RNA Editing Alters miRNA Function in Chronic Lymphocytic Leukemia
Abstract
:1. Introduction
2. Results
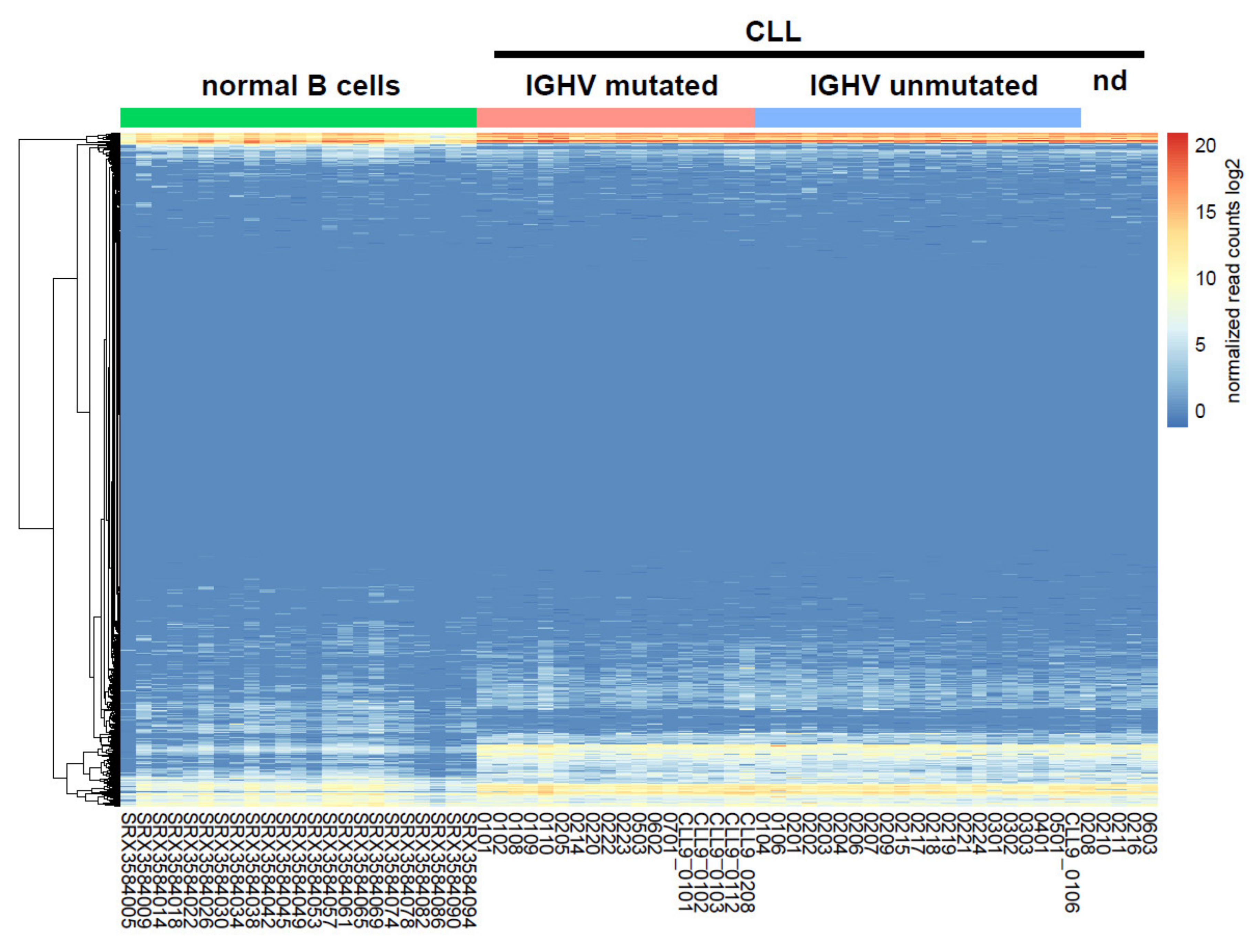
2.1. Identification of miRNAs Differentially Expressed between CLL and Normal B Cells
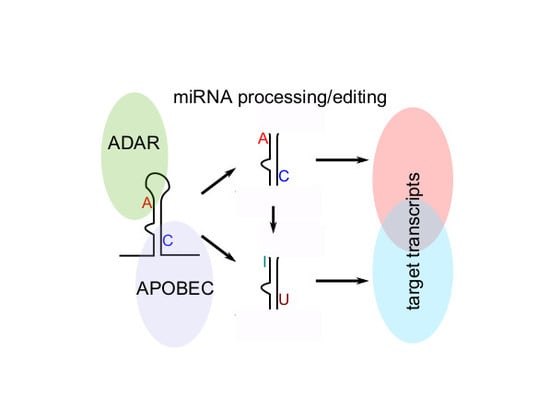
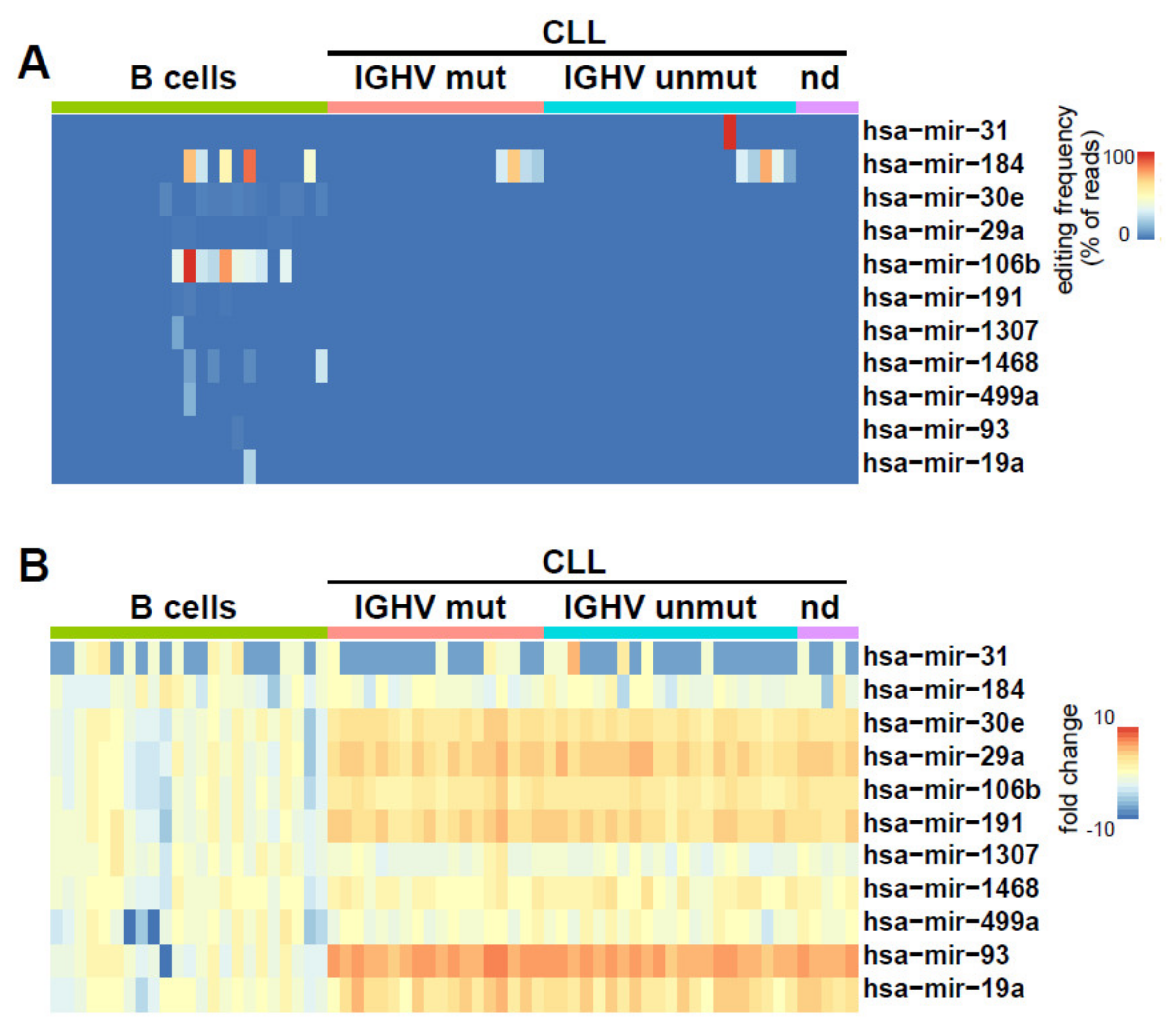
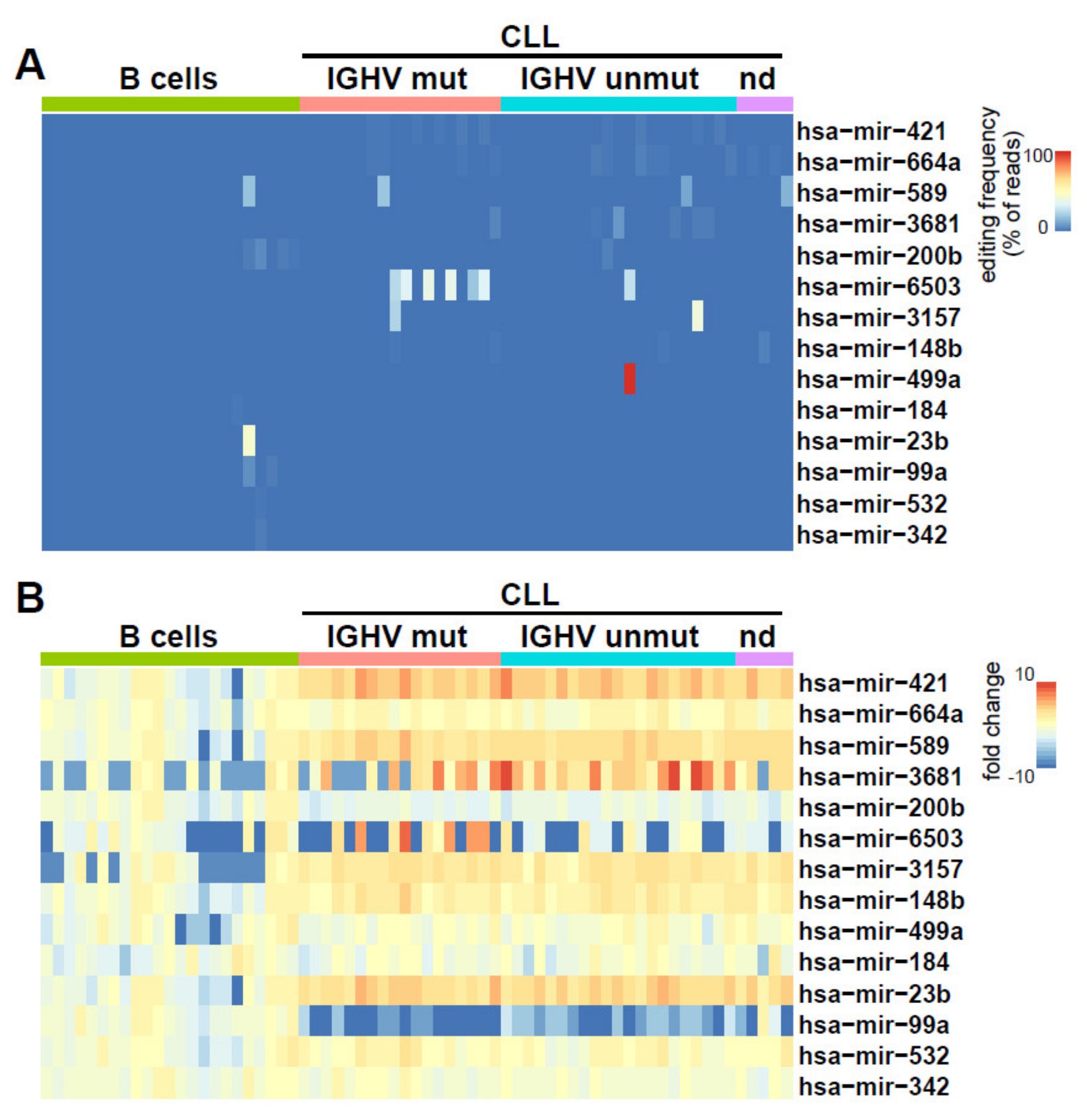
2.2. Identification of miRNAs Editing in CLL and Normal B Cells
2.3. Clinical and Biological Relevance of miRNA Editing in CLL
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. miRNA Sequencing and Bioinformatics
4.3. RNA Editing Analysis and Target Gene Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pleyer, L.; Egle, A.; Hartmann, T.N.; Greil, R. Molecular and cellular mechanisms of CLL: Novel therapeutic approaches. Nat. Rev. Clin. Oncol. 2009, 6, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Hamblin, T.J.; Orchard, J.A.; Gardiner, A.; Oscier, D.; Davis, Z.; Stevenson, F.K. Immunoglobulin V genes and CD38 expression in CLL. Blood 2000, 95, 2455–2457. [Google Scholar] [CrossRef] [PubMed]
- Puente, X.S.; Beà, S.; Valdés-Mas, R.; Villamor, N.; Gutiérrez-Abril, J.; Martin-Subero, J.I.; Munar, M.; Rubio-Perez, C.; Jares, P.; Aymerich, M.; et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature 2015, 526, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Landau, D.A.; Carter, S.L.; Stojanov, P.; McKenna, A.; Stevenson, K.; Lawrence, M.S.; Sougnez, C.; Stewart, C.; Sivachenko, A.; Wang, L.; et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 2013, 152, 714–726. [Google Scholar] [CrossRef] [Green Version]
- Landau, D.A.; Tausch, E.; Taylor-Weiner, A.N.; Stewart, C.; Reiter, J.G.; Bahlo, J.; Kluth, S.; Bozic, I.; Lawrence, M.; Böttcher, S.; et al. Mutations Driving Cll and Their Evolution in Progression and Relapse. Nature 2015, 526, 525–530. [Google Scholar] [CrossRef] [Green Version]
