Extracellular Vesicles Enriched in hsa-miR-301a-3p and hsa-miR-1293 Dynamics in Clear Cell Renal Cell Carcinoma Patients: Potential Biomarkers of Metastatic Disease
Abstract
:1. Introduction
2. Results
2.1. EVs Characterization
2.2. EV-Derived miRNA Levels in ccRCC Patients’ Plasma Samples
2.3. EV-Derived miRNAs Impact on Overall Survival of Metastatic ccRCC Patients
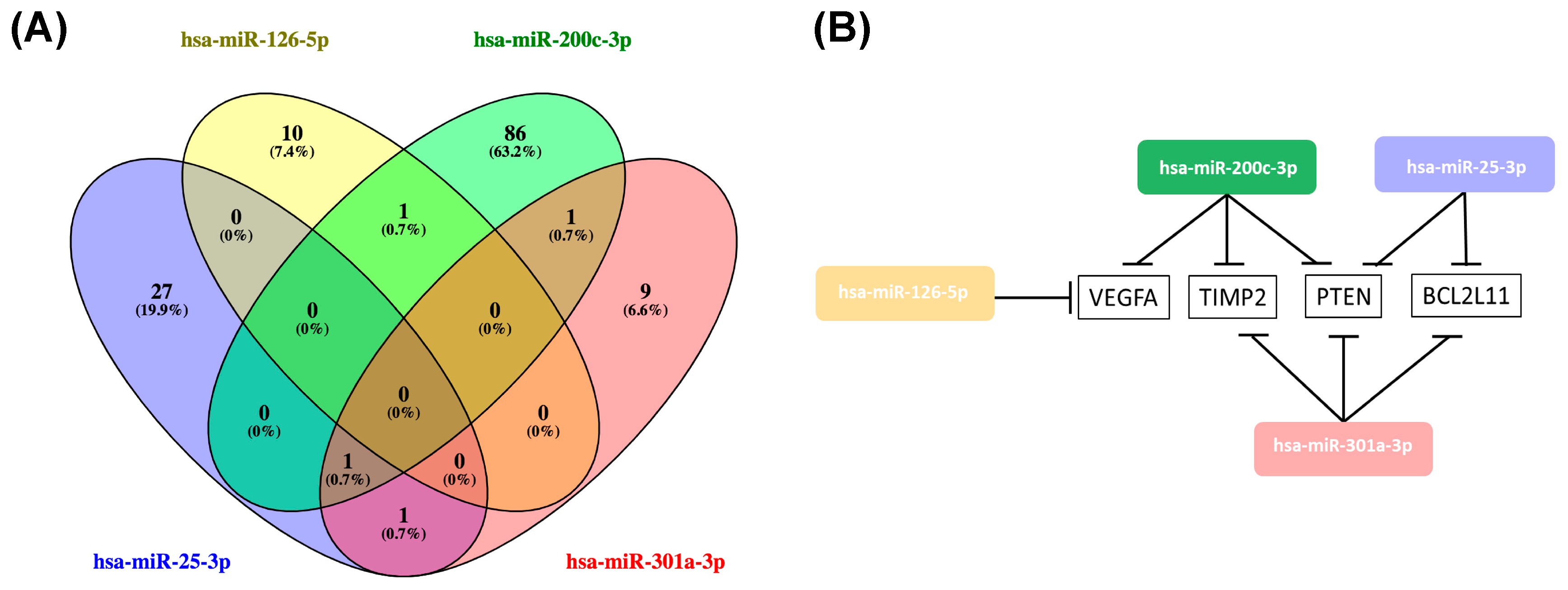
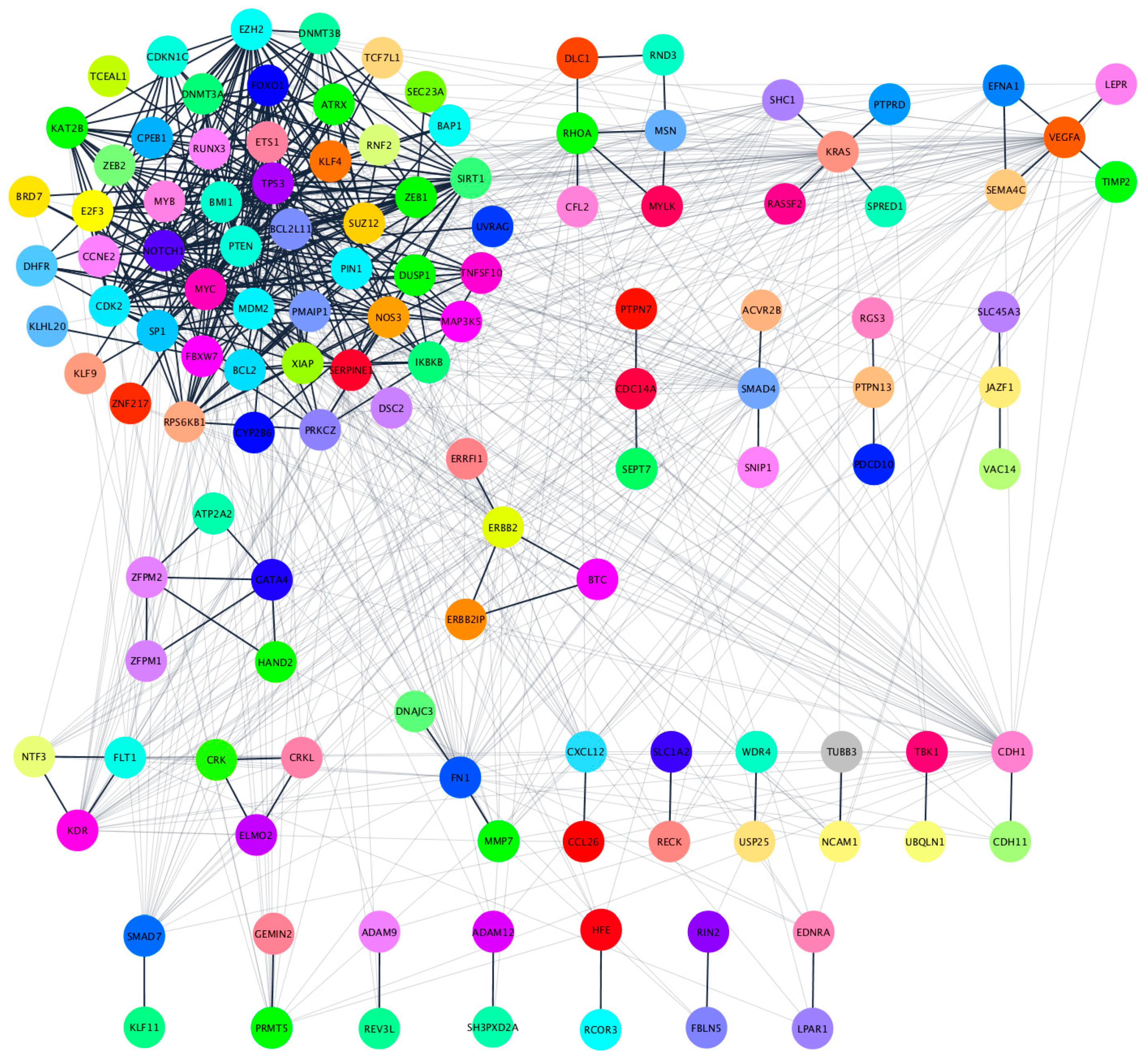
2.4. Hsa-miR-25-3p, hsa-miR-126-5p, hsa-miR-200c-3p, hsa-miR-301a-3p Overlaping Target Genes
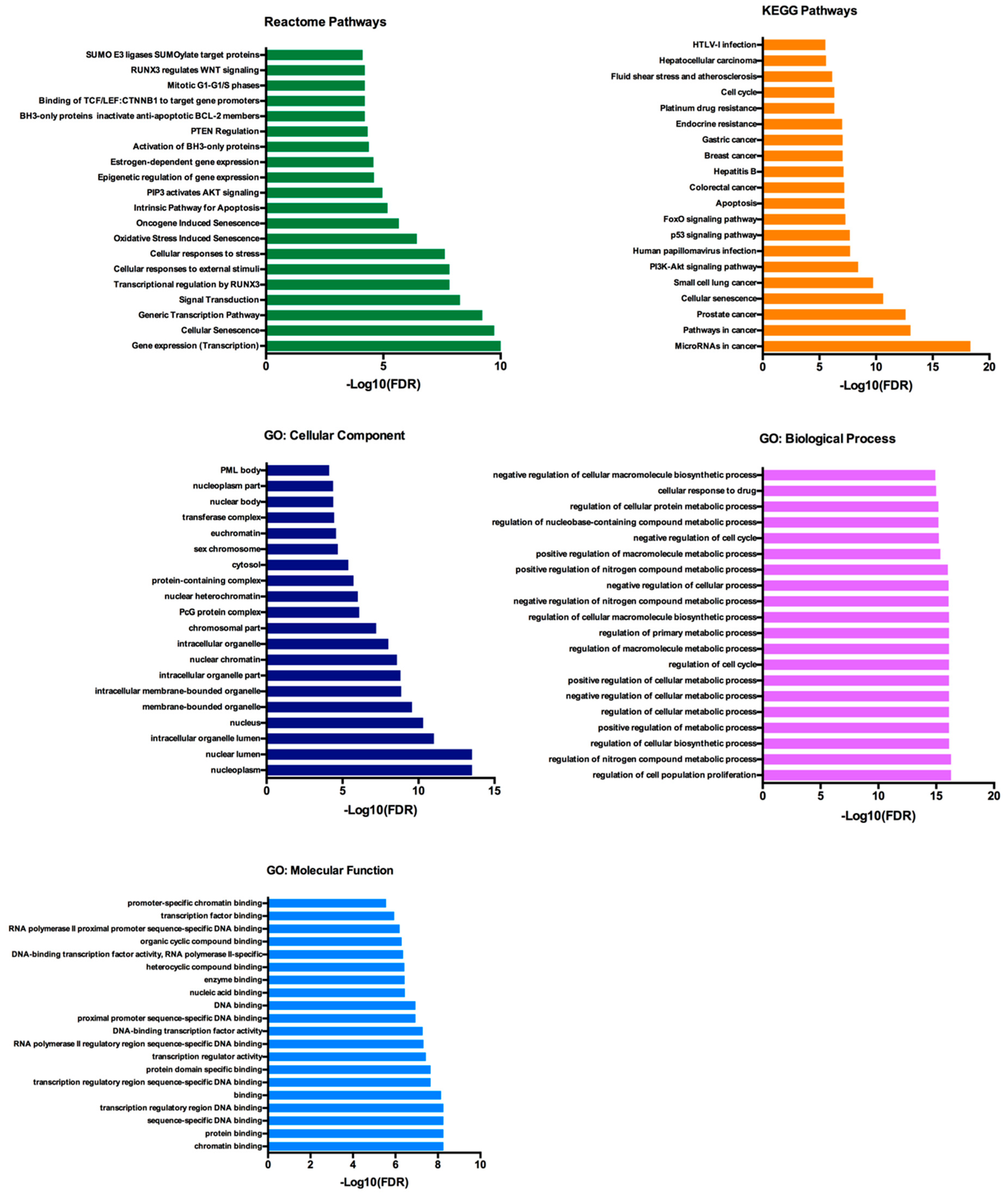
2.5. Functional Annotation and Pathway Enrichment Analysis of has-miR-25-3p, hsa-miR-126-5p, hsa-miR-200c-3p and hsa-miR-301a-3p Targets Network
3. Discussion
4. Materials and Methods
4.1. Ethics Statment
4.2. Study Population
4.3. EVs Isolation
4.4. EVs NTA Analysis
4.5. Quantification of Vesicular Structures by EVs Flow Cytometry
4.6. miRNA Extraction and cDNA Synthesis
4.7. Quantitative Real Time PCR
4.8. Statistical Analysis
4.9. Protein-Protein Interaction (PPI) Network and Cluster Analysis
4.10. Functional Annotation and Pathway Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ricketts, C.J.; De Cubas, A.A.; Fan, H.; Smith, C.C.; Lang, M.; Reznik, E.; Bowlby, R.; Gibb, E.A.; Akbani, R.; Beroukhim, R.; et al. The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma. Cell Rep. 2018, 23, 313–326. [Google Scholar] [CrossRef] [Green Version]
- Capitanio, U.; Montorsi, F. Renal cancer. Lancet 2016, 387, 894–906. [Google Scholar] [CrossRef]
- Moch, H.; Cubilla, A.L.; Humphrey, P.A.; Reuter, V.E.; Ulbright, T.M. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur. Urol. 2016, 70, 93–105. [Google Scholar] [CrossRef]
- Escudier, B.; Porta, C.; Schmidinger, M.; Rioux-Leclercq, N.; Bex, A.; Khoo, V.; Grunwald, V.; Gillessen, S.; Horwich, A. Renal cell carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-updagger. Ann. Oncol. 2019, 30, 706–720. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Ward, P.S.; Thompson, C.B. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell 2012, 21, 297–308. [Google Scholar] [CrossRef] [Green Version]
- Courtney, K.D.; Bezwada, D.; Mashimo, T.; Pichumani, K.; Vemireddy, V.; Funk, A.M.; Wimberly, J.; McNeil, S.S.; Kapur, P.; Lotan, Y.; et al. Isotope Tracing of Human Clear Cell Renal Cell Carcinomas Demonstrates Suppressed Glucose Oxidation In Vivo. Cell Metab. 2018, 28, 793–800. [Google Scholar] [CrossRef] [Green Version]
