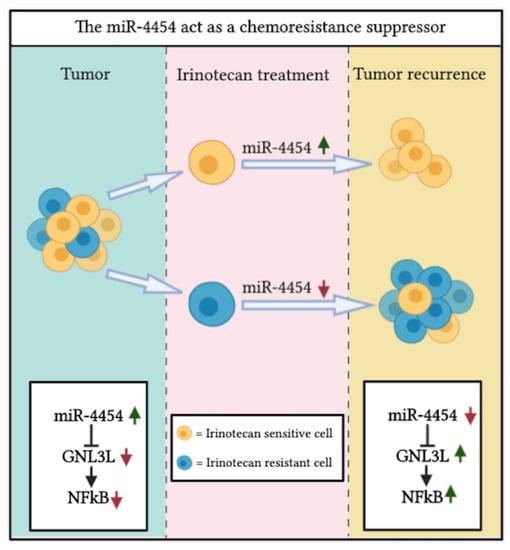
Chemoresistance-Associated Silencing of miR-4454 Promotes Colorectal Cancer Aggression through the GNL3L and NF-κB Pathway
Abstract
1. Introduction
2. Results
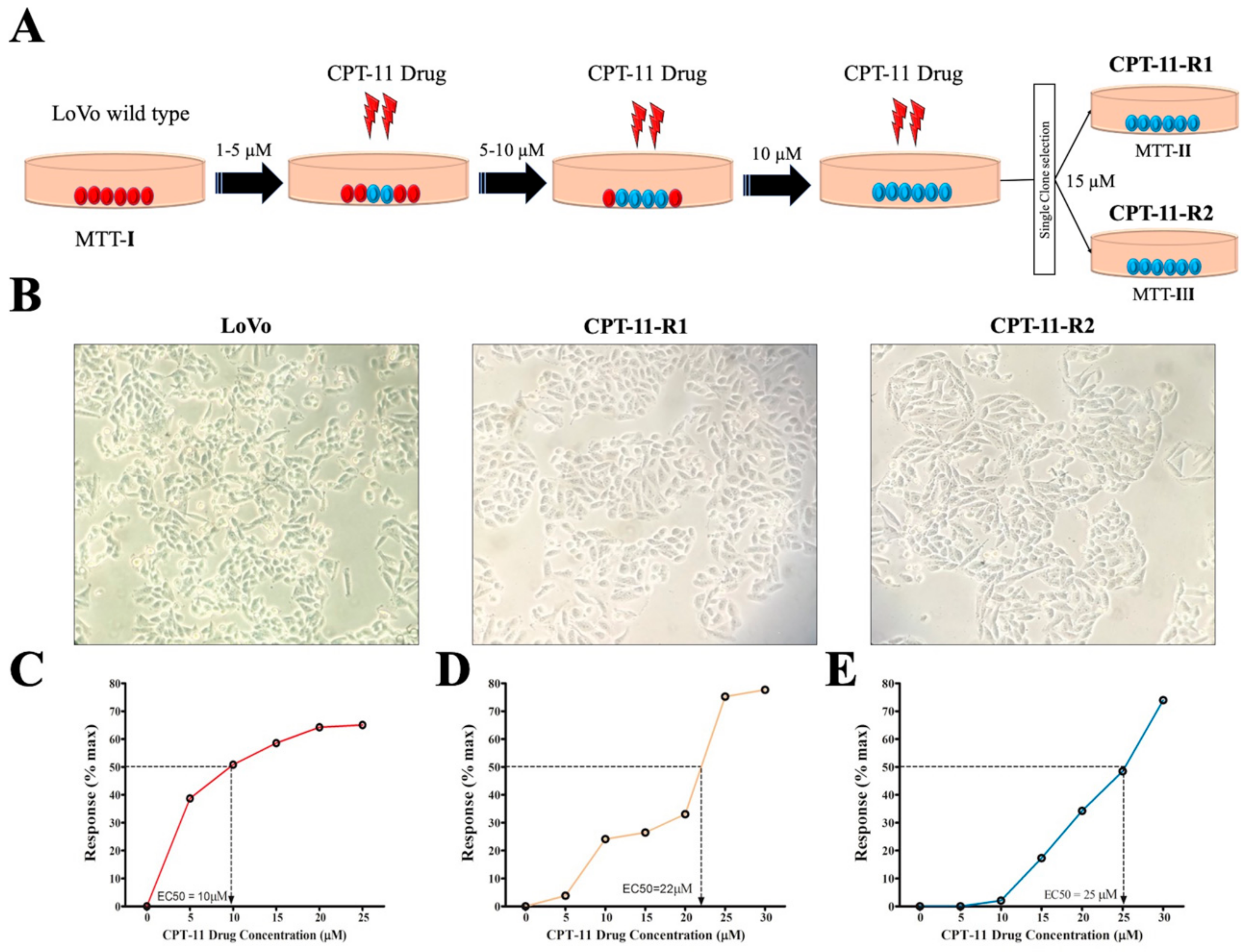
2.1. Generation of Drug-Resistant Cell Lines
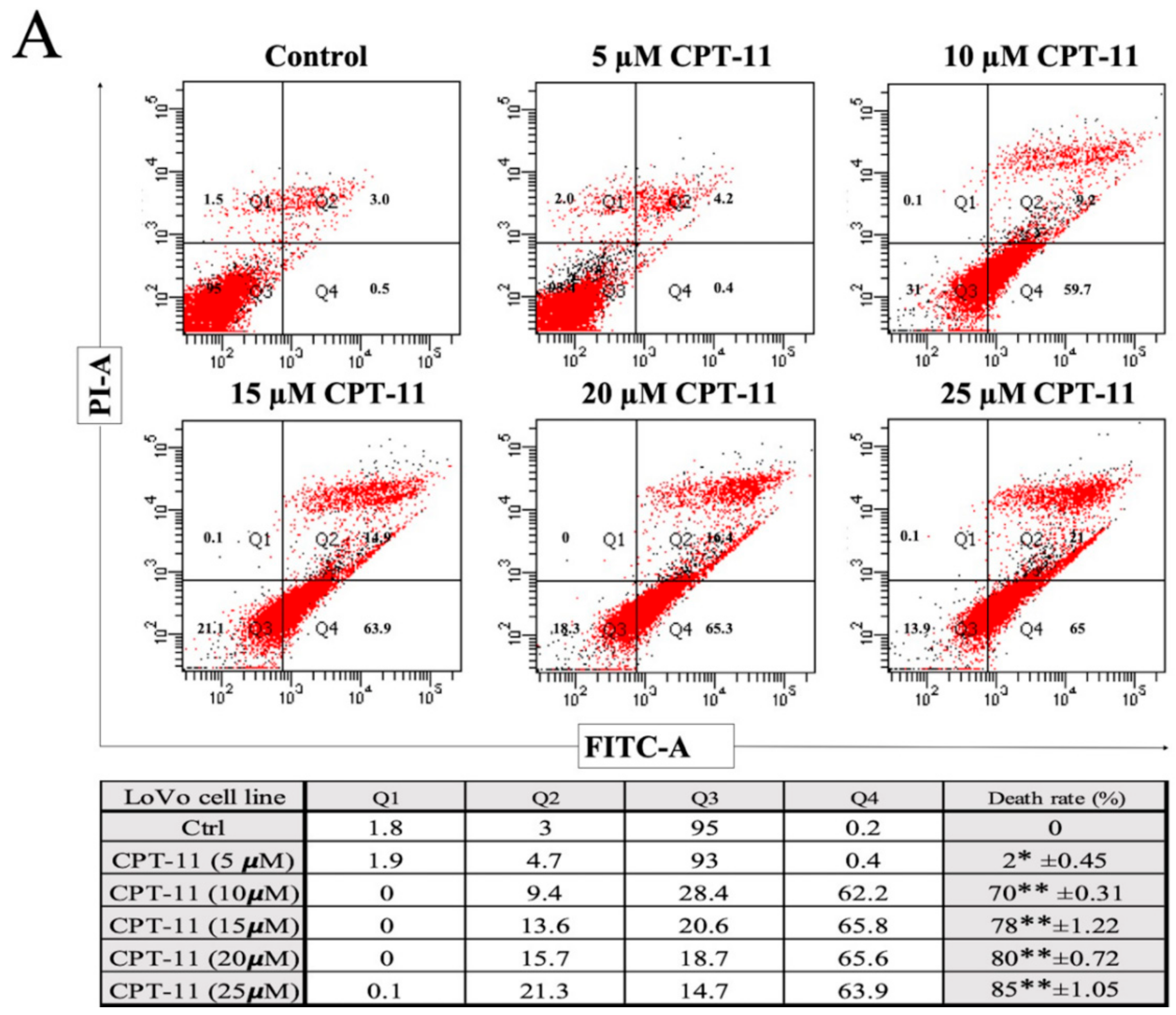
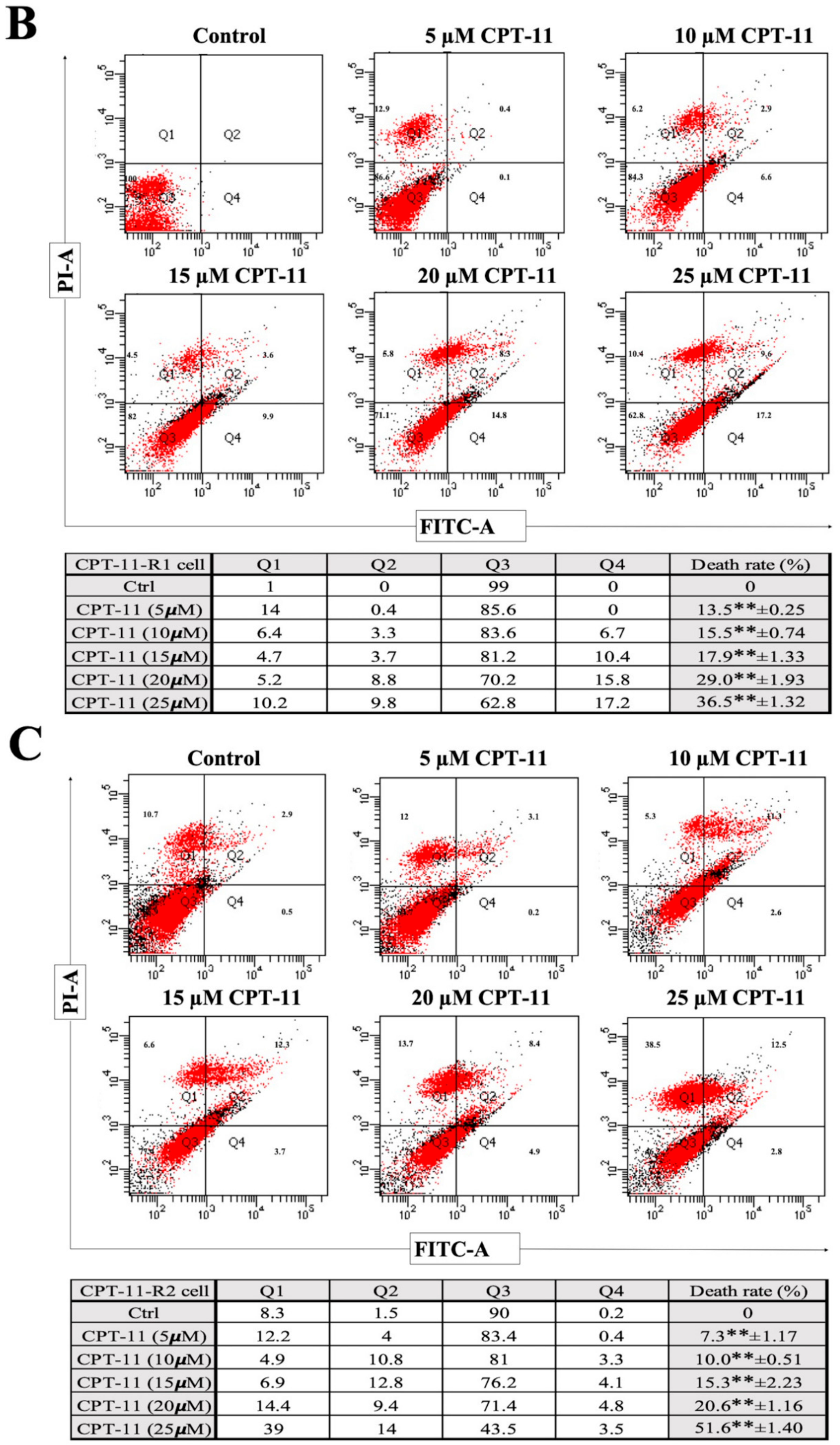
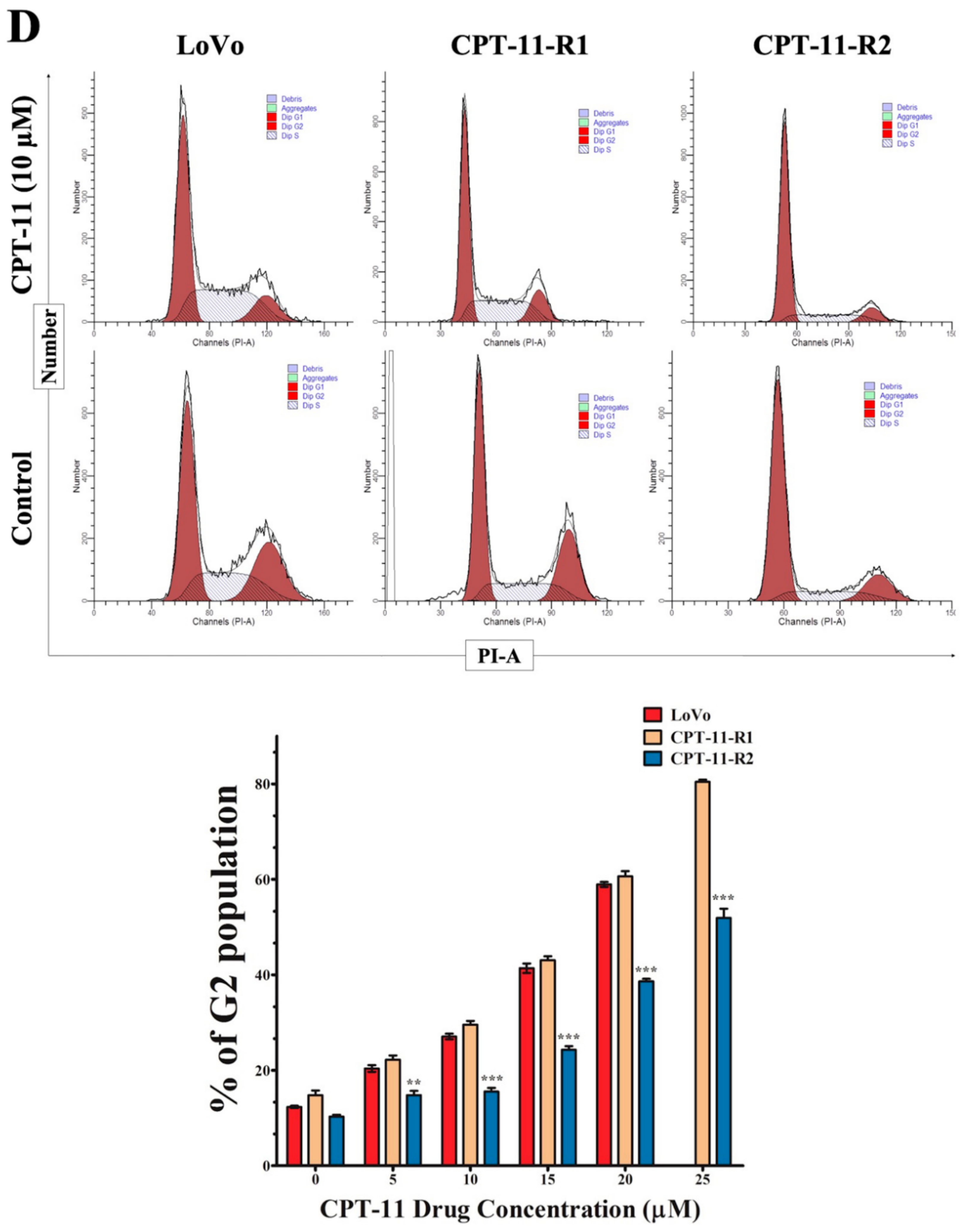
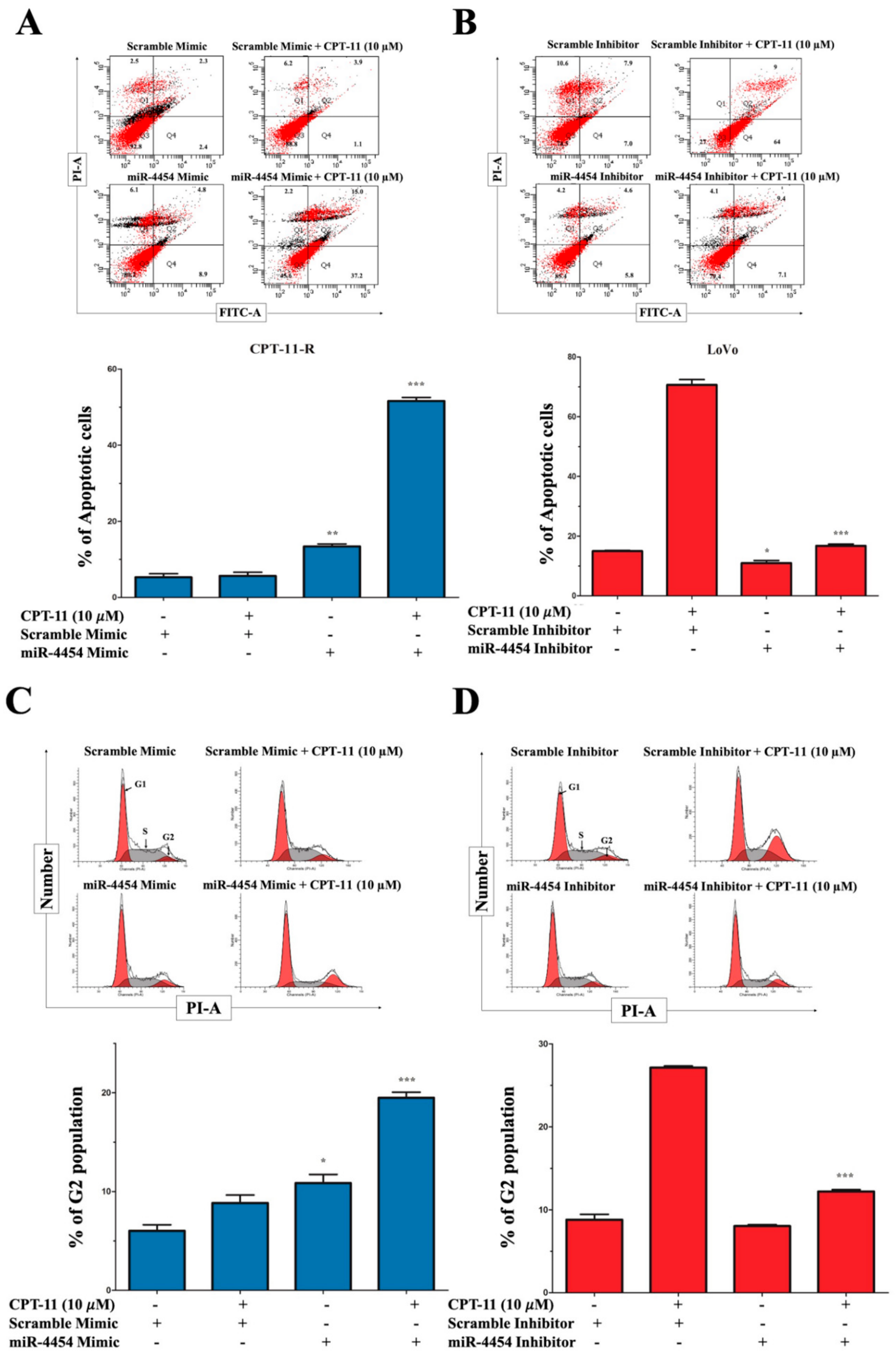
2.2. Apoptotic Reduction and Cell Cycle Alteration Increase CPT-11-R Cell Chemoresistance
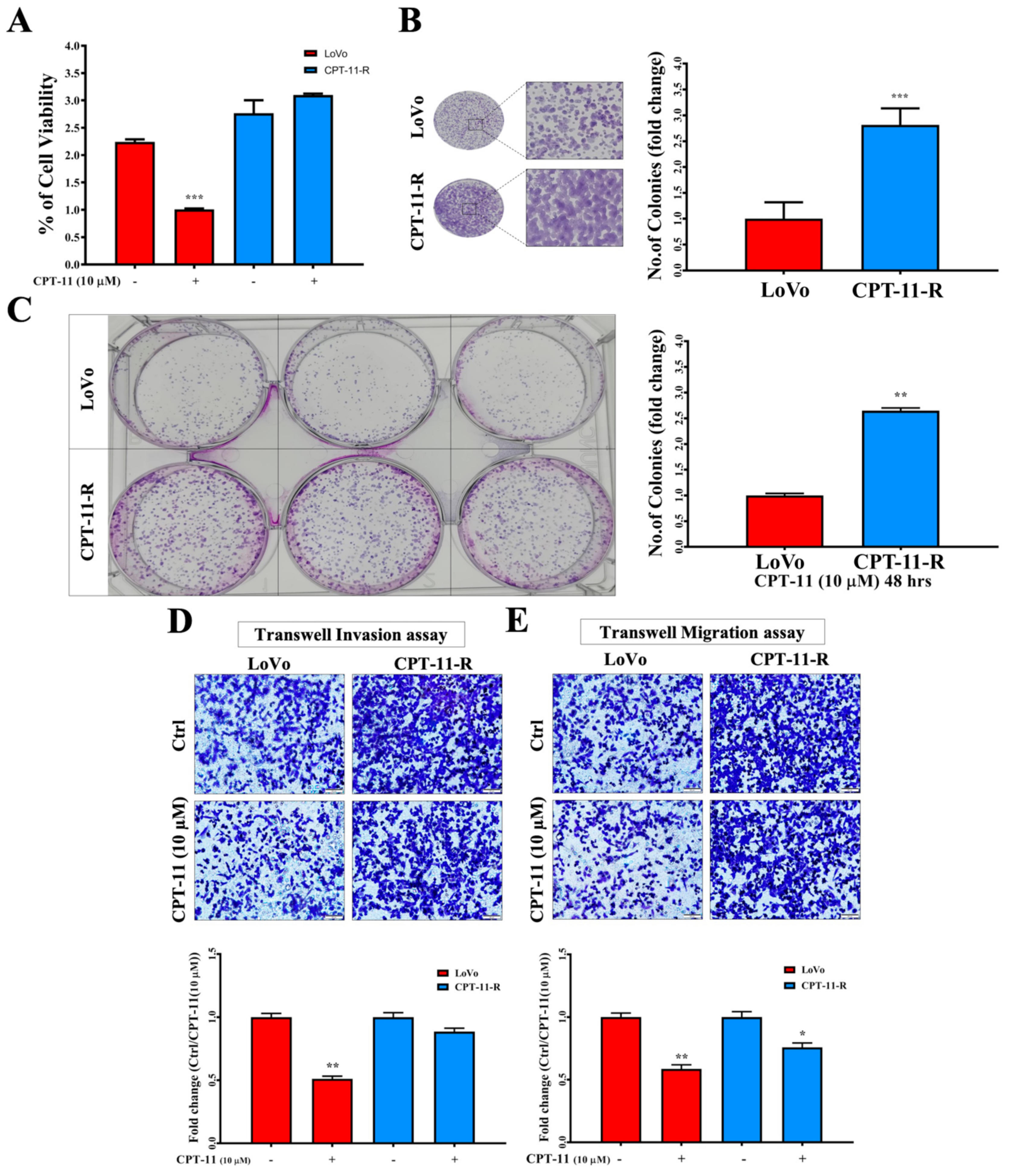
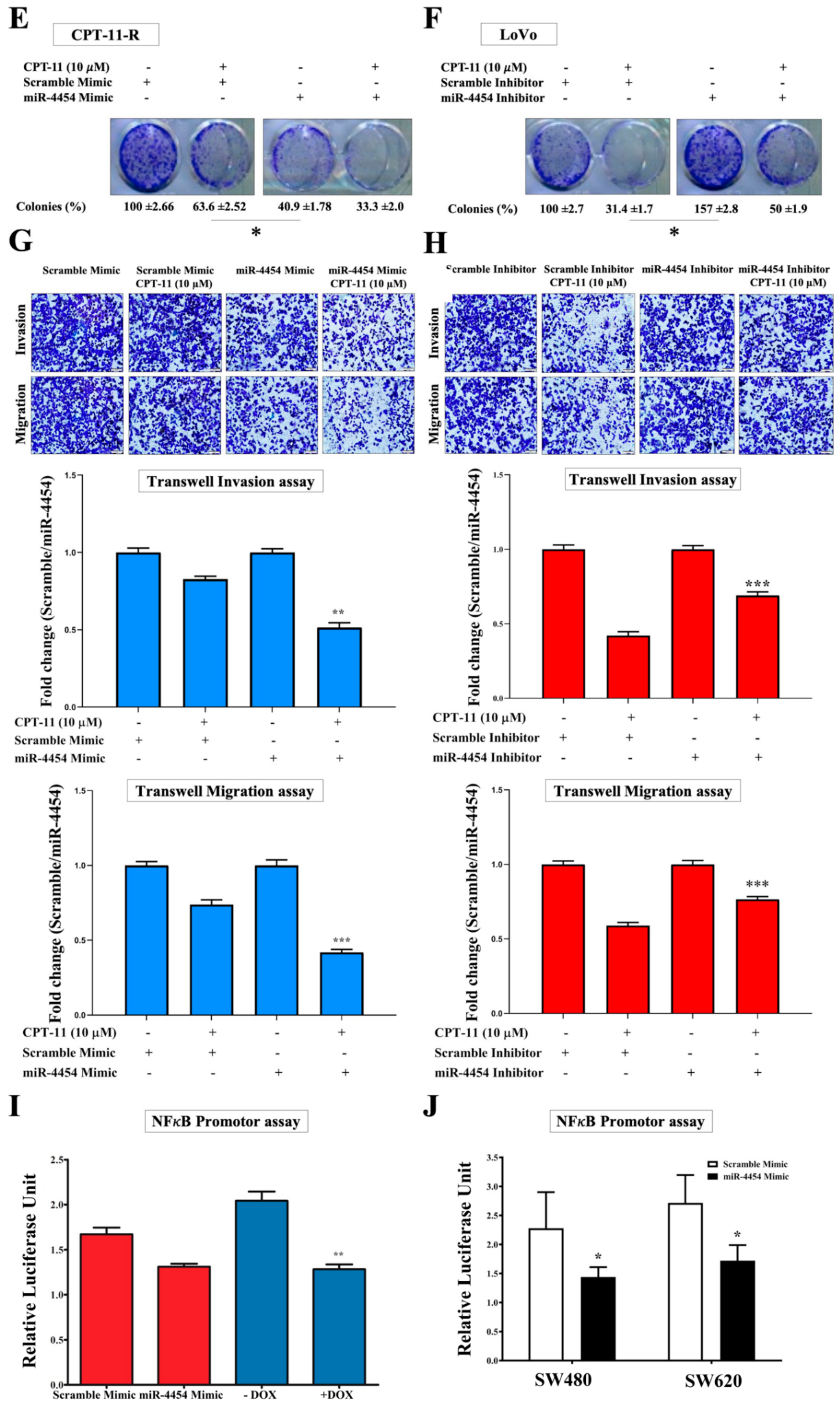
2.3. CPT-11-R Cells Show Enhanced First-Line Chemoresistance, Colony Formation, Invasion, and Migration Ability
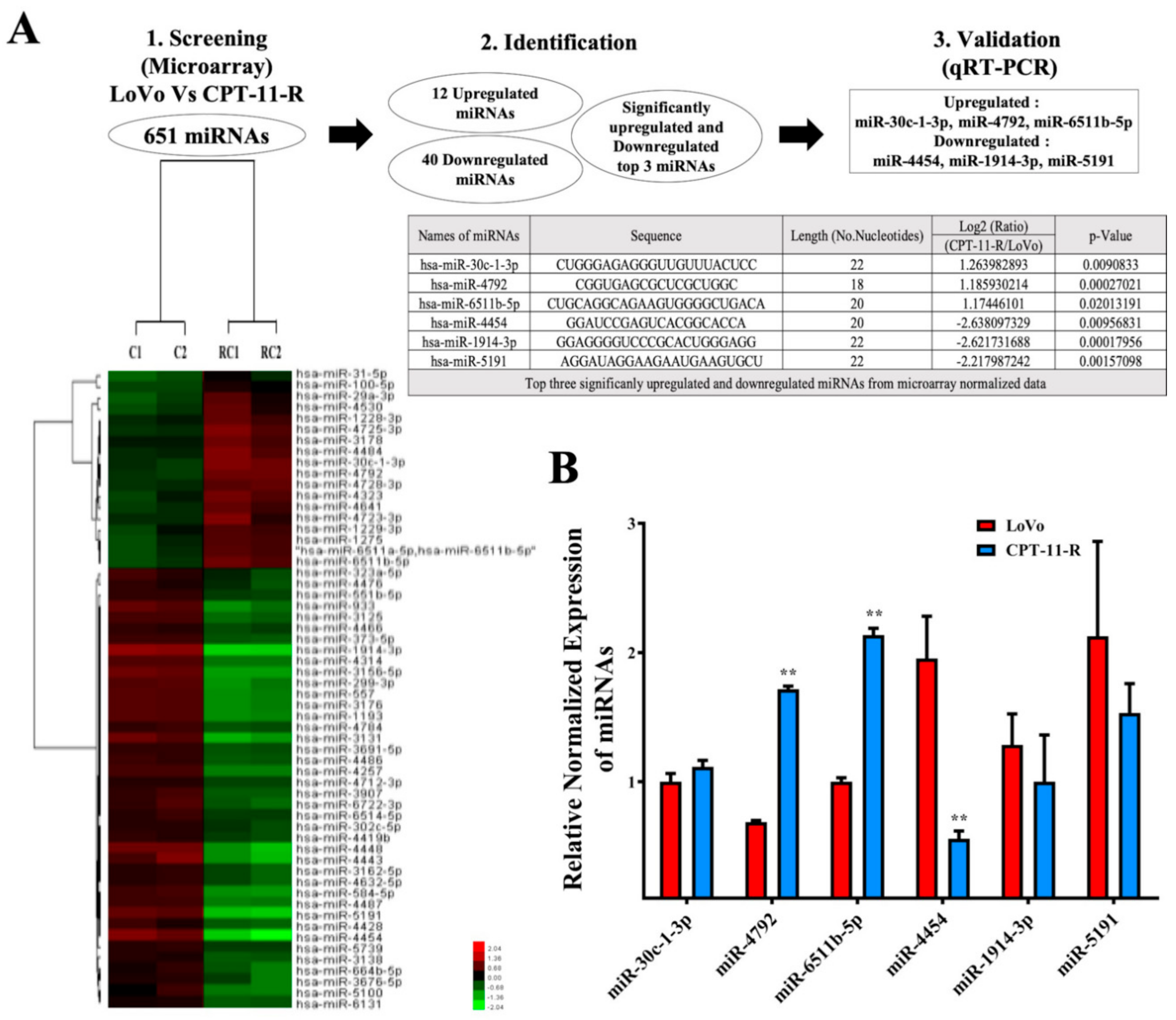
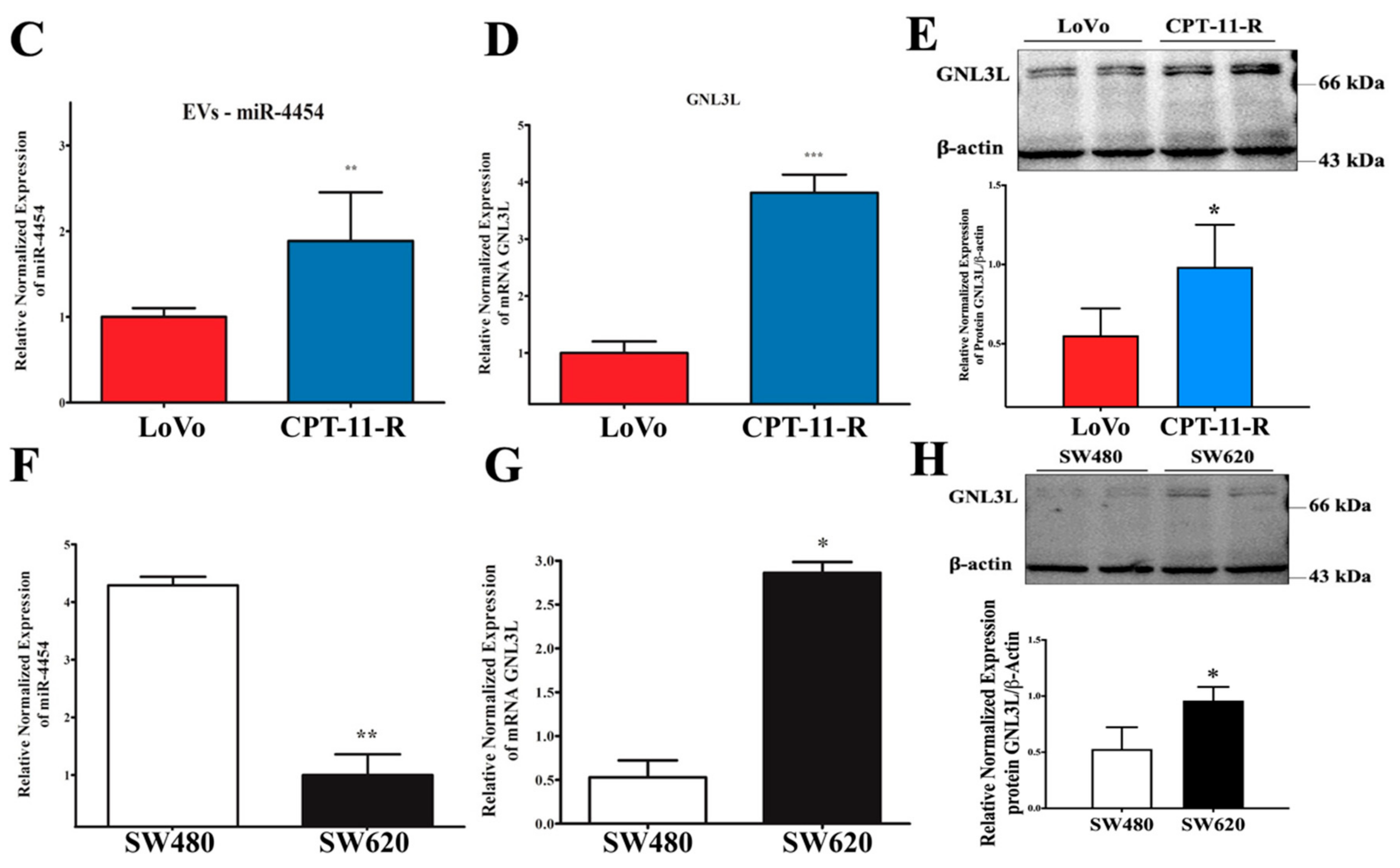
2.4. Downregulation of miR-4454 Is Attributed to Elevated Levels of GNL3L in the CPT-11-R Cells
2.5. GNL3L Is a Functional Target of miR-4454
2.6. MiR-4454 Suppresses Colon Cancer Cell Growth, Invasion, and Migration through the GNL3L and NF-κB Pathways
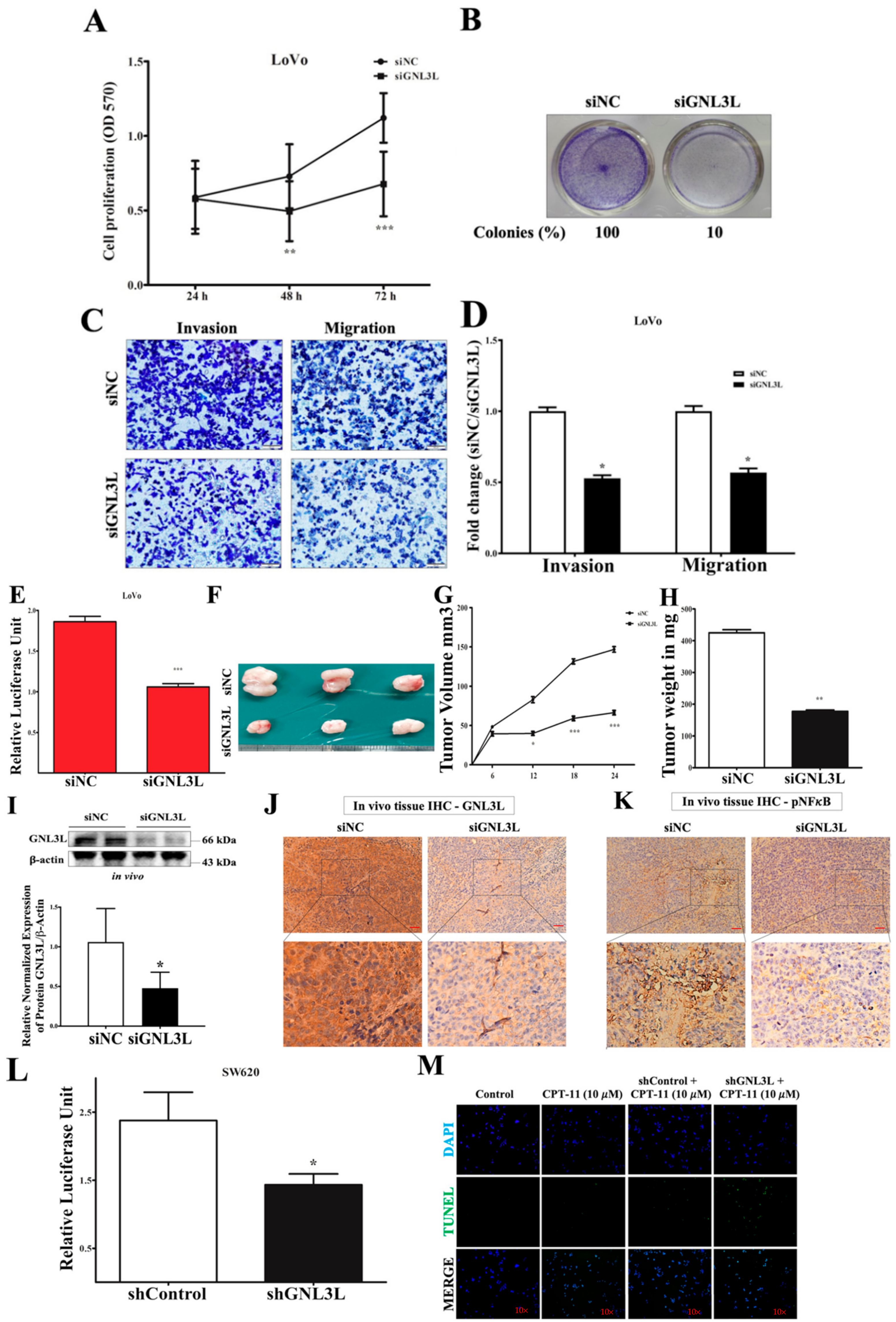
2.7. Silencing GNL3L Attenuates Proliferation, Colony Formation, and Invasion/Migration
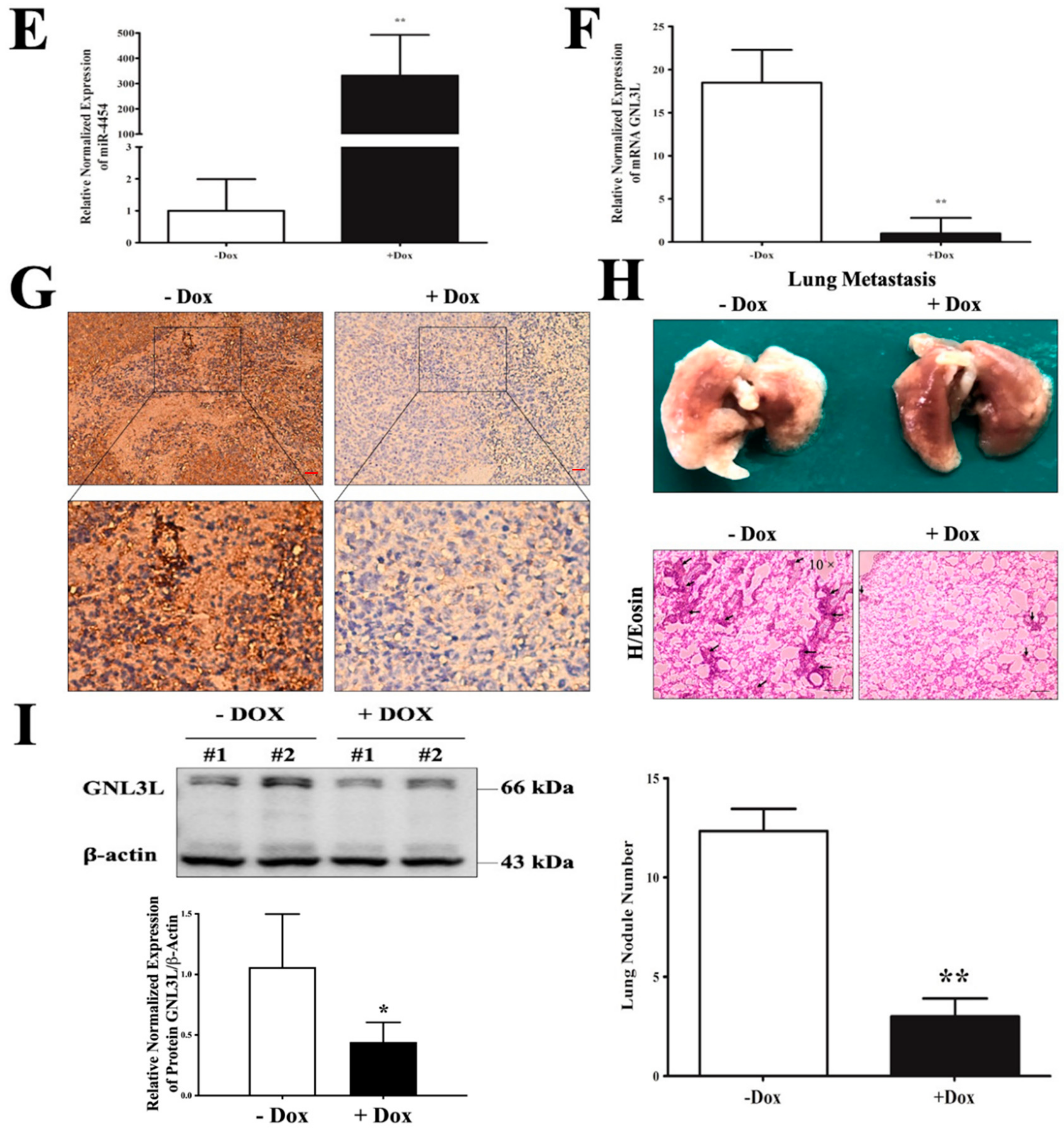
2.8. Doxycycline-Induced Expression of miR-4454 Suppresses Colon Cancer Growth
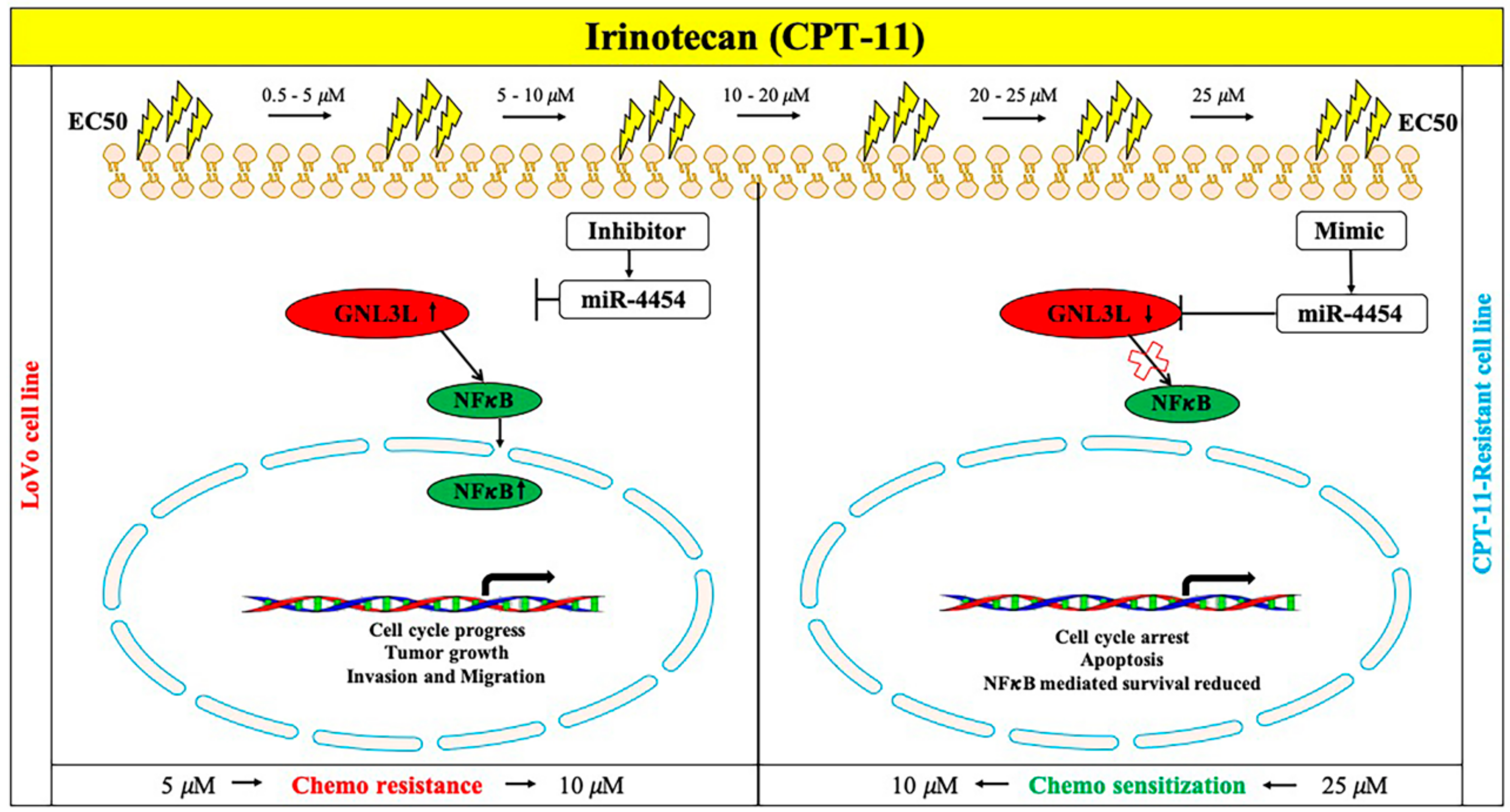
2.9. Schematic Representation of the miR-4454/GNL3L/NFκB Pathway
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. 3-[4,5-Dimethylthiazol-2-yl]-2,5-diphenyltetrazolium Bromide Assay