- Calin, G.A.; Cimmino, A.; Fabbri, M.; Ferracin, M.; Wojcik, S.E.; Shimizu, M.; Taccioli, C.; Zanesi, N.; Garzon, R.; Aqeilan, R.I.; et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc. Natl. Acad. Sci. USA 2008, 105, 5166–5171. [Google Scholar] [CrossRef] [Green Version]
- Zenz, T.; Mertens, D.; Küppers, R.; Döhner, H.; Stilgenbauer, S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat. Rev. Cancer 2009, 10, 37–50. [Google Scholar] [CrossRef]
- Balatti, V.; Acunzo, M.; Pekarky, Y.; Croce, C.M. Novel Mechanisms of Regulation of miRNAs in CLL. Trends Cancer 2016, 2, 134–143. [Google Scholar] [CrossRef] [Green Version]
- Croce, C.M. MicroRNA Dysregulation to Identify Novel Therapeutic Targets. Curr. Top. Microbiol. Immunol. 2017, 407, 191–203. [Google Scholar]
- Rassenti, L.Z.; Balatti, V.; Ghia, E.M.; Palamarchuk, A.; Tomasello, L.; Fadda, P.; Pekarsky, Y.; Widhopf, G.F., 2nd; Kipps, T.J.; Croce, C.M. Microrna Dysregulation to Identify Therapeutic Target Combinations for Chronic Lymphocytic Leukemia. Proc. Natl. Acad. Sci. USA 2017, 114, 10731–10736. [Google Scholar] [CrossRef] [Green Version]
- Javandoost, E.; Majd, E.F.; Rostamian, H.; Koosheh, M.K.-; Mirzaei, H.R. Role of microRNAs in Chronic Lymphocytic Leukemia Pathogenesis. Curr. Med. Chem. 2020, 27, 282–297. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, E.; Levanon, E.Y. A-to-I RNA editing—Immune protector and transcriptome diversifier. Nat. Rev. Genet. 2018, 19, 473–490. [Google Scholar] [CrossRef] [PubMed]
- Rayon-Estrada, V.; Papavasiliou, F.N.; Harjanto, D. RNA Editing Dynamically Rewrites the Cancer Code. Trends Cancer 2015, 1, 211–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenberg, B.R.; Hamilton, C.E.; Mwangi, M.M.; Dewell, S.; Papavasiliou, F.N. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat. Struct. Mol. Biol. 2011, 18, 230–236. [Google Scholar] [CrossRef] [Green Version]
- Tan, M.H.; Li, Q.; Shanmugam, R.; Piskol, R.; Kohler, J.; Young, A.N.; Liu, K.I.; Zhang, R.; Ramaswami, G.; Ariyoshi, K.; et al. Dynamic Landscape and Regulation of Rna Editing in Mammals. Nature 2017, 550, 249–254. [Google Scholar] [CrossRef]
