Triarylpyridine Compounds and Chloroquine Act in Concert to Trigger Lysosomal Membrane Permeabilization and Cell Death in Cancer Cells
Abstract
:1. Introduction
2. Results
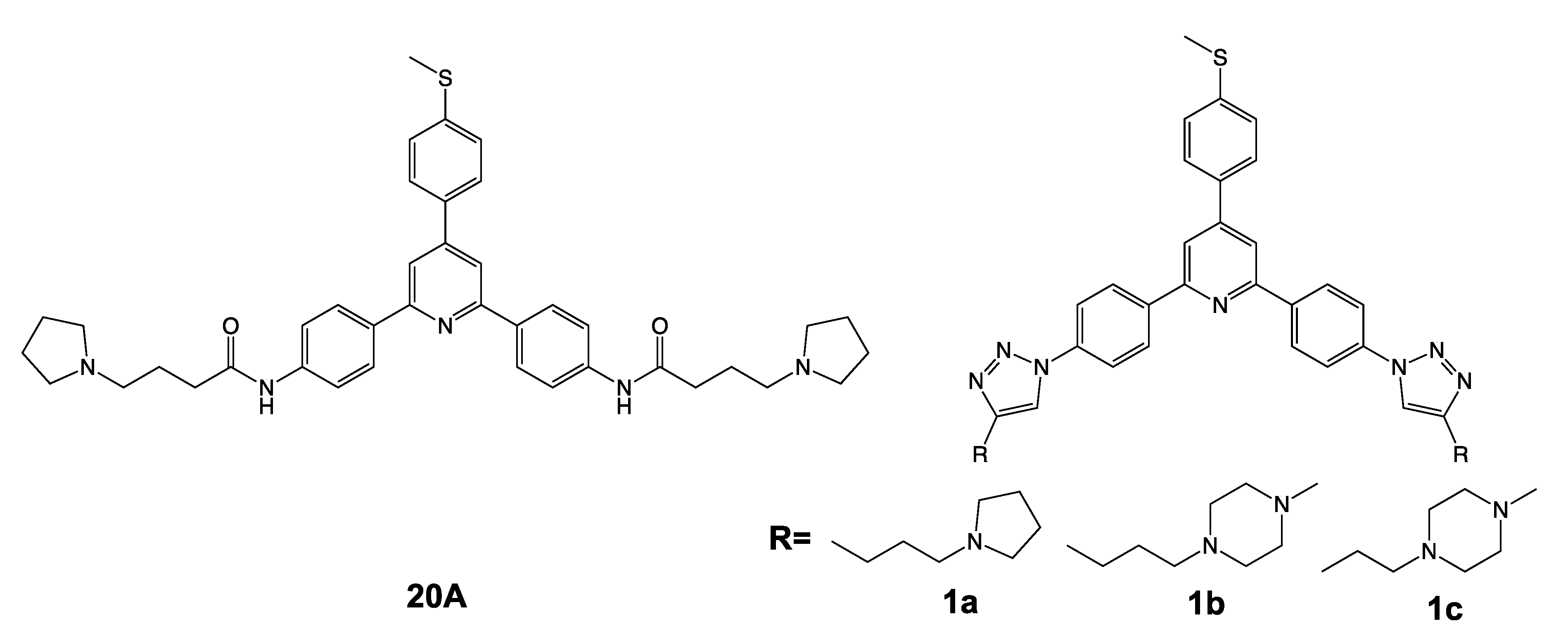
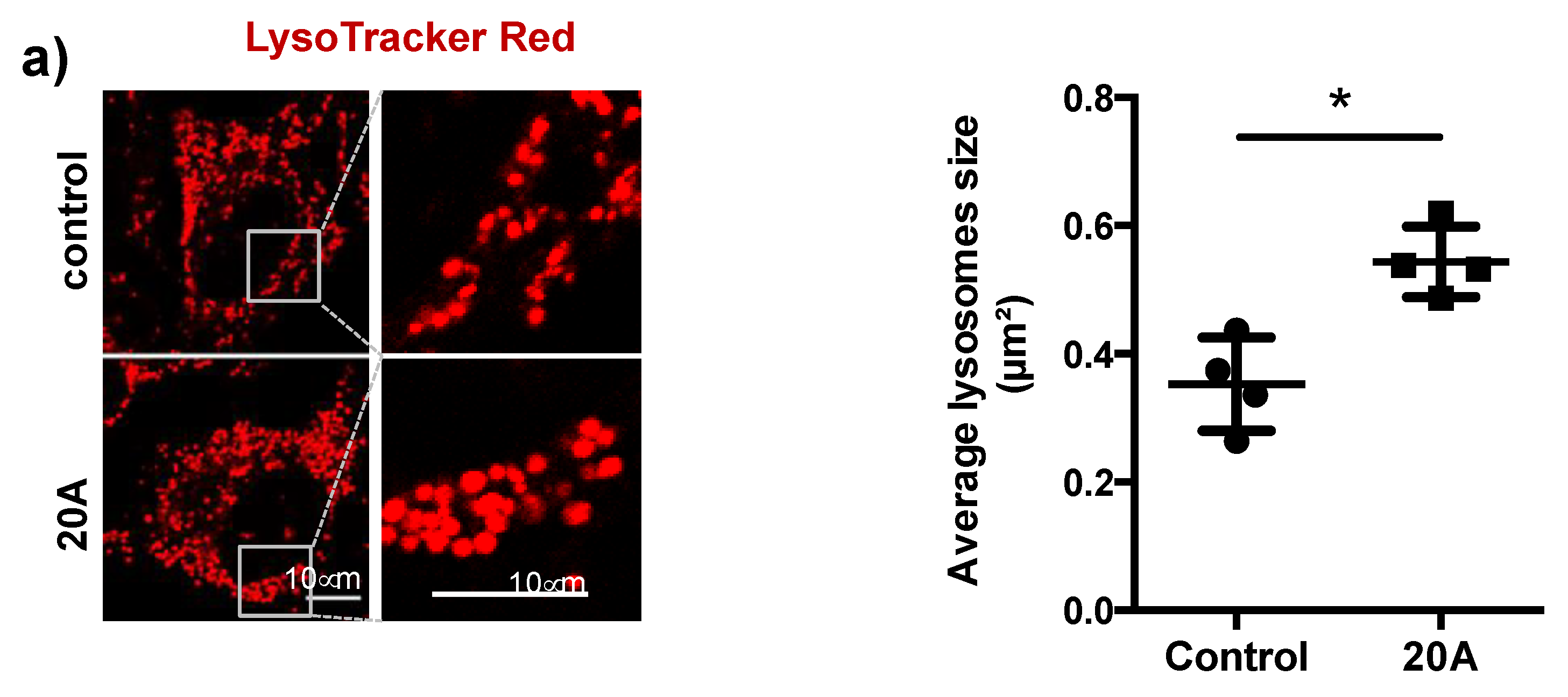
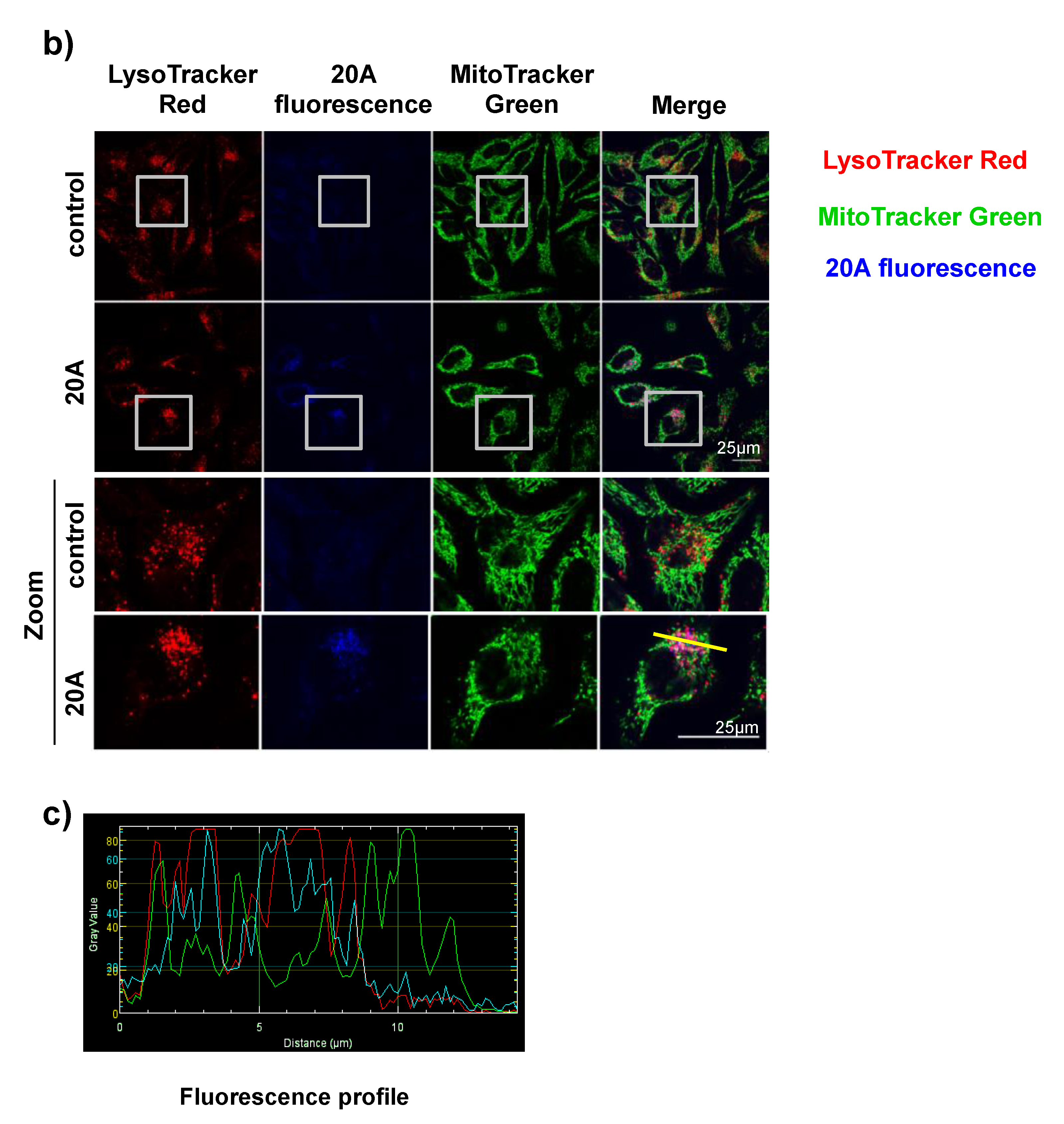
2.1. 20A Accumulates Within the Lysosomal Compartment and Causes Enlargement of the Lysosomes
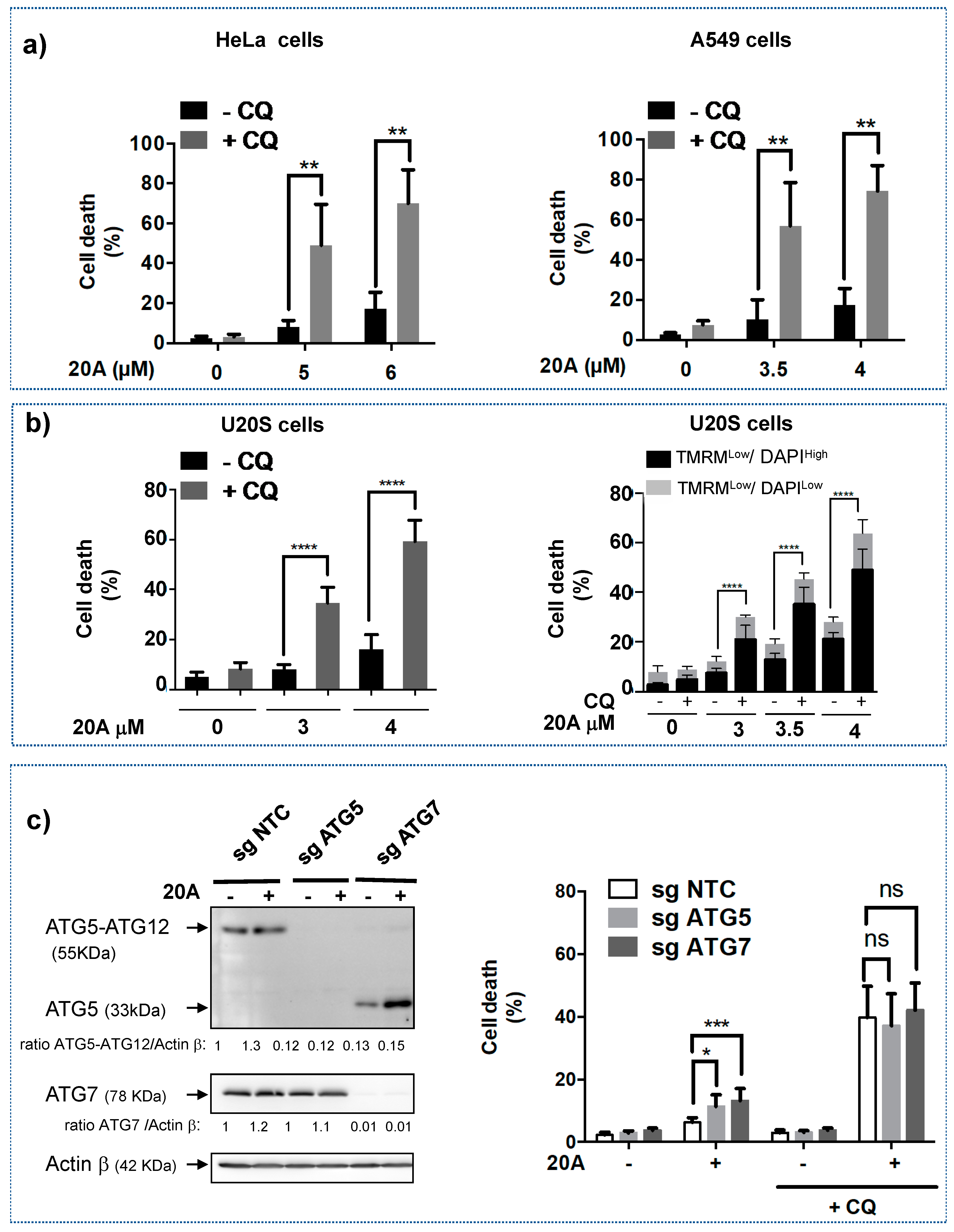
2.2. 20A and Chloroquine Act in Concert to Trigger Cell Death
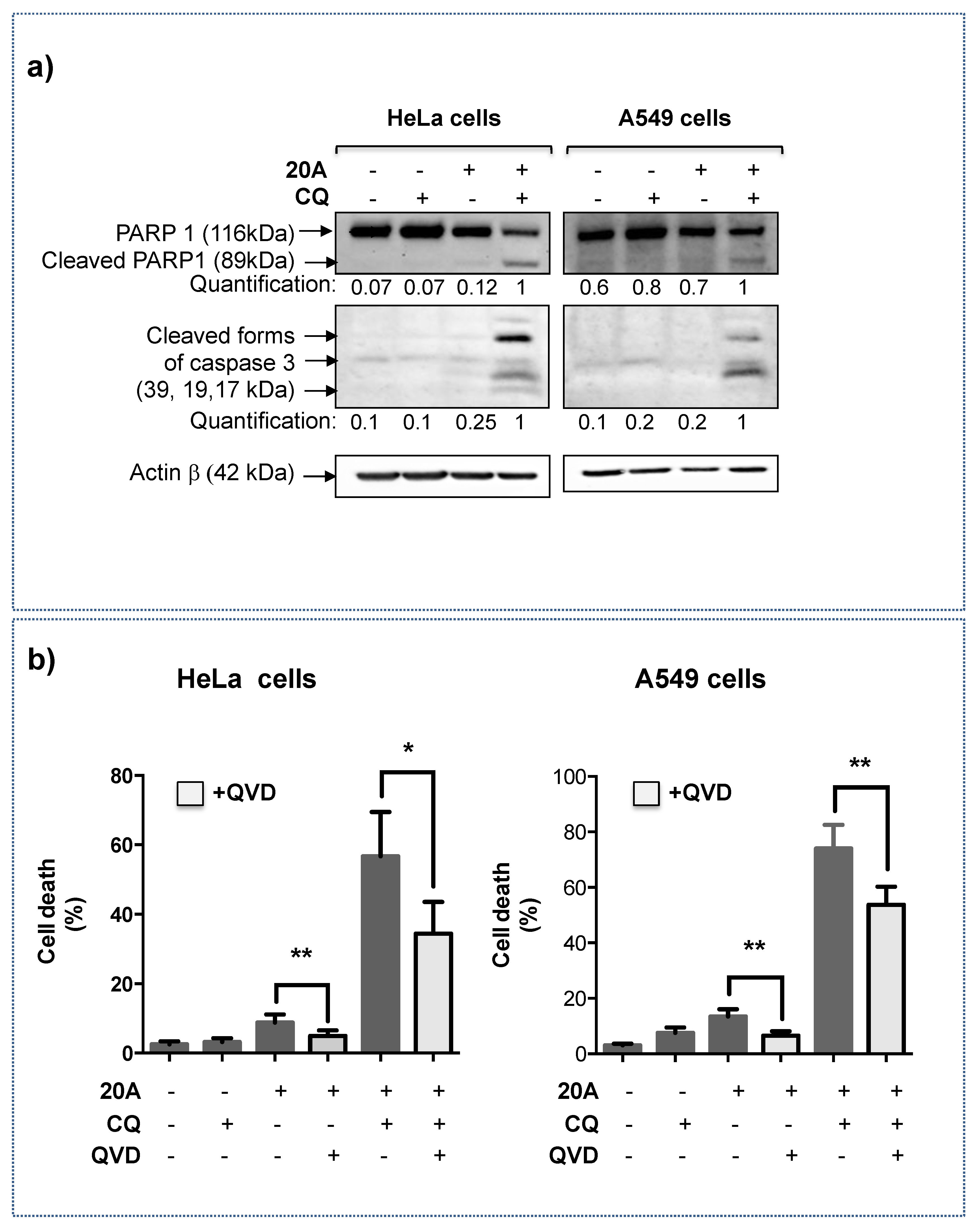
2.3. Apoptosis Is Involved in Cell Death Induced by 20A Plus Chloroquine
2.4. Chloroquine Significantly Activates LMP Triggered by 20A Derivatives
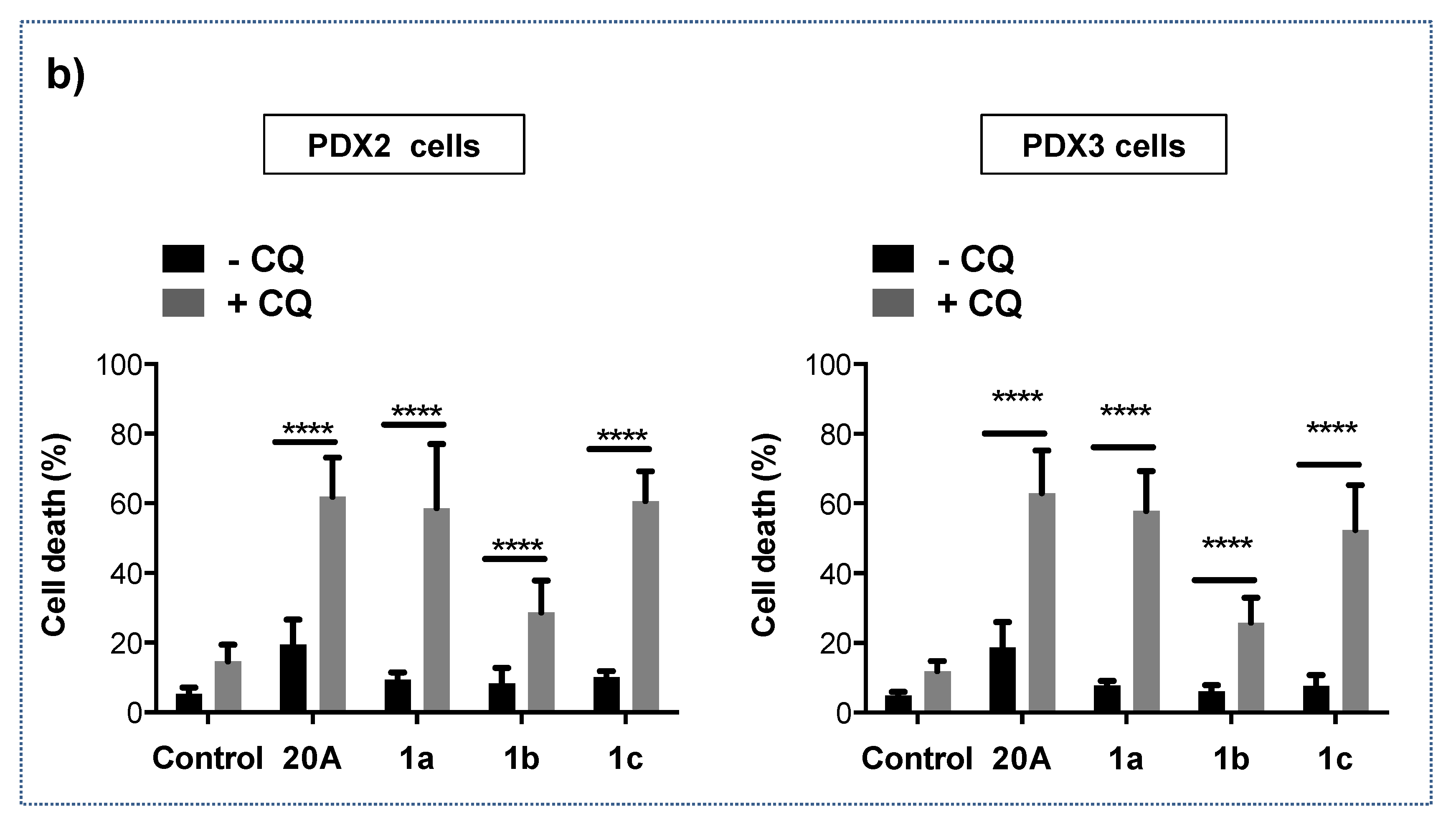
2.5. Combined Treatment with Chloroquine and 20A Derivatives Significantly Activates Cell Death in Lung Adenocarcinoma Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Reagents and Antibodies
4.3. Synthesis of Bis-Triazole Triarylpyridines
4.3.1. Chemicals and Instrumentation for Chemical Synthesis
4.3.2. Synthesis of 2,6-bis(4-aminophenyl)-4-[4-(methylthio)phenyl]pyridine (2) (Figure S2A)
4.3.3. Synthesis of 2,6-bis(4-azidophenyl)-4-[4-(methylthio)phenyl]pyridine (3) (Figure S2A)
4.3.4. General Procedure For The Synthesis of 2,6-bis{4-[4-(substituted-aminoalkyl)-1H-1,2,3-triazol-1-yl]phenyl}-4-[4-(methylthio)phenyl]pyridines 1a–c
4.4. Quadruplex Stabilization
4.5. Mitochondrial and Lysosomal Staining and Evaluation of 20A Localization
4.6. Lysosomal Membrane Permeabilization (LMP) Analysis
4.6.1. Quantification of Galectin 3 Puncta
4.6.2. Co-Localization of Galectin 3 with LAMP1
4.7. Evaluation of Cell Death
4.8. Immunoblot Assay
4.9. Cell viability Assay