- Wettersten, H.I.; Aboud, O.A.; Lara, P.N., Jr.; Weiss, R.H. Metabolic reprogramming in clear cell renal cell carcinoma. Nat. Rev. Nephrol. 2017, 13, 410–419. [Google Scholar] [CrossRef]
- Linehan, W.M.; Srinivasan, R.; Schmidt, L.S. The genetic basis of kidney cancer: A metabolic disease. Nat. Rev. Urol. 2010, 7, 277–285. [Google Scholar] [CrossRef]
- Baldewijns, M.M.; van Vlodrop, I.J.H.; Vermeulen, P.B.; Soetekouw, P.M.M.B.; van Engeland, M.; de Bruine, A.P. VHL and HIF signalling in renal cell carcinogenesis. J. Pathol. 2010, 221, 125–138. [Google Scholar] [CrossRef]
- Haase, V.H. The VHL tumor suppressor: Master regulator of HIF. Curr. Pharm. Des. 2009, 15, 3895–3903. [Google Scholar] [CrossRef] [Green Version]
- Linehan, W.M.; Schmidt, L.S.; Crooks, D.R.; Wei, D.; Srinivasan, R.; Lang, M.; Ricketts, C.J. The Metabolic Basis of Kidney Cancer. Cancer Discov. 2019. [Google Scholar] [CrossRef] [Green Version]
- Morais, M.; Dias, F.; Teixeira, A.L.; Medeiros, R. MicroRNAs and altered metabolism of clear cell renal cell carcinoma: Potential role as aerobic glycolysis biomarkers. Biochim. Biophys. Acta 2017, 1861, 2175–2185. [Google Scholar] [CrossRef]
- Dias, F.; Teixeira, A.L.; Santos, J.I.; Gomes, M.; Nogueira, A.; Assis, J.; Medeiros, R. Renal cell carcinoma development and miRNAs: A possible link to the EGFR pathway. Pharmacogenomics 2013, 14, 1793–1803. [Google Scholar] [CrossRef]
- Dabestani, S.; Thorstenson, A.; Lindblad, P.; Harmenberg, U.; Ljungberg, B.; Lundstam, S. Renal cell carcinoma recurrences and metastases in primary non-metastatic patients: A population-based study. World J. Urol. 2016, 34, 1081–1086. [Google Scholar] [CrossRef]
- Ravaud, A.; Gross-Goupil, M. Overcoming resistance to tyrosine kinase inhibitors in renal cell carcinoma. Cancer Treat. Rev. 2012, 38, 996–1003. [Google Scholar] [CrossRef]
- Auvray, M.; Auclin, E.; Barthelemy, P.; Bono, P.; Kellokumpu-Lehtinen, P.; Gross-Goupil, M.; de Velasco, G.; Powles, T.; Mouillet, G.; Vano, Y.-A.; et al. Second-line targeted therapies after nivolumab-ipilimumab failure in metastatic renal cell carcinoma. Eur. J. Cancer 2019, 108, 33–40. [Google Scholar] [CrossRef]
