and EP.
Our findings suggest that OM, RS, and EP are moderately well correlated across the seven datasets assessed, with an average Pearson’s correlation coefficient r between each pair of signatures (OM vs RS, OM vs EP, and RS vs EP). TILs demonstrated a lower correlation for all datasets with an average r . Our results suggest that OM, RS, EP and TILs were significant predictors of pCR to neoadjuvant chemotherapy in ER+, HER2- breast cancer, with odds ratios .
TILs added significant predictive value to OM with a residual deviance decrease. OM, RS and EP added significant predictive value to TILs and controlling for the differences between the datasets further decreased the residual deviance. The best fit model was RS+TILs+Dataset demonstrating that including additional features provide a better fit to the data.
However, the predicted probability of pCR of the four signatures is affected by missing genes. It is important to highlight that OM is a significant pCR predictor even when only two of its prognostic genes are used (excluding missing genes). Our hypothesis is that if more datasets with complete genes were available then the performance of OM could be improved.
5. Conclusions and Future Work
OM has been clinically validated in the TransATAC cohort and a subset of the TAILORx cohort and has shown superior prognostic performance to RS [7,8,9]. Here, we show that the signature is predictive of treatment response (i.e., pCR response to neoadjuvant chemotherapy treatment). Some of the main differences between OM, RS, EP, and TILs are the number of genes in the assay (genes TILs, genes RS, genes EP, genes OM, when normalization genes are included) and the resulting cost. Some limitations of this study are the approximation of risk scores in micro-array data, the small number of pCR events in the dataset, and the missing genes effect.
Biomarkers that predict patient response to neoadjuvant chemotherapy offer the opportunity for personalised care, improved therapy response rates, reduced adverse effects and decreased costs of unnecessary treatment. In the future, we will extend this work by following two lines of investigation: (i) using more datasets thereby increasing the number of subjects evaluated and reducing the number of missing genes; and (ii) exploring new techniques which combine images with genetic information.
Author Contributions
Conceptualization, C.M. (Claudia Mazo), S.B., C.M. (Catherine Mooney), and W.M.G.; Data curation, C.M. (Claudia Mazo), S.B., C.M. (Catherine Mooney), and W.M.G.; Formal analysis, C.M. (Claudia Mazo), and S.B.; Funding acquisition, C.M. (Claudia Mazo), C.M. (Catherine Mooney), and W.M.G.; Investigation, C.M. (Claudia Mazo), and S.B.; Methodology, C.M. (Claudia Mazo) and S.B.; Project administration, C.M. (Claudia Mazo), C.M. (Catherine Mooney), and W.M.G.; Resources, C.M. (Claudia Mazo), S.B., C.M. (Catherine Mooney), and W.M.G.; Supervision, S.B., C.M. (Catherine Mooney), and W.M.G.; Validation, C.M. (Claudia Mazo), S.B., C.M. (Catherine Mooney), and W.M.G.; Writing—original draft, C.M. (Claudia Mazo); Writing—review and editing, C.M. (Claudia Mazo), S.B., C.M. (Catherine Mooney), and W.M.G. All authors have read and agreed to the published version of the manuscript.
Funding
This project has received funding from Enterprise Ireland (EI) and from the European Union’s Horizon 2020 research and innovation programme under the Marie Slodowska-Curie grant agreement No 713654. This publication has been supported in part by TACTIC—TAiloring the Communication of risk To Individual breast Cancer patients, Irish Research Council (IRC) grant CLNE/2018/1338, CAROLINE Postdoctoral Fellowship. This publication emanated from research supported in part by research grants from Science Foundation Ireland (SFI) under Grant Numbers 15/IA/3104 and 18/SPP/3522. This material is also based upon works supported by the Irish Cancer Society Collaborative Cancer Research Centre BREAST-PREDICT Grant CCRC13GAL.
Conflicts of Interest
W.M.G. is Chief Scientific Officer, shareholder and Director of OncoMark, S.B. is an employee of OncoMark.