4.3. Flow Cytometry for the Annexin V Apoptosis Assay
4.4. Cell Cycle Analysis
4.5. Microarray Profiling and Target Prediction
4.6. Western Blot Analysis
4.7. RNA Extraction, microRNA First-Strand Synthesis, and qRT-PCR
4.8. Extracellular Vehicle RNA Isolation
4.9. MicroRNA and siRNA or shRNA Transfection Experiments
4.10. miR-4454 Dox-Inducible Expression Vector and Gene Reporter Plasmid Construction
4.11. Luciferase Reporter Assay
4.12. Generation of miR-4454 pSBinducer Cells
4.13. Transwell Invasion/Migration Assay
4.14. Colony Formation Assay and Hematoxylin and Eosin Stain
4.15. Immunohistochemistry
4.16. In Situ TUNEL Assay
4.17. Dox Induction In Vivo Tumor Studies
4.18. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Cusack, J.C.; Liu, R.; Baldwin, A.S. NF- kappa B and chemoresistance: Potentiation of cancer drugs via inhibition of NF-kappa B. Drug Resist Updat. 1999, 2, 271–273. [Google Scholar] [CrossRef] [PubMed]
- Cusack, J.C.; Liu, R.; Baldwin, A.S. Inducible chemoresistance to 7-ethyl-10-[4-(1-piperidino)-1-piperidino]-carbonyloxycamptothe cin (CPT-11) in colorectal cancer cells and a xenograft model is overcome by inhibition of nuclear factor-kappaB activation. Cancer Res. 2000, 60, 2323–2330. [Google Scholar] [PubMed]
- Rasheed, Z.A.; Rubin, E.H. Mechanisms of resistance to topoisomerase I-targeting drugs. Oncogene 2003, 22, 7296–7304. [Google Scholar] [CrossRef]
- Yang, A.D. Chronic Oxaliplatin Resistance Induces Epithelial-to-Mesenchymal Transition in Colorectal Cancer Cell Lines. Clin. Cancer Res. 2006, 12, 4147–4153. [Google Scholar] [CrossRef]