- Dabestani, S.; Beisland, C.; Stewart, G.D.; Bensalah, K.; Gudmundsson, E.; Lam, T.B.; Gietzmann, W.; Zakikhani, P.; Marconi, L.; Fernández-Pello, S.; et al. Long-term Outcomes of Follow-up for Initially Localised Clear Cell Renal Cell Carcinoma: RECUR Database Analysis. Eur. Urol. Focus 2019, 5, 857–866. [Google Scholar] [CrossRef] [Green Version]
- Minciacchi, V.R.; Freeman, M.R.; Di Vizio, D. Extracellular vesicles in cancer: Exosomes, microvesicles and the emerging role of large oncosomes. Semin. Cell Dev. Biol. 2015, 40, 41–51. [Google Scholar] [CrossRef] [Green Version]
- Nogues, L.; Benito-Martin, A.; Hergueta-Redondo, M.; Peinado, H. The influence of tumour-derived extracellular vesicles on local and distal metastatic dissemination. Mol. Asp. Med. 2018, 60, 15–26. [Google Scholar] [CrossRef]
- Zhao, H.; Achreja, A.; Iessi, E.; Logozzi, M.; Mizzoni, D.; Di Raimo, R.; Nagrath, D.; Fais, S. The key role of extracellular vesicles in the metastatic process. Biochim. Biophys. Acta Rev. Cancer 2018, 1869, 64–77. [Google Scholar] [CrossRef]
- Jurj, A.; Zanoaga, O.; Braicu, C.; Lazar, V.; Tomuleasa, C.; Irimie, A.; Berindan-Neagoe, I. A Comprehensive Picture of Extracellular Vesicles and Their Contents. Molecular Transfer to Cancer Cells. Cancers 2020, 12, 298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kucharzewska, P.; Belting, M. Emerging roles of extracellular vesicles in the adaptive response of tumour cells to microenvironmental stress. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef]
- Gambari, R.; Fabbri, E.; Borgatti, M.; Lampronti, I.; Finotti, A.; Brognara, E.; Bianchi, N.; Manicardi, A.; Marchelli, R.; Corradini, R. Targeting microRNAs involved in human diseases: A novel approach for modification of gene expression and drug development. Biochem. Pharm. 2011, 82, 1416–1429. [Google Scholar] [CrossRef]
- Mohr, A.M.; Mott, J.L. Overview of microRNA biology. Semin. Liver Dis. 2015, 35, 3–11. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef] [Green Version]
- Lai, X.; Wolkenhauer, O.; Vera, J. Understanding microRNA-mediated gene regulatory networks through mathematical modelling. Nucleic Acids Res. 2016, 44, 6019–6035. [Google Scholar] [CrossRef] [Green Version]