Abbreviations
The following abbreviations are used in this manuscript:
| BCI | Breast Cancer Index |
| CI | Confidence Interval |
| EP | EndoPredict |
| GEO | Gene Expression Omnibus |
| MTRs | Master Transcriptional Regulators |
| OM | OncoMasTR |
| pCR | Pathological Complete Response |
| RS | Oncotype DX |
| TILs | Tumor Infiltrating Lymphocytes |
| EI | Enterprise Ireland |
| IRC | Irish Research Council |
| SFI | Science Foundation Ireland |
References
- American Cancer Society (ACS). 2019. Available online: https://www.cancer.org/cancer/breast-cancer.html (accessed on 26 November 2019).
- International Agency for Research on Cancer (IARC). 2019. Available online: https://www.iarc.fr/ (accessed on 26 November 2019).
- Mazo, C.; Kearns, C.; Mooney, C.; Gallagher, W.M. Clinical Decision Support Systems in Breast Cancer: A Systematic Review. Cancers 2020, 12, 369. [Google Scholar] [CrossRef] [PubMed]
- Paraskevi, T. Quality of life outcomes in patients with breast cancer. Oncol. Rev. 2012, 6, e2. [Google Scholar] [CrossRef] [PubMed]
- Lanigan, F.; Brien, G.L.; Fan, Y.; Madden, S.F.; Jerman, E.; Maratha, A.; Aloraifi, F.; Hokamp, K.; Dunne, E.J.; Lohan, A.J.; et al. Delineating transcriptional networks of prognostic gene signatures refines treatment recommendations for lymph node-negative breast cancer patients. FEBS J. 2015, 282, 3455–3473. [Google Scholar] [CrossRef] [PubMed]
- Moran, B.; Rahman, A.; Palonen, K.; Lanigan, F.T.; Gallagher, W.M. Master transcriptional regulators in cancer: Discovery via reverse engineering approaches and subsequent validation. Cancer Res. 2017, 77, 2186–2190. [Google Scholar] [CrossRef]
- Loughman, T.; Chan-Ju Wang, A.; Dynoodt, P.; Fender, B.; Lopez Ruiz, C.; Barron, S.; Stapleton, S.; O’Leary, D.; Fabre, A.; Quinn, C.; et al. 204P Analytical validation of OncoMasTR, a multigene test for predicting risk of distant recurrence in hormone receptor-positive early stage breast cancer. Ann. Oncol. 2018, 29. [Google Scholar] [CrossRef]
- Buus, R.; Sestak, I.; Barron, S.; Loughman, T.; Fender, B.; Ruiz, C.L.; Dynoodt, P.; Wang, C.J.A.; O’Leary, D.; Gallagher, W.M.; et al. Validation of the OncoMasTR Risk Score in Estrogen Receptor–Positive/HER2-Negative Patients: A TransATAC study. Clin. Cancer Res. 2020, 26, 623–631. [Google Scholar] [CrossRef]
- O’Connor, D.; Kelly, C.M.; Crown, J.; Russell, N.; Barron, S.; Loughman, T.; Lynch, S.; O’Grady, A.; Sheehan, K.M.; Fay, J.; et al. Additional prognostic value of OncoMasTR multigene prognostic signature to clinicopathological information in patients with HR-positive, HER2-negative, lymph node-negative breast cancer from the TAILORx Tissue Bank, Ireland. J. Clin. Oncol. 2019, 37, 535. [Google Scholar] [CrossRef]
- Paik, S.; Shak, S.; Tang, G.; Kim, C.; Baker, J.; Cronin, M.; Baehner, F.L.; Walker, M.G.; Watson, D.; Park, T.; et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004, 351, 2817–2826. [Google Scholar] [CrossRef]
- Filipits, M.; Rudas, M.; Jakesz, R.; Dubsky, P.; Fitzal, F.; Singer, C.F.; Dietze, O.; Greil, R.; Jelen, A.; Sevelda, P.; et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin. Cancer Res. 2011, 17, 6012–6020. [Google Scholar] [CrossRef]