- Ghosh, R.D.; Ghuwalewala, S.; Das, P.; Mandloi, S.; Alam, S.K.; Chakraborty, J.; Sarkar, S.; Chakrabarti, S.; Panda, C.K.; Roychoudhury, S. MicroRNA profiling of cisplatin-resistant oral squamous cell carcinoma cell lines enriched with cancer-stem-cell-like and epithelial-mesenchymal transition-type features. Sci. Rep. 2016, 6, 23932. [Google Scholar] [CrossRef]
- Luqmani, Y. Mechanisms of Drug Resistance in Cancer Chemotherapy. Med Princ. Pr. 2005, 14, 35–48. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Cusack, J.C.; Liu, R.; Baldwin, A.S., Jr. Control of inducible chemoresistance: Enhanced anti-tumor therapy through increased apoptosis by inhibition of NF-κB. Nat. Med. 1999, 5, 412–417. [Google Scholar] [CrossRef]
- Xu, Y.; Villalona-Calero, M.A. Irinotecan: Mechanisms of tumor resistance and novel strategies for modulating its activity. Ann. Oncol. 2002, 13, 1841–1851. [Google Scholar] [CrossRef]
- Chen, M.-C.; Lee, N.-H.; Ho, T.-J.; Hsu, H.-H.; Kuo, C.-H.; Kuo, W.-W.; Lin, Y.-M.; Tsai, F.-J.; Tsai, C.-H.; Huang, C.-Y. Resistance to irinotecan (CPT-11) activates epidermal growth factor receptor/nuclear factor kappa B and increases cellular metastasis and autophagy in LoVo colon cancer cells. Cancer Lett. 2014, 349, 51–60. [Google Scholar] [CrossRef]
- Huang, T.T.; Wuerzberger-Davis, S.M.; Seufzer, B.J.; Shumway, S.D.; Kurama, T.; Boothman, D.A.; Miyamoto, S. NF-kappaB activation by camptothecin. A linkage between nuclear DNA damage and cytoplasmic signaling events. J. Boil. Chem. 2000, 275, 9501–9509. [Google Scholar] [CrossRef] [PubMed]