- Paz, N.; Levanon, E.Y.; Amariglio, N.; Heimberger, A.B.; Ram, Z.; Constantini, S.; Barbash, Z.S.; Adamsky, K.; Safran, M.; Hirschberg, A.; et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007, 17, 1586–1595. [Google Scholar] [CrossRef] [Green Version]
- Paz-Yaacov, N.; Bazak, L.; Buchumenski, I.; Porath, H.; Danan-Gotthold, M.; Knisbacher, B.A.; Eisenberg, E.; Levanon, E.Y. Elevated RNA Editing Activity Is a Major Contributor to Transcriptomic Diversity in Tumors. Cell Rep. 2015, 13, 267–276. [Google Scholar] [CrossRef] [Green Version]
- Han, L.; Diao, L.; Yu, S.; Xu, X.; Li, J.; Zhang, R.; Yang, Y.; Werner, H.M.; Eterovic, A.K.; Yuan, Y.; et al. The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer Cell 2015, 28, 515–528. [Google Scholar] [CrossRef] [Green Version]
- Peng, X.; Xu, X.; Wang, Y.; Hawke, D.H.; Yu, S.; Han, L.; Zhou, Z.; Mojumdar, K.; Jeong, K.J.; Labrie, M.; et al. A-to-I RNA Editing Contributes to Proteomic Diversity in Cancer. Cancer Cell 2018, 33, 817–828. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Lin, C.H.; Chan, T.H.M.; Chow, R.K.K.; Song, Y.; Liu, M.; Yuan, Y.; Fu, L.; Kong, K.L.; Qi, L.; et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 2013, 19, 209–216. [Google Scholar] [CrossRef]
- Hu, X.; Chen, J.; Shi, X.; Feng, F.; Lau, K.W.; Chen, Y.; Chen, Y.; Jiang, L.; Cui, F.; Zhang, Y.; et al. RNA editing of AZIN1 induces the malignant progression of non-small-cell lung cancers. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shigeyasu, K.; Okugawa, Y.; Toden, S.; Miyoshi, J.; Toiyama, Y.; Nagasaka, T.; Takahashi, N.; Kusunoki, M.; Takayama, T.; Yamada, Y.; et al. AZIN1 RNA editing confers cancer stemness and enhances oncogenic potential in colorectal cancer. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Egle, A.; Steurer, M.; Melchardt, T.; Weiss, L.; Gassner, F.J.; Zaborsky, N.; Geisberger, R.; Catakovic, K.; Hartmann, T.N.; Pleyer, L.; et al. Fludarabine and Rituximab with Escalating Doses of Lenalidomide Followed by Lenalidomide/Rituximab Maintenance in Previously Untreated Chronic Lymphocytic Leukaemia (Cll): The Revlirit Cll-5 Agmt Phase I/Ii Study. Ann. Hematol. 2018, 97, 1825–1839. [Google Scholar] [CrossRef] [PubMed]
- Pallasch, C.; Patz, M.; Park, Y.J.; Hagist, S.; Eggle, D.; Claus, R.; Debey-Pascher, S.; Schulz, A.; Frenzel, L.P.; Claasen, J.; et al. miRNA deregulation by epigenetic silencing disrupts suppression of the oncogene PLAG1 in chronic lymphocytic leukemia. Blood 2009, 114, 3255–3264. [Google Scholar] [CrossRef] [Green Version]
- Calin, G.A.; Liu, C.-G.; Sevignani, C.; Ferracin, M.; Felli, N.; Dumitru, C.D.; Shimizu, M.; Cimmino, A.; Zupo, S.; Dono, M.; et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. USA 2004, 101, 11755–11760. [Google Scholar] [CrossRef] [Green Version]
- Lewis, B.P.; Shih, I.-H.; Jones-Rhoades, M.W.; Bartel, B.; Burge, C.B. Prediction of Mammalian MicroRNA Targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef] [Green Version]
- Bazak, L.; Haviv, A.; Barak, M.; Jacob-Hirsch, J.; Deng, P.; Zhang, R.; Isaacs, F.J.; Rechavi, G.; Li, J.B.; Eisenberg, E.; et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2013, 24, 365–376. [Google Scholar] [CrossRef] [Green Version]
- Liddicoat, B.J.; Piskol, R.; Chalk, A.M.; Ramaswami, G.; Higuchi, M.; Hartner, J.C.; Li, J.B.; Seeburg, P.H.; Walkley, C.R. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 2015, 349, 1115–1120. [Google Scholar] [CrossRef] [Green Version]
- Ishizuka, J.J.; Manguso, R.T.; Cheruiyot, C.K.; Bi, K.; Panda, A.; Iracheta-Vellve, A.; Miller, B.C.; Du, P.P.; Yates, K.B.; Dubrot, J.; et al. Loss of Adar1 in Tumours Overcomes Resistance to Immune Checkpoint Blockade. Nature 2019, 565, 43–48. [Google Scholar] [CrossRef]
- De Sousa, M.C.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef] [Green Version]