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Xu, H.; Ren, D. Lysosomal physiology. Annu. Rev. Physiol. 2015, 77, 57–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Appelqvist, H.; Wäster, P.; Kågedal, K.; Öllinger, K. The lysosome: From waste bag to potential therapeutic target. J. Mol. Cell Biol. 2013, 5, 214–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kroemer, G.; Jäättelä, M. Lysosomes and autophagy in cell death control. Nat. Rev. Cancer 2005, 5, 886–897. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 21, 2861–2873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boya, P. Lysosomal function and dysfunction: Mechanism and disease. Antioxid. Redox Signal. 2012, 17, 766–774. [Google Scholar] [CrossRef] [Green Version]
- Kallunki, T.; Olsen, O.D.; Jäättelä, M. Cancer-associated lysosomal changes: Friends or foes? Oncogene 2013, 32, 1995–2004. [Google Scholar] [CrossRef] [Green Version]
- Gong, Y.; Duvvuri, M.; Duncan, M.B.; Liu, J.; Krise, J.P. Niemann-Pick C1 protein facilitates the efflux of the anticancer drug daunorubicin from cells according to a novel vesicle-mediated pathway. J. Pharmacol. Exp. Ther. 2006, 316, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Herlevsen, M.; Oxford, G.; Owens, C.R.; Conaway, M.; Theodorescu, D. Depletion of major vault protein increases doxorubicin sensitivity and nuclear accumulation and disrupts its sequestration in lysosomes. Mol. Cancer Ther. 2007, 6, 1804–1813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, P.J.; Sykes, H.R.; Fox, M.E.; Furlong, I.J. Subcellular distribution of the anticancer drug mitoxantrone in human and drug-resistant murine cells analyzed by flow cytometry and confocal microscopy and its relationship to the induction of DNA damage. Cancer Res. 1992, 52, 4000–4008. [Google Scholar]
