Cancer-Associated Substitutions in RNA Recognition Motifs of PUF60 and U2AF65 Reveal Residues Required for Correct Folding and 3′ Splice-Site Selection
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Preparations
2.2. Cell Cultures and Transfections
2.3. Detection of Spliced Products
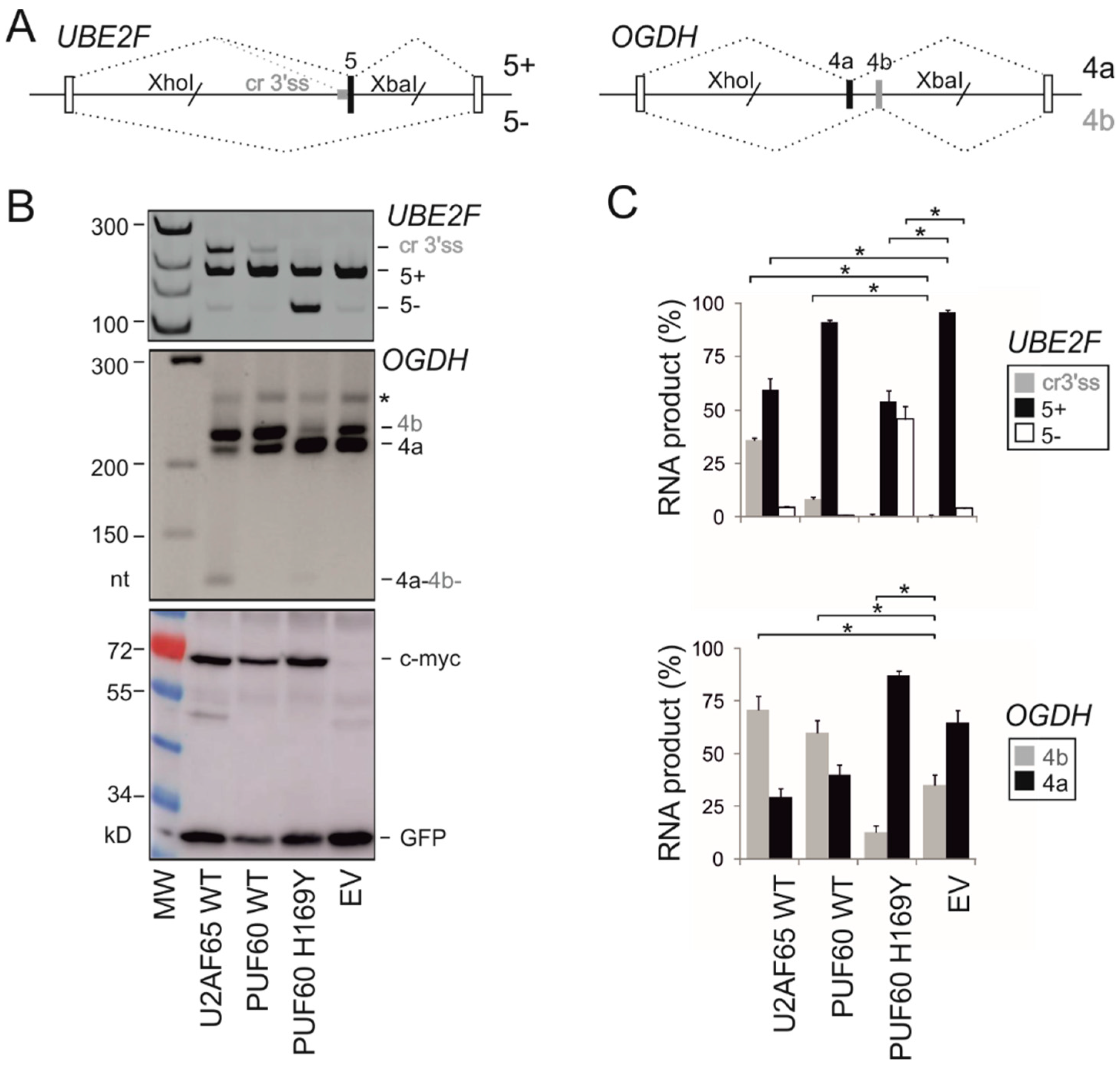
2.4. Utilization of Cryptic 3′ss of UBE2F Exon 5 in Human Endogenous Transcripts
2.5. Immunoblotting
2.6. Protein Expression and Purification
2.7. Electrophoretic Mobility Shift Assay (EMSA)
2.8. Differential Scanning Fluorimetry (DSF)
2.9. Solubility and Stability Predictions
3. Results
3.1. Selection of Functional Assays for Cancer-associated Substitutions in PUF60 and U2AF65 RRMs