- Miska, E.A. How microRNAs control cell division, differentiation and death. Curr. Opin. Genet. Dev. 2005, 15, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Ameres, S.L.; Zamore, P. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Boil. 2013, 14, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Oom, A.; Humphries, B.A.; Yang, C. MicroRNAs: Novel Players in Cancer Diagnosis and Therapies. BioMed Res. Int. 2014, 2014, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Drewinko, B.; Romsdahl, M.M.; Yang, L.Y.; Ahearn, M.J.; Trujillo, J.M. Establishment of a human carcinoembryonic antigen-producing colon adenocarcinoma cell line. Cancer Res. 1976, 36, 467–475. [Google Scholar]
- Rao, M.S.; Kumari, G.; Balasundaram, D.; Sankaranarayanan, R.; Mahalingam, S. A Novel Lysine-rich Domain and GTP Binding Motifs Regulate the Nucleolar Retention of Human Guanine Nucleotide Binding Protein, GNL3L. J. Mol. Boil. 2006, 364, 637–654. [Google Scholar] [CrossRef]
- Meng, L.; Hsu, J.K.; Tsai, R.Y. GNL3L depletion destabilizes MDM2 and induces p53-dependent G2/M arrest. Oncogene 2011, 30, 1716–1726. [Google Scholar] [CrossRef]
- Thoompumkal, I.J.; Rehna, K.; Anbarasu, K.; Mahalingam, S. Leucine Zipper Down-regulated in Cancer-1 (LDOC1) interacts with Guanine nucleotide binding protein-like 3-like (GNL3L) to modulate Nuclear Factor-kappa B (NF-κB) signaling during cell proliferation. Cell Cycle 2016, 15, 3251–3267. [Google Scholar] [CrossRef]