- Grange, C.; Tapparo, M.; Collino, F.; Vitillo, L.; Damasco, C.; Deregibus, M.C.; Tetta, C.; Bussolati, B.; Camussi, G. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 2011, 71, 5346–5356. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, A.L.; Dias, F.; Gomes, M.; Fernandes, M.; Medeiros, R. Circulating biomarkers in renal cell carcinoma: The link between microRNAs and extracellular vesicles, where are we now? J. Kidney Cancer VHL 2014, 1, 84. [Google Scholar] [CrossRef]
- Grange, C.; Brossa, A.; Bussolati, B. Extracellular Vesicles and Carried miRNAs in the Progression of Renal Cell Carcinoma. Int. J. Mol. Sci. 2019, 20, 1832. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Yang, G.; Zhao, D.; Wang, J.; Bai, Y.; Peng, Q.; Wang, H.; Fang, R.; Chen, G.; Wang, Z.; et al. CD103-positive CSC exosome promotes EMT of clear cell renal cell carcinoma: Role of remote MiR-19b-3p. Mol. Cancer 2019, 18, 86. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Long, M.; Yu, G.; Cheng, Y.; Yang, Q.; Liu, J.; Wang, Y.; Sheng, J.; Wang, L.; Wang, Z.; et al. Urinary exosome miR-30c-5p as a biomarker of clear cell renal cell carcinoma that inhibits progression by targeting HSPA5. J. Cell Mol. Med. 2019, 23, 6755–6765. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Ni, M.; Su, Y.; Wang, H.; Zhu, S.; Zhao, A.; Li, G. MicroRNAs in Serum Exosomes as Potential Biomarkers in Clear-cell Renal Cell Carcinoma. Eur. Urol. Focus 2018, 4, 412–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Wang, T.; Chen, C.; Wu, Z.; Bai, P.; Li, S.; Chen, B.; Liu, R.; Zhang, K.; Li, W.; et al. Serum exosomal miR-210 as a potential biomarker for clear cell renal cell carcinoma. J. Cell Biochem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Fujii, N.; Hirata, H.; Ueno, K.; Mori, J.; Oka, S.; Shimizu, K.; Kawai, Y.; Inoue, R.; Yamamoto, Y.; Matsumoto, H.; et al. Extracellular miR-224 as a prognostic marker for clear cell renal cell carcinoma. Oncotarget 2017, 8, 109877–109888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; He, J.; Min, L.; He, Y.; Guan, H.; Wang, J.; Peng, X. Extracellular vesicles transmitted miR-31-5p promotes sorafenib resistance by targeting MLH1 in renal cell carcinoma. Int. J. Cancer 2019. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, W.; Chen, S.; Wang, Y.; Xia, S. Distinct non-invasive evaluation values of tumor-derived cell-free microRNAs, circulating microvesicles and exosomal microRNAs after renal carcinoma cryoablation. Biochem. Biophys. Res. Commun. 2018, 503, 1278–1283. [Google Scholar] [CrossRef]