- Zhitomirsky, B.; Assaraf, Y.G. Lysosomal sequestration of hydrophobic weak base chemotherapeutics triggers lysosomal biogenesis and lysosome-dependent cancer multidrug resistance. Oncotarget 2015, 6, 1143–1156. [Google Scholar] [CrossRef] [Green Version]
- Goldman, S.D.B.; Funk, R.S.; Rajewski, R.A.; Krise, J.P. Mechanisms of amine accumulation in, and egress from, lysosomes. Bioanalysis 2009, 1, 1445–1459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrao, P.; Sincock, P.; Cole, S.; Ashman, L. Intracellular P-gp contributes to functional drug efflux and resistance in acute myeloid leukaemia. Leuk. Res. 2001, 25, 395–405. [Google Scholar] [CrossRef]
- Repnik, U.; Hafner Česen, M.; Turk, B. Lysosomal membrane permeabilization in cell death: Concepts and challenges. Mitochondrion 2014, 19 Pt A, 49–57. [Google Scholar] [CrossRef]
- Fehrenbacher, N.; Gyrd-Hansen, M.; Poulsen, B.; Felbor, U.; Kallunki, T.; Boes, M.; Weber, E.; Leist, M.; Jäättelä, M. Sensitization to the lysosomal cell death pathway upon immortalization and transformation. Cancer Res. 2004, 64, 5301–5310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, N.H.T.; Olsen, O.D.; Groth-Pedersen, L.; Ellegaard, A.-M.; Bilgin, M.; Redmer, S.; Ostenfeld, M.S.; Ulanet, D.; Dovmark, T.H.; Lønborg, A.; et al. Transformation-associated changes in sphingolipid metabolism sensitize cells to lysosomal cell death induced by inhibitors of acid sphingomyelinase. Cancer Cell 2013, 24, 379–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boya, P.; Andreau, K.; Poncet, D.; Zamzami, N.; Perfettini, J.-L.; Metivier, D.; Ojcius, D.M.; Jäättelä, M.; Kroemer, G. Lysosomal membrane permeabilization induces cell death in a mitochondrion-dependent fashion. J. Exp. Med. 2003, 197, 1323–1334. [Google Scholar] [CrossRef]