- Okamoto, N.; Yasukawa, M.; Nguyen, C.; Kasim, V.; Maida, Y.; Possemato, R.; Shibata, T.; Ligon, K.L.; Fukami, K.; Hahn, W.C.; et al. Maintenance of tumor initiating cells of defined genetic composition by nucleostemin. Proc. Natl. Acad. Sci. USA 2011, 108, 20388–20393. [Google Scholar] [CrossRef]
- Hur, K.; Toiyama, Y.; Schetter, A.J.; Okugawa, Y.; Harris, C.C.; Boland, C.R.; Goel, A. Identification of a Metastasis-Specific MicroRNA Signature in Human Colorectal Cancer. J. Natl. Cancer Inst. 2015, 107. [Google Scholar] [CrossRef]
- Ji, H.; Chen, M.; Greening, D.; He, W.; Rai, A.; Zhang, W.; Simpson, R. Deep Sequencing of RNA from Three Different Extracellular Vesicle (EV) Subtypes Released from the Human LIM1863 Colon Cancer Cell Line Uncovers Distinct Mirna-Enrichment Signatures. PLoS ONE 2014, 9, e110314. [Google Scholar] [CrossRef] [PubMed]
- Lulla, A.R.; Slifker, M.J.; Zhou, Y.; Lev, A.; Einarson, M.B.; Dicker, D.T.; El-Deiry, W.S. miR-6883 Family miRNAs Target CDK4/6 to Induce G1 Phase Cell-Cycle Arrest in Colon Cancer Cells. Cancer Res. 2017, 77, 6902–6913. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.C.; Lee, N.H.; Hsu, H.H.; Ho, T.J.; Tu, C.C.; Chen, R.J.; Lin, Y.M.; Viswanadha, V.P.; Kuo, W.W.; Huang, C.Y. Inhibition of NF-kappaB and metastasis in irinotecan (CPT-11)-resistant LoVo colon cancer cells by thymoquinone via JNK and p38. Environ. Toxicol. 2017, 32, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Rothenberg, M.L. Irinotecan (CPT-11): Recent Developments and Future Directions–Colorectal Cancer and Beyond. Oncology 2001, 6, 66–80. [Google Scholar] [CrossRef]