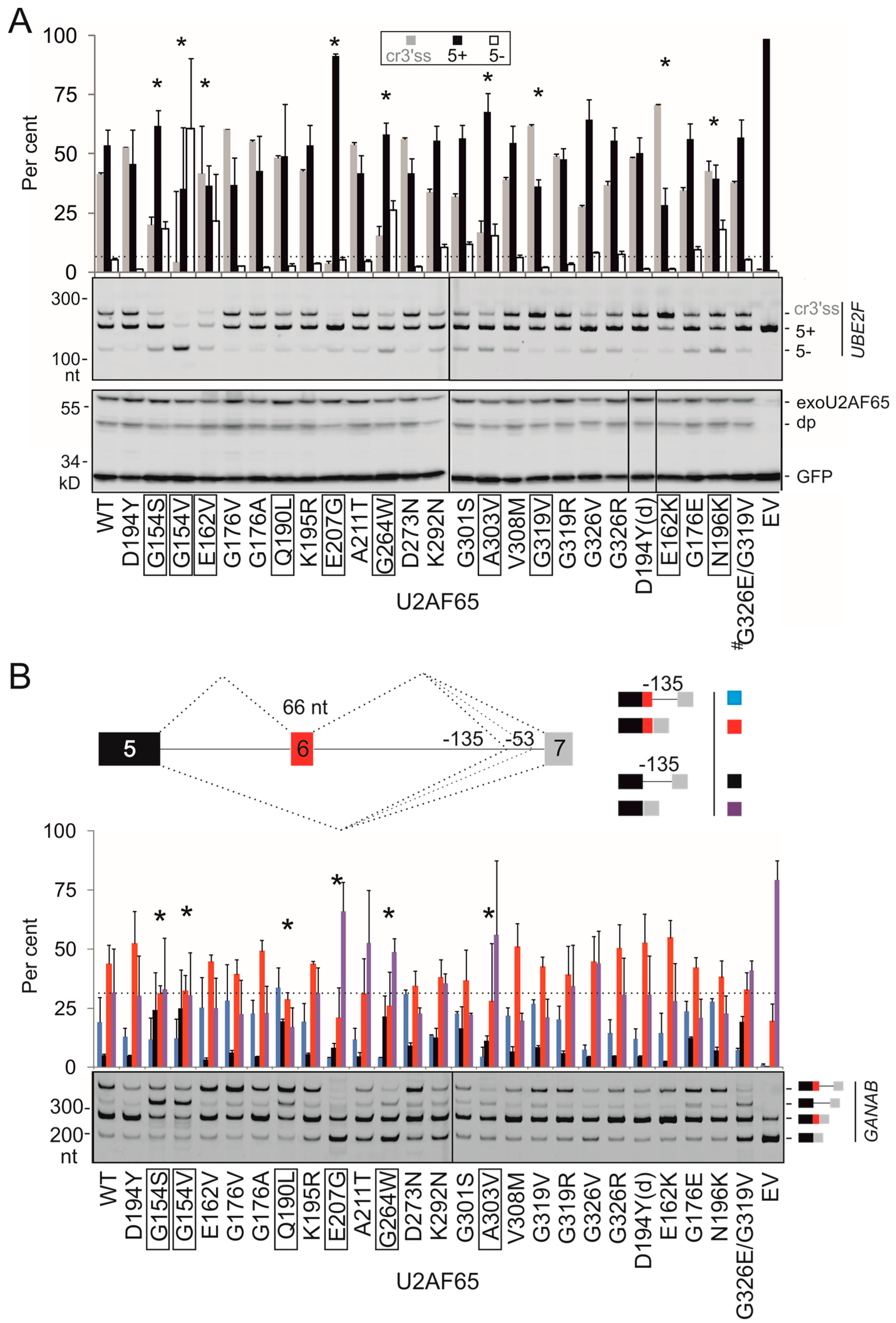
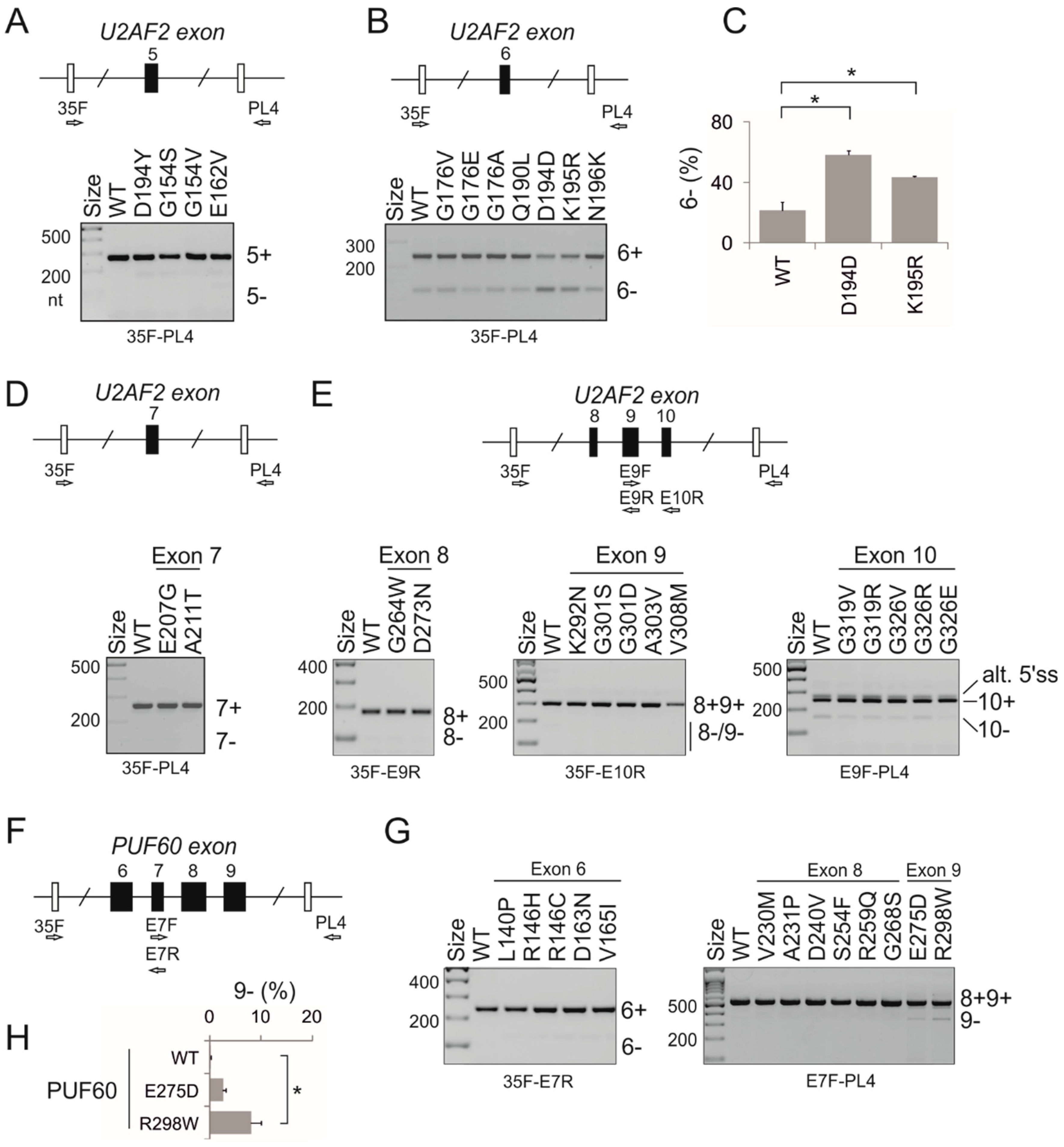
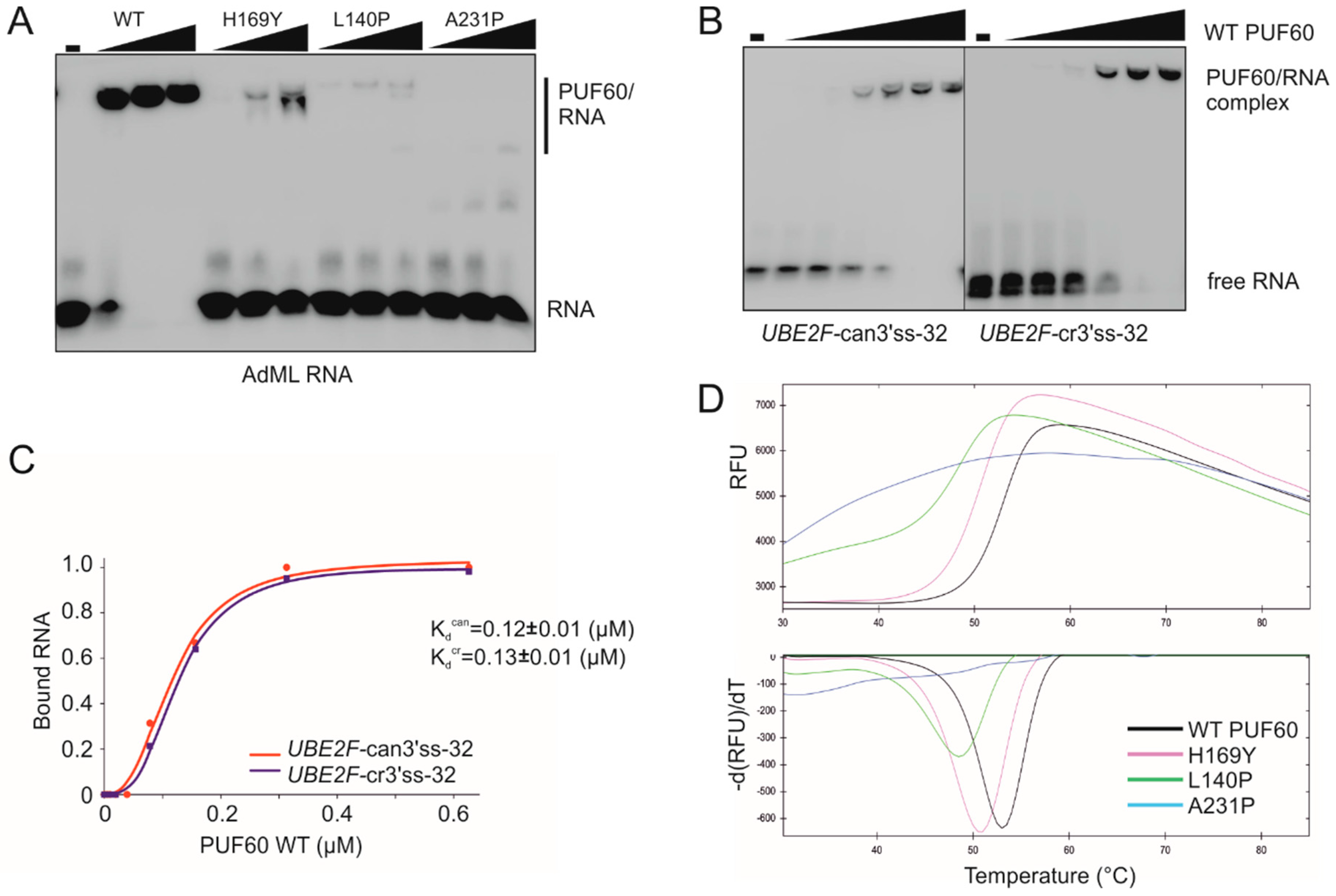
3.2. PUF60 and U2AF65 RRM Substitutions that Alter 3′ss Usage
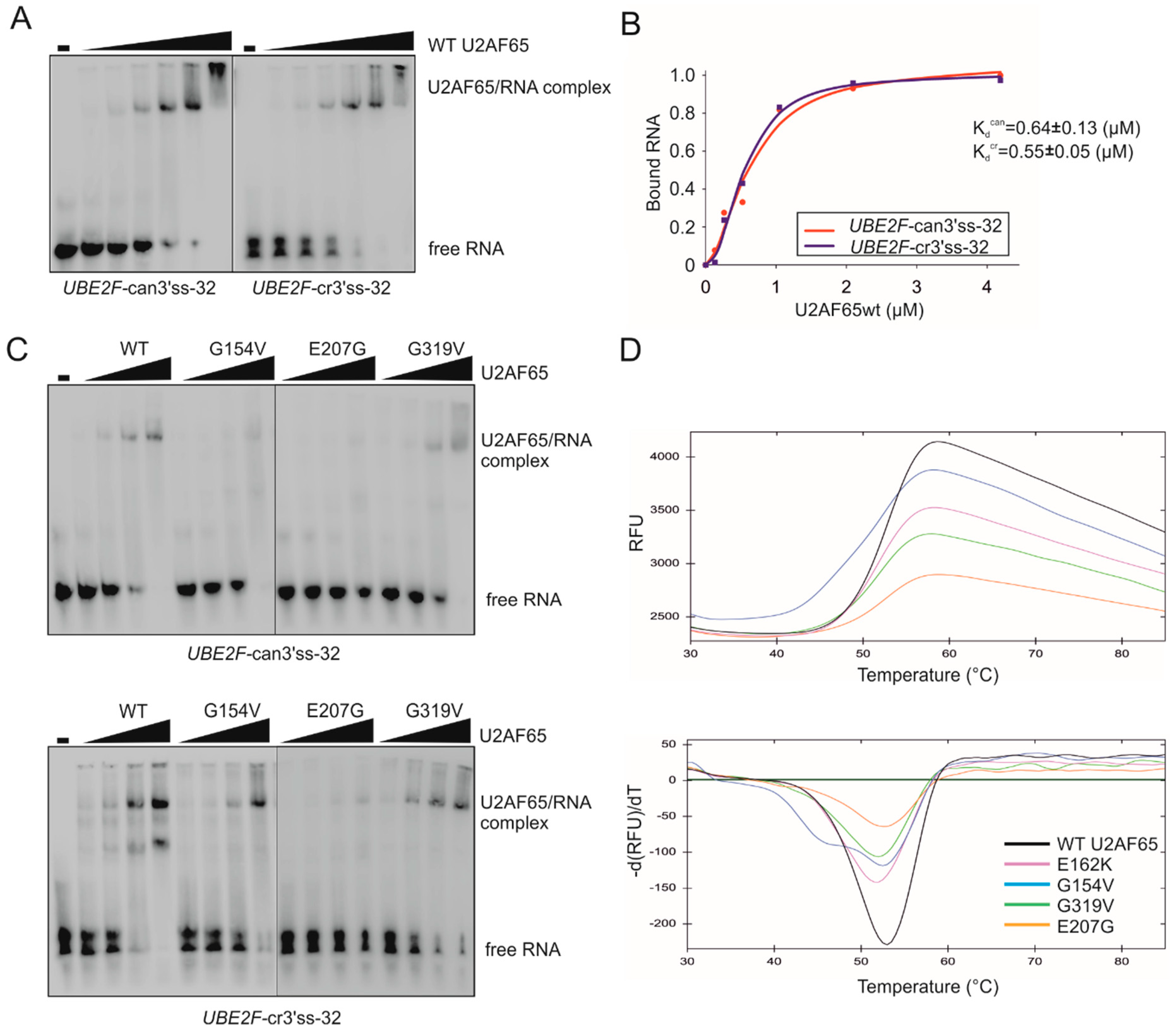
3.3. Functional Consequences of Cancer-Associated RRM Substitutions in U2AF65 and PUF60
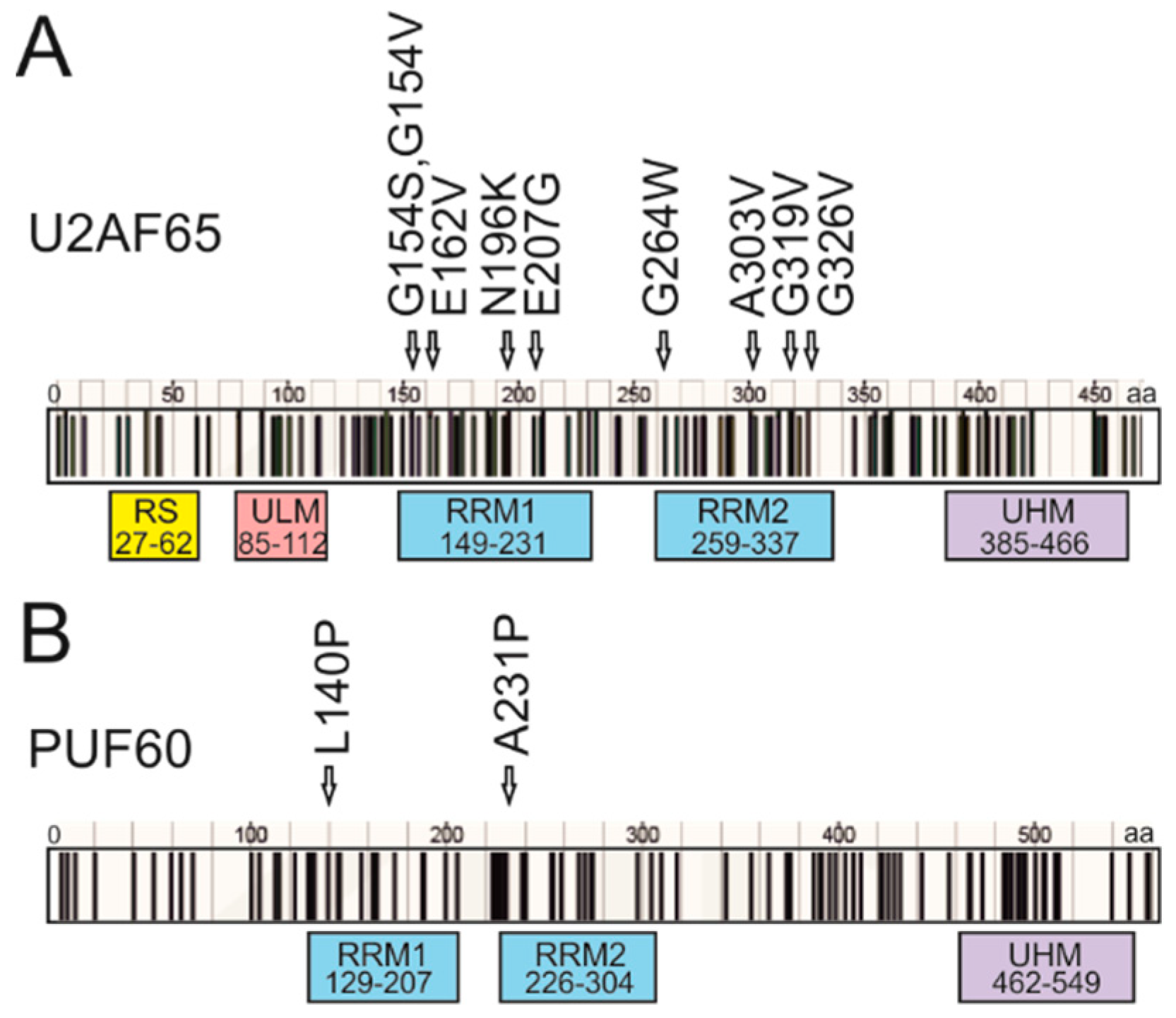
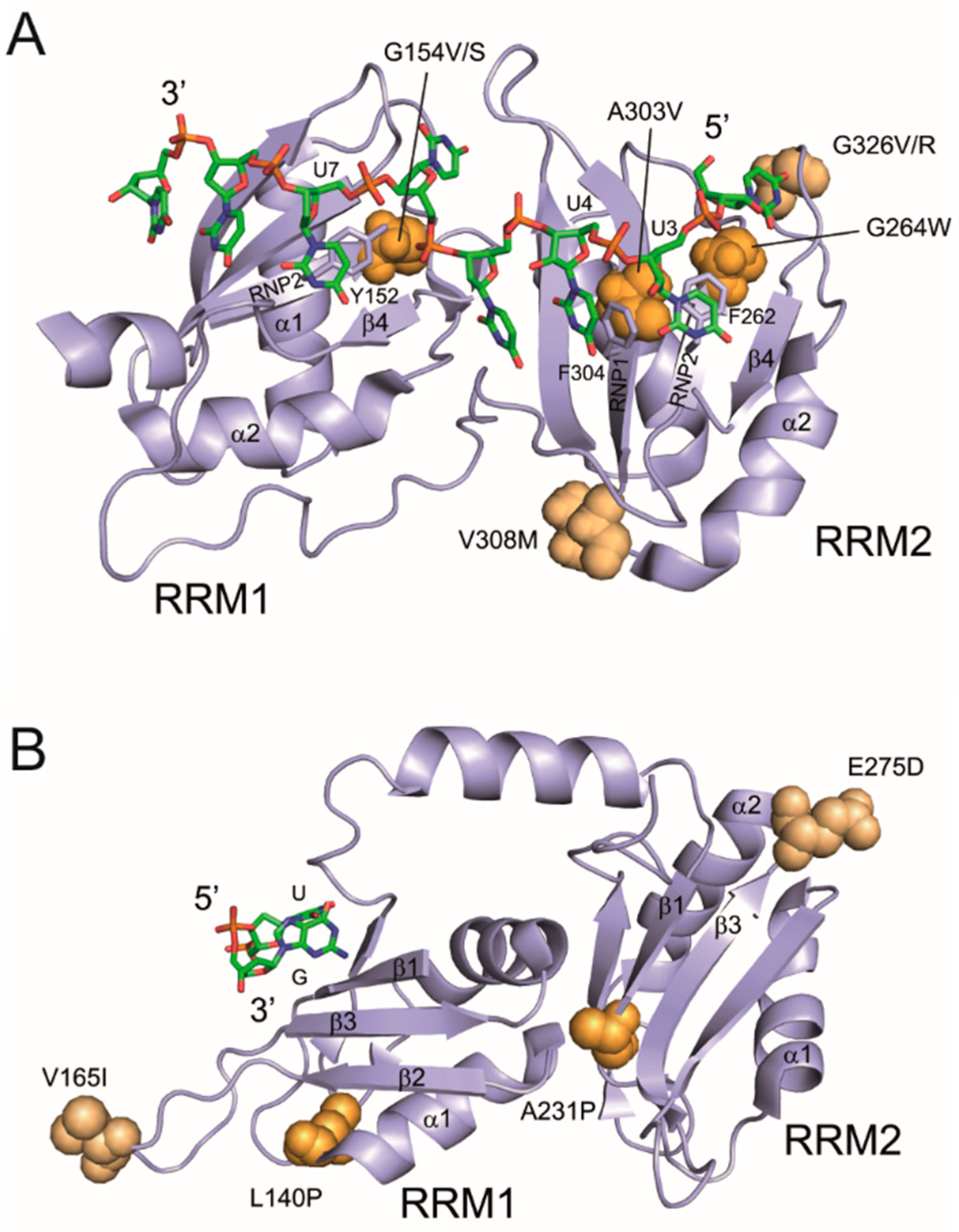
3.4. Mapping of Cancer-Associated RRM Substitutions on to High-Resolution PUF60 and U2AF65 Structures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Yoshida, K.; Sanada, M.; Shiraishi, Y.; Nowak, D.; Nagata, Y.; Yamamoto, R.; Sato, Y.; Sato-Otsubo, A.; Kon, A.; Nagasaki, M.; et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011, 478, 64–69. [Google Scholar] [CrossRef]