- Butz, H.; Nofech-Mozes, R.; Ding, Q.; Khella, H.W.Z.; Szabó, P.M.; Jewett, M.; Finelli, A.; Lee, J.; Ordon, M.; Stewart, R.; et al. Exosomal MicroRNAs Are Diagnostic Biomarkers and Can Mediate Cell–Cell Communication in Renal Cell Carcinoma. Eur. Urol. Focus 2016, 2, 210–218. [Google Scholar] [CrossRef]
- Maia, J.; Batista, S.; Couto, N.; Gregorio, A.; Bodo, C.; Elzanowska, J.; Moraes, M.; Costa-Silva, B. Population Analysis of Extracellular Vesicles in Microvolumes of Biofluids. bioRxiv 2020. [Google Scholar] [CrossRef]
- Chou, C.-H.; Shrestha, S.; Yang, C.-D.; Chang, N.-W.; Lin, Y.-L.; Liao, K.-W.; Huang, W.-C.; Sun, T.-H.; Tu, S.-J.; Lee, W.-H.; et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018, 46, D296–D302. [Google Scholar] [CrossRef]
- Brücher, B.L.D.M.; Jamall, I.S. Cell-cell communication in the tumor microenvironment, carcinogenesis, and anticancer treatment. Cell Physiol. Biochem. 2014, 34, 213–243. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Li, Y.; Pan, Y.; Lan, X.; Song, F.; Sun, J.; Zhou, K.; Liu, X.; Ren, X.; Wang, F.; et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat. Commun. 2018, 9, 5395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Luo, G.; Zhang, K.; Cao, J.; Huang, C.; Jiang, T.; Liu, B.; Su, L.; Qiu, Z. Hypoxic Tumor-Derived Exosomal miR-301a Mediates M2 Macrophage Polarization via PTEN/PI3Kγ to Promote Pancreatic Cancer Metastasis. Cancer Res. 2018, 78, 4586–4598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawakubo-Yasukochi, T.; Morioka, M.; Hazekawa, M.; Yasukochi, A.; Nishinakagawa, T.; Ono, K.; Kawano, S.; Nakamura, S.; Nakashima, M. miR-200c-3p spreads invasive capacity in human oral squamous cell carcinoma microenvironment. Mol. Carcinog. 2018, 57, 295–302. [Google Scholar] [CrossRef]
- Mormile, R. Prevention and management of allograft rejection in heart transplantation: Are miR-126-5p and miR-126-5p-expressing mesenchymal stem cell exosomes the Ariadne’s thread? Cardiovasc. Pathol. 2020, 47, 107208. [Google Scholar] [CrossRef]
- Baek, D.; Villén, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef] [Green Version]
- Maehama, T.; Taylor, G.S.; Dixon, J.E. PTEN and myotubularin: Novel phosphoinositide phosphatases. Annu. Rev. Biochem. 2001, 70, 247–279. [Google Scholar] [CrossRef]