- Aits, S.; Jäättelä, M. Lysosomal cell death at a glance. J. Cell Sci. 2013, 126, 1905–1912. [Google Scholar] [CrossRef] [Green Version]
- Piao, S.; Amaravadi, R.K. Targeting the lysosome in cancer. Ann. N. Y. Acad. Sci. 2016, 1371, 45. [Google Scholar] [CrossRef] [Green Version]
- Kwok, C.K.; Merrick, C.J. G-Quadruplexes: Prediction, characterization, and biological application. Trends Biotechnol. 2017, 35, 997–1013. [Google Scholar] [CrossRef]
- Dixon, I.M.; Lopez, F.; Tejera, A.M.; Estève, J.-P.; Balasco, M.A.; Patviel, G.; Meunier, B. A G-Quadruplex ligand with 10000-fold selectivity over duplex DNA. J. Am. Chem. Soc. 2007, 129, 1502–1503. [Google Scholar] [CrossRef]
- Nicoludis, J.M.; Barrett, S.P.; Mergny, J.L.; Yatsunyk, L.A. Interaction of human telomeric DNA with N-methyl mesoporphyrin IX. Nucleic Acids Res. 2012, 40, 5432–5447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Urso, A.; Randazzo, R.; Rizzo, V.; Gangemi, C.; Romanucci, V.; Zarrelli, A.; Tomaselli, G.; Milardi, D.; Borbone, N.; Purrello, R.; et al. Stabilization vs. destabilization of G-quadruplex superstructures: The role of the porphyrin derivative having spermine arms. Phys. Chem. Chem. Phys. 2017, 19, 17404–17410. [Google Scholar] [CrossRef] [PubMed]
- Cimino-Reale, G.; Zaffaroni, N.; Folini, M. Emerging role of G-quadruplex DNA as target in anticancer therapy. Curr. Pharm. Des. 2016, 22, 6612–6624. [Google Scholar] [CrossRef] [PubMed]
- Beauvarlet, J.; Bensadoun, P.; Darbo, E.; Labrunie, G.; Rousseau, B.; Richard, E.; Draskovic, I.; Londono-Vallejo, A.; Dupuy, J.-W.; Das, R.N.; et al. Modulation of the ATM/autophagy pathway by a G-quadruplex ligand tips the balance between senescence and apoptosis in cancer cells. Nucleic Acids Res. 2019, 47, 2739–2756. [Google Scholar] [CrossRef]