- Danaher, P.; Warren, S.; Dennis, L.; D’Amico, L.; White, A.; Disis, M.L.; Geller, M.A.; Odunsi, K.; Beechem, J.; Fling, S.P. Gene expression markers of tumor infiltrating leukocytes. J. Immunother. Cancer 2017, 5, 18. [Google Scholar] [CrossRef]
- Bianchini, G.; Qi, Y.; Alvarez, R.H.; Iwamoto, T.; Coutant, C.; Ibrahim, N.K.; Valero, V.; Cristofanilli, M.; Green, M.C.; Radvanyi, L.; et al. Molecular Anatomy of Breast Cancer Stroma and Its Prognostic Value in Estrogen Receptor_Positive and _Negative Cancers. J. Clin. Oncol. 2010, 28, 4316–4323. [Google Scholar] [CrossRef] [PubMed]
- Karn, T.; Pusztai, L.; Holtrich, U.; Iwamoto, T.; Shiang, C.Y.; Schmidt, M.; Miller, V.; Solbach, C.; Gaetje, R.; Hanker, L.; et al. Homogeneous Datasets of Triple Negative Breast Cancers Enable the Identification of Novel Prognostic and Predictive Signatures. PLoS ONE 2011, 6, e0028403. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.; Gray, R.J.; Demaria, S.; Goldstein, L.; Perez, E.A.; Shulman, L.N.; Martino, S.; Wang, M.; Jones, V.E.; Saphner, T.J.; et al. Prognostic Value of Tumor-Infiltrating Lymphocytes in Triple-Negative Breast Cancers From Two Phase III Randomized Adjuvant Breast Cancer Trials: ECOG 2197 and ECOG 1199. J. Clin. Oncol. 2014, 32, 2959–2966. [Google Scholar] [CrossRef] [PubMed]
- Loi, S.; Sirtaine, N.; Piette, F.; Salgado, R.; Viale, G.; Van Eenoo, F.; Rouas, G.; Francis, P.; Crown, J.P.; Hitre, E.; et al. Prognostic and Predictive Value of Tumor-Infiltrating Lymphocytes in a Phase III Randomized Adjuvant Breast Cancer Trial in Node-Positive Breast Cancer Comparing the Addition of Docetaxel to Doxorubicin With Doxorubicin-Based Chemotherapy: BIG 02-98. J. Clin. Oncol. 2013, 31, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, T.; Bianchini, G.; Booser, D.; Qi, Y.; Coutant, C.; Ya-Hui Shiang, C.; Santarpia, L.; Matsuoka, J.; Hortobagyi, G.N.; Symmans, W.F.; et al. Gene Pathways Associated With Prognosis and Chemotherapy Sensitivity in Molecular Subtypes of Breast Cancer. JNCI J. Natl. Cancer Inst. 2010, 103, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Von Minckwitz, G.; Brase, J.C.; Sinn, B.V.; Gade, S.; Kronenwett, R.; Pfitzner, B.M.; Salat, C.; Loi, S.; Schmitt, W.D.; et al. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J. Clin. Oncol. 2015, 33, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Wimberly, H.; Brown, J.R.; Schalper, K.; Haack, H.; Silver, M.R.; Nixon, C.; Bossuyt, V.; Pusztai, L.; Lannin, D.R.; Rimm, D.L. PD-L1 expression correlates with tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy in breast cancer. Cancer Immunol. Res. 2015, 3, 326–332. [Google Scholar] [CrossRef]
- Mazo, C.; Barron, S.; Mooney, C.; Gallagher, W. 257P Multi-gene prognostic signatures and prediction of pathological complete response of ER-Positive HER2-negative breast cancer patients to neo-adjuvant chemotherapy. Ann. Oncol. 2019, 30. [Google Scholar] [CrossRef]
- Altman, D.G. Practical Statistics for Medical Research; CRC Press: Boca Raton, FL, USA, 1990. [Google Scholar]