- Geva, G.A.; Gielchinsky, I.; Aviv, N.; Max, K.E.A.; Gofrit, O.N.; Gur-Wahnon, D.; Ben-Dov, I.Z. Urine cell-free microRNA as biomarkers for transitional cell carcinoma. BMC Res. Notes 2017, 10, 641. [Google Scholar] [CrossRef] [Green Version]
- Fan, C.; Zhao, C.; Wang, F.; Li, S.; Wang, J. Significance of PTEN Mutation in Cellular Process, Prognosis, and Drug Selection in Clear Cell Renal Cell Carcinoma. Front. Oncol. 2019, 9, 357. [Google Scholar] [CrossRef]
- Guo, S.-P.; Zhai, Y.-Q.; Wang, W.-L.; Ma, F.-C. Expression and significance of a new tumor suppression gene PTEN in primary renal cell carcinoma. Ai Zheng 2002, 21, 582–587. [Google Scholar]
- Zhang, L.; Zhang, S.; Yao, J.; Lowery, F.J.; Zhang, Q.; Huang, W.-C.; Li, P.; Li, M.; Wang, X.; Zhang, C.; et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 2015, 527, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Akbani, R.; Ng, P.K.S.; Werner, H.M.J.; Shahmoradgoli, M.; Zhang, F.; Ju, Z.; Liu, W.; Yang, J.-Y.; Yoshihara, K.; Li, J.; et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat. Commun. 2014, 5, 3887. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; German, P.; Bai, S.; Barnes, S.; Guo, W.; Qi, X.; Lou, H.; Liang, J.; Jonasch, E.; Mills, G.B.; et al. The PI3K/AKT Pathway and Renal Cell Carcinoma. J. Genet. Genom. 2015, 42, 343–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Lu, Y.; Akbani, R.; Ju, Z.; Roebuck, P.L.; Liu, W.; Yang, J.-Y.; Broom, B.M.; Verhaak, R.G.W.; Kane, D.W.; et al. TCPA: A resource for cancer functional proteomics data. Nat. Methods 2013, 10, 1046–1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Gao, W.; Zhang, C.; Wen, S.; Huangfu, H.; Kang, J.; Wang, B. Hsa-miR-301a-3p Acts as an Oncogene in Laryngeal Squamous Cell Carcinoma via Target Regulation of Smad4. J. Cancer 2015, 6, 1260–1275. [Google Scholar] [CrossRef] [Green Version]
- Takagawa, Y.; Gen, Y.; Muramatsu, T.; Tanimoto, K.; Inoue, J.; Harada, H.; Inazawa, J. miR-1293, a Candidate for miRNA-Based Cancer Therapeutics, Simultaneously Targets BRD4 and the DNA Repair Pathway. Mol. Ther. 2020, in press. [Google Scholar] [CrossRef]