- Shukla, G.C.; Singh, J.; Barik, S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol. Cell. Pharmacol. 2011, 3, 83–92. [Google Scholar] [PubMed]
- Warf, M.B.; Shepherd, B.A.; Johnson, W.E.; Bass, B.L. Effects of ADARs on small RNA processing pathways in C. elegans. Genome Res. 2012, 22, 1488–1498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allegra, D.; Bilan, V.; Garding, A.; Döhner, H.; Stilgenbauer, S.; Kuchenbauer, F.; Mertens, D. Defective Drosha Processing Contributes to Downregulation of Mir-15/-16 in Chronic Lymphocytic Leukemia. Leukemia 2014, 28, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Shoshan, E.; Mobley, A.K.; Braeuer, R.R.; Kamiya, T.; Huang, L.; Vasquez, M.E.; Salameh, A.; Lee, H.J.; Kim, S.J.; Ivan, C.; et al. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nature 2015, 17, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Blow, M.; Grocock, R.J.; Van Dongen, S.; Enright, A.; Dicks, E.; Futreal, P.A.; Wooster, R.; Stratton, M.R. RNA editing of human microRNAs. Genome Biol. 2006, 7, R27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez, J.G.; Hourcade, J.D.D.; Del Mazo, J. Reprogramming of microRNAs by adenosine-to-inosine editing and the selective elimination of edited microRNA precursors in mouse oocytes and preimplantation embryos. Nucleic Acids Res. 2013, 41, 5483–5493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, J.T.; Patterson, D.G.; King, V.M.; Amin, S.V.; Polska, C.J.; Houserova, D.; Crucello, A.; Barnhill, E.C.; Miller, M.M.; Sherman, T.D.; et al. ADAR Mediated RNA Editing Modulates MicroRNA Targeting in Human Breast Cancer. Processes 2018, 6, 42. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Chendrimada, T.P.; Wang, Q.; Higuchi, M.; Seeburg, P.H.; Shiekhattar, R.; Nishikura, K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2005, 13, 13–21. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Song, Y.; Shi, X.; Liu, J.; Xiong, S.; Chen, W.; Fu, Q.; Huang, Z.; Gu, N.; Zhang, R. The landscape of miRNA editing in animals and its impact on miRNA biogenesis and targeting. Genome Res. 2017, 28, 132–143. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, P.; Jares, P.; Rico, D.; Gómez-López, G.; Martínez-Trillos, A.; Villamor, N.; Ecker, S.; Gonzalez-Perez, A.; Knowles, D.G.; Monlong, J.; et al. Transcriptome characterization by RNA sequencing identifies a major molecular and clinical subdivision in chronic lymphocytic leukemia. Genome Res. 2013, 24, 212–226. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Baysal, B.E. Stem-loop structure preference for site-specific RNA editing by APOBEC3A and APOBEC3G. Peer J. 2017, 5, e4136. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Patnaik, S.K.; Kemer, Z.; Baysal, B.E. Transient overexpression of exogenous APOBEC3A causes C-to-U RNA editing of thousands of genes. RNA Biol. 2016, 14, 603–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, S.; Patnaik, S.K.; Taggart, R.T.; Baysal, B.E. The double-domain cytidine deaminase APOBEC3G is a cellular site-specific RNA editing enzyme. Sci. Rep. 2016, 6, 39100. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Patnaik, S.K.; Taggart, R.T.; Kannisto, E.D.; Enriquez, S.M.; Gollnick, P.; Baysal, B.E. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat. Commun. 2015, 6, 6881. [Google Scholar] [CrossRef]
- Rebhandl, S.; Huemer, M.; Greil, R.; Geisberger, R. AID/APOBEC deaminases and cancer. Oncoscience 2015, 2, 320–333. [Google Scholar] [CrossRef] [Green Version]
- Rebhandl, S.; Huemer, M.; Gassner, F.J.; Zaborsky, N.; Hebenstreit, D.; Catakovic, K.; Grössinger, E.M.; Greil, R.; Geisberger, R. APOBEC3 signature mutations in chronic lymphocytic leukemia. Leukemia 2014, 28, 1929–1932. [Google Scholar] [CrossRef] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2012, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Alon, S.; Mor, E.; Vigneault, F.; Church, G.M.; Locatelli, F.; Galeano, F.; Gallo, A.; Shomron, N.; Eisenberg, E. Systematic