- Okeyo-Owuor, T.; White, B.S.; Chatrikhi, R.; Mohan, D.R.; Kim, S.; Griffith, M.; Ding, L.; Ketkar-Kulkarni, S.; Hundal, J.; Laird, K.M.; et al. U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing. Leukemia 2015, 29, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Inoue, D.; Bradley, R.K.; Abdel-Wahab, O. Spliceosomal gene mutations in myelodysplasia: Molecular links to clonal abnormalities of hematopoiesis. Genes Dev. 2016, 30, 989–1001. [Google Scholar] [CrossRef]
- Yoshida, H.; Park, S.Y.; Oda, T.; Akiyoshi, T.; Sato, M.; Shirouzu, M.; Tsuda, K.; Kuwasako, K.; Unzai, S.; Muto, Y.; et al. A novel 3′ splice site recognition by the two zinc fingers in the U2AF small subunit. Genes Dev. 2015, 29, 1649–1660. [Google Scholar] [CrossRef]
- Cretu, C.; Schmitzova, J.; Ponce-Salvatierra, A.; Dybkov, O.; De Laurentiis, E.I.; Sharma, K.; Will, C.L.; Urlaub, H.; Luhrmann, R.; Pena, V. Molecular Architecture of SF3b and Structural Consequences of Its Cancer-Related Mutations. Mol. Cell 2016, 64, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Alsafadi, S.; Houy, A.; Battistella, A.; Popova, T.; Wassef, M.; Henry, E.; Tirode, F.; Constantinou, A.; Piperno-Neumann, S.; Roman-Roman, S.; et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 2016, 7, 10615. [Google Scholar] [CrossRef] [PubMed]