- Cunningham, D.; Pyrhönen, S.; James, R.D.; Punt, C.J.; Hickish, T.; Heikkilä, R.; Johannesen, T.B.; Starkhammar, H.; Topham, C.A.; Awad, L.; et al. Randomised trial of irinotecan plus supportive care versus supportive care alone after fluorouracil failure for patients with metastatic colorectal cancer. Lancet 1998, 352, 1413–1418. [Google Scholar] [CrossRef]
- Tergaonkar, V.; Pando, M.; Vafa, O.; Wahl, G.; Verma, I. p53 stabilization is decreased upon NFkappaB activation: A role for NFkappaB in acquisition of resistance to chemotherapy. Cancer Cell 2002, 1, 493–503. [Google Scholar] [CrossRef]
- Tang, X.; Zha, L.; Li, H.; Liao, G.; Huang, Z.; Peng, X.; Wang, Z. Upregulation of GNL3 expression promotes colon cancer cell proliferation, migration, invasion and epithelial-mesenchymal transition via the Wnt/beta-catenin signaling pathway. Oncol. Rep. 2017, 38, 2023–2032. [Google Scholar] [CrossRef]
- Allen, K.E.; Weiss, G.J. Resistance May Not Be Futile: microRNA Biomarkers for Chemoresistance and Potential Therapeutics. Mol. Cancer Ther. 2010, 9, 3126–3136. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Yu, J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pander, A.; Chinnaiyan, A.M. ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform. Neoplasia 2004, 6, 1–6. [Google Scholar] [CrossRef]
- Wu, K.-M.; Hsu, Y.-M.; Ying, M.-C.; Tsai, F.-J.; Tsai, C.-H.; Chung, J.; Yang, J.-S.; Tang, C.-H.; Cheng, L.-Y.; Su, P.-H.; et al. High-density lipoprotein ameliorates palmitic acid-induced lipotoxicity and oxidative dysfunction in H9c2 cardiomyoblast cells via ROS suppression. Nutr. Metab. 2019, 16, 36. [Google Scholar] [CrossRef]