- Smith, N.M.; Labrunie, G.; Corry, B.; Tran, P.L.T.; Norret, M.; Djavaheri-Mergny, M.; Raston, C.L.; Mergny, J.-L. Unraveling the relationship between structure and stabilization of triarylpyridines as G-quadruplex binding ligands. Org. Biomol. Chem. 2011, 9, 6154–6162. [Google Scholar] [CrossRef]
- Kazmi, F.; Hensley, T.; Pope, C.; Funk, R.S.; Loewen, G.J.; Buckley, D.B.; Parkinson, A. Lysosomal Sequestration (trapping) of lipophilic amine (cationic amphiphilic) drugs in immortalized human hepatocytes (Fa2N-4 Cells). Drug Metab. Dispos. 2013, 41, 897–905. [Google Scholar] [CrossRef] [Green Version]
- Boya, P.; Gonzalez-Polo, R.-A.; Poncet, D.; Andreau, K.; Vieira, H.L.A.; Roumier, T.; Perfettini, J.-L.; Kroemer, G. Mitochondrial membrane permeabilization is a critical step of lysosome-initiated apoptosis induced by hydroxychloroquine. Oncogene 2003, 22, 3927–3936. [Google Scholar] [CrossRef] [Green Version]
- Aits, S.; Kricker, J.; Liu, B.; Ellegaard, A.-M.; Hämälistö, S.; Tvingsholm, S.; Corcelle-Termeau, E.; Høgh, S.; Farkas, T.; Jonassen, A.H.; et al. Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy 2015, 11, 1408–1424. [Google Scholar] [CrossRef] [Green Version]
- Ambrogio, C.; Gómez-López, G.; Falcone, M.; Vidal, A.; Nadal, E.; Crosetto, N.; Blasco, R.B.; Fernández-Marcos, P.J.; Sánchez-Céspedes, M.; Ren, X.; et al. Combined inhibition of DDR1 and Notch signaling is a therapeutic strategy for KRAS-driven lung adenocarcinoma. Nat. Med. 2016, 22, 270–277. [Google Scholar] [CrossRef]
- Cao, Q.; Zhong, X.Z.; Zou, Y.; Murrell-Lagnado, R.; Zhu, M.X.; Dong, X.-P. Calcium release through P2X4 activates calmodulin to promote endolysosomal membrane fusion. J. Cell Biol. 2015, 209, 879–894. [Google Scholar] [CrossRef] [Green Version]