- Kawata, N.; Yamaguchi, K.; Igarashi, T.; Takahashi, S. TIMP-1 as well as Microvessel Invasion and High Nuclear Grade Is a Significant Determinant Factor for Extension of Tumor Diameter in Localized RCC. J. Oncol. 2016, 2016, 5035127. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Ma, Y.; Wang, Y.; Chen, T.; Wang, H.; Chu, H.; Zhao, G.; Zhang, G. Identification of miR-1293 potential target gene: TIMP-1. Mol. Cell Biochem. 2013, 384, 1–6. [Google Scholar] [CrossRef]
- Srigley, J.R.; Delahunt, B.; Eble, J.N.; Egevad, L.; Epstein, J.I.; Grignon, D.; Hes, O.; Moch, H.; Montironi, R.; Tickoo, S.K.; et al. The International Society of Urological Pathology (ISUP) Vancouver Classification of Renal Neoplasia. Am. J. Surg. Pathol. 2013, 37, 1469–1489. [Google Scholar] [CrossRef]
- Zeringer, E.; Li, M.; Barta, T.; Schageman, J.; Pedersen, K.W.; Neurauter, A.; Magdaleno, S.; Setterquist, R.; Vlassov, A.V. Methods for the extraction and RNA profiling of exosomes. World J. Methodol. 2013, 3, 11–18. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, J.H.; Apeltsin, L.; Newman, A.M.; Baumbach, J.; Wittkop, T.; Su, G.; Bader, G.D.; Ferrin, T.E. clusterMaker: A multi-algorithm clustering plugin for Cytoscape. BMC Bioinform. 2011, 12, 436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enright, A.J.; Van Dongen, S.; Ouzounis, C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002, 30, 1575–1584. [Google Scholar] [CrossRef]



| Clinical-Pathological Characteristics | Group A (N = 32) | Group B (N = 37) | |||
|---|---|---|---|---|---|
| N | % | N | % | ||
| Gender | |||||
| Male | 24 | 75.0 | 26 | 70.3 | |
| Female | 8 | 25.0 | 11 | 29.7 | |
| Age | |||||
| Average ± SD | 61.9 ± 12.4 | 62.4 ± 9.9 | |||
| Type of surgery | |||||
| Partial nephrectomy | 18 | 56.3 | 0 | 0.0 | |
| Radical nephrectomy | 14 | 43.8 | 34 | 91.9 | |
| No surgery | 0 | 0 | 3 | 8.1 | |
| Tumor size | |||||
| <7 cm | 24 | 75.0 | 9 | 24.32 | |
| ≥7 cm | 8 | 25.0 | 22 | 59.46 | |
| Undetermined | 0 | 0 | 6 | 16.22 | |
| T | |||||
| T1 | 17 | 53.1 | 10 | 27.03 | |
| T2 | 1 | 3.1 | 6 | 16.22 | |
| T3 | 13 | 40.6 | 16 | 43.24 | |
| T4 | 0 | 0 | 3 | 8.12 | |
| Tx | 1 | 3.1 | 2 | 5.40 | |
| N | |||||
| N0 | 0 | 0 | 15 | 40.54 | |
| N1–N2 | 0 | 0 | 5 | 13.51 | |
| Nx | 0 | 0 | 17 | 45.95 | |
| M | |||||
| M0 | 27 | 100 | 22 | 59.46 | |
| M1 | 0 | 0 | 15 | 40.54 | |
| Mx | 5 | 0 | 0 | 0.0 | |
| Clinical Stage | |||||
| I | 17 | 53.1 | 10 | 27.03 | |
| II | 1 | 3.1 | 6 | 16.22 | |
| III | 13 | 40.6 | 16 | 43.24 | |
| IV | 0 | 0 | 3 | 8.12 | |