- Rasmussen, M.H.; Lyskjær, I.; Jersie-Christensen, R.R.; Tarpgaard, L.; Primdal-Bengtson, B.; Nielsen, M.M.; Pedersen, J.S.; Hansen, T.P.; Hansen, F.; Olsen, J.V.; et al. miR-625-3p regulates oxaliplatin resistance by targeting MAP2K6-p38 signalling in human colorectal adenocarcinoma cells. Nat. Commun. 2016, 7, 12436. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Chen, K.B.; Tsai, C.H.; Tsai, F.J.; Huang, C.Y.; Tang, C.H.; Yang, J.S.; Hsu, Y.M.; Peng, S.F.; Chung, J.G. Casticin inhibits human prostate cancer DU 145 cell migration and invasion via Ras/Akt/NF-kappaB signaling pathways. J. Food Biochem. 2019, 43, e12902. [Google Scholar] [CrossRef] [PubMed]
- Hubner, E.K.; Lechler, C.; Rösner, T.N.; Kohnke-Ertel, B.; Schmid, R.M.; Ehmer, U. Constitutive and Inducible Systems for Genetic In Vivo Modification of Mouse Hepatocytes Using Hydrodynamic Tail Vein Injection. J. Vis. Exp. 2018, 132, e56613. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Liu, X.; Mak, C.S.-L.; Yung, M.M.-H.; Leung, T.H.-Y.; Xu, D.; Ngu, S.F.; Chan, K.K.-L.; Yang, H.; Ngan, H.; et al. Methylation-associated silencing of miR-193a-3p promotes ovarian cancer aggressiveness by targeting GRB7 and MAPK/ERK pathways. Theranostics 2018, 8, 423–436. [Google Scholar] [CrossRef]

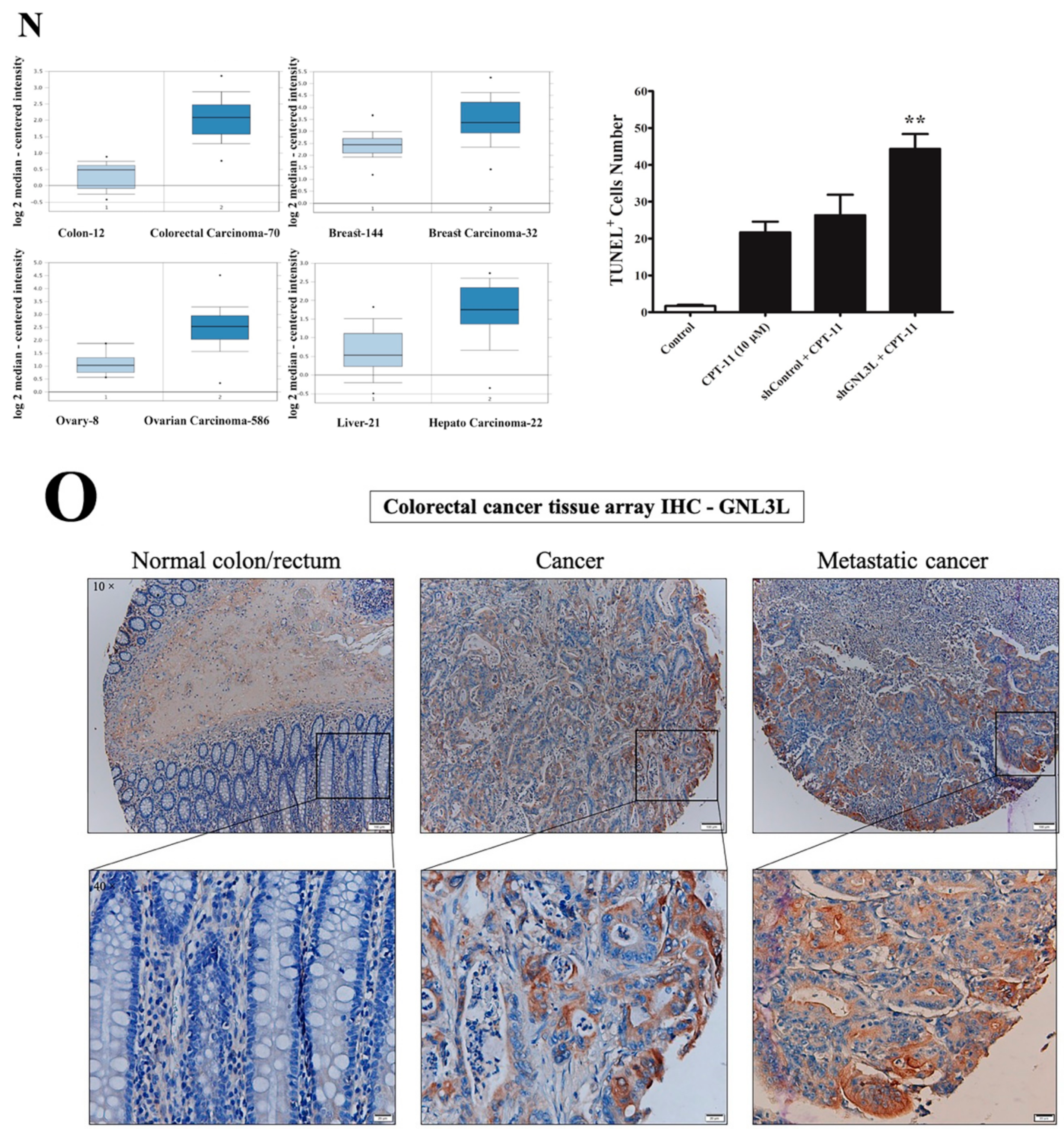

| Group | Cases (n) | GNL3L Expression | 𝝌2 | * p Value | |
|---|---|---|---|---|---|
| Low | High | ||||
| Expression | Expression | ||||
| Normal colon/rectum | 33 | 25 (76%) | 8 (24%) | ||