identification of edited microRNAs in the human brain. Genome Res. 2012, 22, 1533–1540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reczko, M.; Maragkakis, M.; Alexiou, P.; Grosse, I.; Hatzigeorgiou, A.G. Functional Microrna Targets in Protein Coding Sequences. Bioinformatics 2012, 28, 771–776. [Google Scholar] [CrossRef] [PubMed]

| Parameters | Total Numbers (%) |
|---|---|
| CLL samples | 44 (100) |
| Male (%) | 24 (55) |
| Female (%) | 20 (45) |
| Age (years) | |
| Mean | 65.9 |
| Median | 66.9 |
| Range | 43.3–79.8 |
| Duration of disease (years) | |
| Mean | 3.8 |
| Range | 0–10.3 |
| RAI stage at diagnosis | |
| nda | 2 (5) |
| I | 7 (16) |
| II | 15 (34) |
| III | 12 (27) |
| IV | 8 (18) |
| Molecular risk parameters | |
| Unmutated Ig VH | 21 (48) |
| IGHV nda | 5 (11) |
| FISH karyotype | |
| del11q | 9 (20) |
| del13q | 19 (43) |
| del17p | 4 (9) |
| Trisomy 12 | 6 (14) |
| Normal karyotype | 5 (11) |
| Karyotype nda | 1 (2) |
| Treatment status | |
| Untreated at sampling | 44 (100) |
| Untreated at last follow up | 0 (0) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gassner, F.J.; Zaborsky, N.; Feldbacher, D.; Greil, R.; Geisberger, R. RNA Editing Alters miRNA Function in Chronic Lymphocytic Leukemia. Cancers 2020, 12, 1159. https://doi.org/10.3390/cancers12051159
Gassner FJ, Zaborsky N, Feldbacher D, Greil R, Geisberger R. RNA Editing Alters miRNA Function in Chronic Lymphocytic Leukemia. Cancers. 2020; 12(5):1159. https://doi.org/10.3390/cancers12051159
Chicago/Turabian StyleGassner, Franz J., Nadja Zaborsky, Daniel Feldbacher, Richard Greil, and Roland Geisberger. 2020. "RNA Editing Alters miRNA Function in Chronic Lymphocytic Leukemia" Cancers 12, no. 5: 1159. https://doi.org/10.3390/cancers12051159
APA StyleGassner, F. J., Zaborsky, N., Feldbacher, D., Greil, R., & Geisberger, R. (2020). RNA Editing Alters miRNA Function in Chronic Lymphocytic Leukemia. Cancers, 12(5), 1159. https://doi.org/10.3390/cancers12051159