- Cao, Q.; Yang, Y.; Zhong, X.Z.; Dong, X.-P. The lysosomal Ca2+ release channel TRPML1 regulates lysosome size by activating calmodulin. J. Biol. Chem. 2017, 292, 8424–8435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martina, J.A.; Chen, Y.; Gucek, M.; Puertollano, R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012, 8, 903–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Settembre, C.; Di Malta, C.; Polito, V.A.; Garcia Arencibia, M.; Vetrini, F.; Erdin, S.; Erdin, S.U.; Huynh, T.; Medina, D.; Colella, P.; et al. TFEB links autophagy to lysosomal biogenesis. Science 2011, 332, 1429–1433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beauvarlet, J.; Mergny, J.-L.; Djavaheri-Mergny, M. Activation of the Ataxia Telangiectasia Mutated/Autophagy pathway by a G-quadruplex ligand links senescence with apoptosis. Mol. Cell Oncol. 2019, 6. [Google Scholar] [CrossRef] [PubMed]
- Boya, P.; Reggiori, F.; Codogno, P. Emerging regulation and functions of autophagy. Nat. Cell Biol. 2013, 15, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Boya, P.; Kroemer, G. Lysosomal membrane permeabilization in cell death. Oncogene 2008, 27, 6434–6451. [Google Scholar] [CrossRef] [Green Version]
- Kang, C.-C.; Huang, W.-C.; Kouh, C.-W.; Wang, Z.-F.; Cho, C.-C.; Chang, C.-C.; Wang, C.-L.; Chang, T.-C.; Seemann, J.; Huang, L.J. Chemical principles for the design of a novel fluorescent probe with high cancer-targeting selectivity and sensitivity. Integr. Biol. 2013, 5, 1217–1228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shivalingam, A.; Izquierdo, M.A.; Le Marois, A.; Vyšniauskas, A.; Suhling, K.; Kuimova, M.K.; Vilar, R. The interactions between a small molecule and G-quadruplexes are visualized by fluorescence lifetime imaging microscopy. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Drygin, D.; Siddiqui-Jain, A.; O’Brien, S.; Schwaebe, M.; Lin, A.; Bliesath, J.; Ho, C.B.; Proffitt, C.; Trent, K.; Whitten, J.P.; et al. Anticancer activity of CX-3543: A direct inhibitor of rRNA biogenesis. Cancer Res. 2009, 69, 7653–7661. [Google Scholar] [CrossRef] [Green Version]
- Faudale, M.; Cogoi, S.; Xodo, L.E. Photoactivated cationic alkyl-substituted porphyrin binding to G4-RNA in the 5’-UTR of KRAS oncogene represses translation. Chem. Commun. 2012, 48, 874–876. [Google Scholar] [CrossRef]
- Montespan, C.; Marvin, S.A.; Austin, S.; Burrage, A.M.; Roger, B.; Rayne, F.; Faure, M.; Campell, E.M.; Schneider, C.; Reimer, R.; et al. Multi-layered control of Galectin-8 mediated autophagy during adenovirus cell entry through a conserved PPxY motif in the viral capsid. PLoS Pathog. 2017, 13, e1006217. [Google Scholar] [CrossRef]
- Smith, N.M.; Raston, C.; Smith, C.B.; Sobolev, A.N. PEG mediated synthesis of amino-functionalised 2,4,6-triarylpyridines. Green Chem. 2007, 9, 1185–1190. [Google Scholar] [CrossRef]
- Zahmatkesh, S. Ionic liquid catalyzed synthesis and characterization of heterocyclic and optically active poly (amide-imide)s incorporating L-amino acids. Amino Acids 2011, 40, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Weiss, M. Acetic acid—Ammonium acetate reactions. An improved chichibabin pyridine synthesis. J. Am. Chem. Soc. 1952, 74, 200–202. [Google Scholar] [CrossRef]