| Not available | 1 | 3.1 | 3 | 5.40 | |
| ISUP classification | |||||
| 1 | 2 | 6.3 | 1 | 2.70 | |
| 2 | 18 | 56.3 | 14 | 37.84 | |
| 3 | 11 | 34.3 | 8 | 21.62 | |
| 4 | 1 | 3.1 | 9 | 24.32 | |
| Not available | 0 | 0 | 5 | 13.51 | |
| Smoking status | |||||
| Smoker | 6 | 18.8 | 3 | 8.11 | |
| Ex-smoker | 11 | 34.4 | 7 | 18.92 | |
| Non-smoker | 14 | 43.8 | 27 | 72,97 | |
| Not available | 1 | 3.1 | 0 | 0.0 | |
| Hypertension | |||||
| Yes | 20 | 62.5 | 15 | 40.54 | |
| No | 11 | 34.4 | 22 | 59.45 | |
| Not available | 1 | 3.1 | 0 | 0.0 | |
| Diabetes mellitus | |||||
| DM I | 0 | 0.0 | 0 | 0.0 | |
| DM II | 9 | 28.1 | 13 | 35.14 | |
| No | 22 | 68.8 | 23 | 62.16 | |
| Not available | 1 | 3.1 | 1 | 2.70 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dias, F.; Teixeira, A.L.; Nogueira, I.; Morais, M.; Maia, J.; Bodo, C.; Ferreira, M.; Silva, A.; Vilhena, M.; Lobo, J.; et al. Extracellular Vesicles Enriched in hsa-miR-301a-3p and hsa-miR-1293 Dynamics in Clear Cell Renal Cell Carcinoma Patients: Potential Biomarkers of Metastatic Disease. Cancers 2020, 12, 1450. https://doi.org/10.3390/cancers12061450
Dias F, Teixeira AL, Nogueira I, Morais M, Maia J, Bodo C, Ferreira M, Silva A, Vilhena M, Lobo J, et al. Extracellular Vesicles Enriched in hsa-miR-301a-3p and hsa-miR-1293 Dynamics in Clear Cell Renal Cell Carcinoma Patients: Potential Biomarkers of Metastatic Disease. Cancers. 2020; 12(6):1450. https://doi.org/10.3390/cancers12061450
Chicago/Turabian StyleDias, Francisca, Ana Luísa Teixeira, Inês Nogueira, Mariana Morais, Joana Maia, Cristian Bodo, Marta Ferreira, Alexandra Silva, Manuela Vilhena, João Lobo, and et al. 2020. "Extracellular Vesicles Enriched in hsa-miR-301a-3p and hsa-miR-1293 Dynamics in Clear Cell Renal Cell Carcinoma Patients: Potential Biomarkers of Metastatic Disease" Cancers 12, no. 6: 1450. https://doi.org/10.3390/cancers12061450
APA StyleDias, F., Teixeira, A. L., Nogueira, I., Morais, M., Maia, J., Bodo, C., Ferreira, M., Silva, A., Vilhena, M., Lobo, J., Sequeira, J. P., Maurício, J., Oliveira, J., Kok, K., Costa-Silva, B., & Medeiros, R. (2020). Extracellular Vesicles Enriched in hsa-miR-301a-3p and hsa-miR-1293 Dynamics in Clear Cell Renal Cell Carcinoma Patients: Potential Biomarkers of Metastatic Disease. Cancers, 12(6), 1450. https://doi.org/10.3390/cancers12061450