- Campbell, M.J.; Swinscow, T.D.V. Statistics at Square One; John Wiley & Sons: New York, NY, USA, 2011. [Google Scholar]
- Kirkwood, B.R.; Sterne, J.A. Essential Medical Statistics; John Wiley & Sons: New York, NY, USA, 2010. [Google Scholar]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Kolesnikov, N.; Hastings, E.; Keays, M.; Melnichuk, O.; Tang, Y.A.; Williams, E.; Dylag, M.; Kurbatova, N.; Brandizi, M.; Burdett, T.; et al. ArrayExpress update—Simplifying data submissions. Nucleic Acids Res. 2015, 43, D1113–D1116. [Google Scholar] [CrossRef]
- Zhang, J.; Baran, J.; Cros, A.; Guberman, J.M.; Haider, S.; Hsu, J.; Liang, Y.; Rivkin, E.; Wang, J.; Whitty, B.; et al. International Cancer Genome Consortium Data Portal—A one-stop shop for cancer genomics data. Database 2011. [Google Scholar] [CrossRef] [PubMed]
- Lappalainen, I.; Almeida-King, J.; Kumanduri, V.; Senf, A.; Spalding, J.D.; Saunders, G.; Kandasamy, J.; Caccamo, M.; Leinonen, R.; Vaughan, B.; et al. The European Genome-phenome Archive of human data consented for biomedical research. Nature Genet. 2015, 47, 692–695. [Google Scholar] [CrossRef] [PubMed]
- Soliman, H.H.; Robson, M.E.; Wagner, S.; Flake, D.D.; Schwartzberg, L.S.; Sharma, P.; Magliocco, A.M.; Kronenwett, R.; Lancaster, J.M.; Lanchbury, J.S.; et al. In silico evaluation of the 12-gene molecular score (EndoPredict) and the recurrence score (Oncotype DX) as predictors of response to neo-adjuvant chemotherapy in estrogen receptor positive (ER+), HER2 negative (HER2-) breast cancer. J. Clin. Oncol. 2018, 36, 539. [Google Scholar] [CrossRef]
- Cardoso, F.; van’t Veer, L.J.; Bogaerts, J.; Slaets, L.; Viale, G.; Delaloge, S.; Pierga, J.Y.; Brain, E.; Causeret, S.; DeLorenzi, M.; et al. 70-gene signature as an aid to treatment decisions in early-stage breast cancer. N. Engl. J. Med. 2016, 375, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, T.O.; Parker, J.S.; Leung, S.; Voduc, D.; Ebbert, M.; Vickery, T.; Davies, S.R.; Snider, J.; Stijleman, I.J.; Reed, J.; et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor–positive breast cancer. Clin. Cancer Res. 2010, 16, 5222–5232. [Google Scholar] [CrossRef] [PubMed]
- Jerevall, P.L.; Ma, X.J.; Li, H.; Salunga, R.; Kesty, N.C.; Erlander, M.G.; Sgroi, D.; Holmlund, B.; Skoog, L.; Fornander, T.; et al. Prognostic utility of HOXB13: IL17BR and molecular grade index in early-stage breast cancer patients from the Stockholm trial. Br. J. Cancer 2011, 104, 1762–1769. [Google Scholar] [CrossRef] [PubMed]
- Cuzick, J.; Dowsett, M.; Pineda, S.; Wale, C.; Salter, J.; Quinn, E.; Zabaglo, L.; Mallon, E.; Green, A.R.; Ellis, I.O.; et al. Prognostic value of a combined estrogen receptor, progesterone receptor, Ki-67, and human epidermal growth factor receptor 2 immunohistochemical score and comparison with the Genomic Health recurrence score in early breast cancer. J. Clin. Oncol. 2011, 29, 4273–4278. [Google Scholar] [CrossRef]
- Sestak, I.; Dowsett, M.; Zabaglo, L.; Lopez-Knowles, E.; Ferree, S.; Cowens, J.W.; Cuzick, J. Factors predicting late recurrence for estrogen receptor–positive breast cancer. J. Natl. Cancer Inst. 2013, 105, 1504–1511. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).