| Cancer | 33 | 11 (33%) | 22 (67%) | 11.98 | <0.0003 |
| Metastatic cancer | 33 | 5 (15%) | 28 (85%) | 24.44 | <0.0001 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kannathasan, T.; Kuo, W.-W.; Chen, M.-C.; Viswanadha, V.P.; Shen, C.-Y.; Tu, C.-C.; Yeh, Y.-L.; Bharath, M.; Shibu, M.A.; Huang, C.-Y. Chemoresistance-Associated Silencing of miR-4454 Promotes Colorectal Cancer Aggression through the GNL3L and NF-κB Pathway. Cancers 2020, 12, 1231. https://doi.org/10.3390/cancers12051231
Kannathasan T, Kuo W-W, Chen M-C, Viswanadha VP, Shen C-Y, Tu C-C, Yeh Y-L, Bharath M, Shibu MA, Huang C-Y. Chemoresistance-Associated Silencing of miR-4454 Promotes Colorectal Cancer Aggression through the GNL3L and NF-κB Pathway. Cancers. 2020; 12(5):1231. https://doi.org/10.3390/cancers12051231
Chicago/Turabian StyleKannathasan, Thetchinamoorthy, Wei-Wen Kuo, Ming-Cheng Chen, Vijaya Padma Viswanadha, Chia-Yao Shen, Chuan-Chou Tu, Yu-Lan Yeh, Mahalakshmi Bharath, Marthandam Asokan Shibu, and Chih-Yang Huang. 2020. "Chemoresistance-Associated Silencing of miR-4454 Promotes Colorectal Cancer Aggression through the GNL3L and NF-κB Pathway" Cancers 12, no. 5: 1231. https://doi.org/10.3390/cancers12051231
APA StyleKannathasan, T., Kuo, W.-W., Chen, M.-C., Viswanadha, V. P., Shen, C.-Y., Tu, C.-C., Yeh, Y.-L., Bharath, M., Shibu, M. A., & Huang, C.-Y. (2020). Chemoresistance-Associated Silencing of miR-4454 Promotes Colorectal Cancer Aggression through the GNL3L and NF-κB Pathway. Cancers, 12(5), 1231. https://doi.org/10.3390/cancers12051231