- De Rache, A.; Mergny, J.-L. Assessment of selectivity of G-quadruplex ligands via an optimised FRET melting assay. Biochimie 2015, 115, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Trapp, S.; Rosania, G.R.; Horobin, R.W.; Kornhuber, J. Quantitative modeling of selective lysosomal targeting for drug design. Eur. Biophys. J. 2008, 37, 1317–1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beauvarlet, J.; Nath Das, R.; Alvarez-Valadez, K.; Martins, I.; Muller, A.; Darbo, E.; Richard, E.; Soubeyran, P.; Kroemer, G.; Guillon, J.; et al. Triarylpyridine Compounds and Chloroquine Act in Concert to Trigger Lysosomal Membrane Permeabilization and Cell Death in Cancer Cells. Cancers 2020, 12, 1621. https://doi.org/10.3390/cancers12061621
Beauvarlet J, Nath Das R, Alvarez-Valadez K, Martins I, Muller A, Darbo E, Richard E, Soubeyran P, Kroemer G, Guillon J, et al. Triarylpyridine Compounds and Chloroquine Act in Concert to Trigger Lysosomal Membrane Permeabilization and Cell Death in Cancer Cells. Cancers. 2020; 12(6):1621. https://doi.org/10.3390/cancers12061621
Chicago/Turabian StyleBeauvarlet, Jennifer, Rabindra Nath Das, Karla Alvarez-Valadez, Isabelle Martins, Alexandra Muller, Elodie Darbo, Elodie Richard, Pierre Soubeyran, Guido Kroemer, Jean Guillon, and et al. 2020. "Triarylpyridine Compounds and Chloroquine Act in Concert to Trigger Lysosomal Membrane Permeabilization and Cell Death in Cancer Cells" Cancers 12, no. 6: 1621. https://doi.org/10.3390/cancers12061621
APA StyleBeauvarlet, J., Nath Das, R., Alvarez-Valadez, K., Martins, I., Muller, A., Darbo, E., Richard, E., Soubeyran, P., Kroemer, G., Guillon, J., Mergny, J.-L., & Djavaheri-Mergny, M. (2020). Triarylpyridine Compounds and Chloroquine Act in Concert to Trigger Lysosomal Membrane Permeabilization and Cell Death in Cancer Cells. Cancers, 12(6), 1621. https://doi.org/10.3390/cancers12061621