- Carrocci, T.J.; Zoerner, D.M.; Paulson, J.C.; Hoskins, A.A. SF3b1 mutations associated with myelodysplastic syndromes alter the fidelity of branchsite selection in yeast. Nucleic Acids Res. 2017, 45, 4837–4852. [Google Scholar] [CrossRef] [PubMed]
- Darman, R.B.; Seiler, M.; Agrawal, A.A.; Lim, K.H.; Peng, S.; Aird, D.; Bailey, S.L.; Bhavsar, E.B.; Chan, B.; Colla, S.; et al. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3′ Splice Site Selection through Use of a Different Branch Point. Cell Rep. 2015, 13, 1033–1045. [Google Scholar] [CrossRef] [PubMed]
- Forbes, S.A.; Beare, D.; Boutselakis, H.; Bamford, S.; Bindal, N.; Tate, J.; Cole, C.G.; Ward, S.; Dawson, E.; Ponting, L.; et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017, 45, D777–D783. [Google Scholar] [CrossRef]
- Page-McCaw, P.S.; Amonlirdviman, K.; Sharp, P.A. PUF60: A novel U2AF65-related splicing activity. RNA 1999, 5, 1548–1560. [Google Scholar] [CrossRef]
- Hastings, M.L.; Allemand, E.; Duelli, D.M.; Myers, M.P.; Krainer, A.R. Control of pre-mRNA splicing by the general splicing factors PUF60 and U2AF. PLoS ONE 2007, 2, e538. [Google Scholar] [CrossRef] [PubMed]
- Corsini, L.; Hothorn, M.; Stier, G.; Rybin, V.; Scheffzek, K.; Gibson, T.J.; Sattler, M. Dimerization and protein binding specificity of the U2AF homology motif of the splicing factor PUF60. J. Biol. Chem. 2009, 284, 630–639. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Yang, B.; Wu, T.; Huang, J.; Tang, P.; Zhou, Y.; Zhou, J.; Qiu, J.; Jiang, L.; Li, H.; et al. Mechanisms for U2AF to define 3′ splice sites and regulate alternative splicing in the human genome. Nat. Struct. Mol. Biol. 2014, 21, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Kralovicova, J.; Sevcikova, I.; Stejskalova, E.; Obuca, M.; Hiller, M.; Stanek, D.; Vorechovsky, I. PUF60-activated exons uncover altered 3′ splice-site selection by germline missense mutations in a single RRM. Nucleic Acids Res. 2018, 46, 6166–6187. [Google Scholar] [CrossRef]
- Glasser, E.; Agrawal, A.A.; Jenkins, J.L.; Kielkopf, C.L. Cancer-Associated Mutations Mapped on High-Resolution Structures of the U2AF2 RNA Recognition Motifs. Biochemistry 2017, 56, 4757–4761. [Google Scholar] [CrossRef]
- Afroz, T.; Cienikova, Z.; Clery, A.; Allain, F.H. One, Two, Three, Four! How Multiple RRMs Read the Genome Sequence. Methods Enzymol. 2015, 558, 235–278. [Google Scholar] [CrossRef]
- Zamore, P.D.; Green, M.R. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc. Natl. Acad. Sci. USA 1989, 86, 9243–9247. [Google Scholar] [CrossRef]
- Zamore, P.D.; Patton, J.G.; Green, M.R. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 1992, 355, 609–614. [Google Scholar] [CrossRef]
- Dauber, A.; Golzio, C.; Guenot, C.; Jodelka, F.M.; Kibaek, M.; Kjaergaard, S.; Leheup, B.; Martinet, D.; Nowaczyk, M.J.; Rosenfeld, J.A.; et al. SCRIB and PUF60 are primary drivers of the multisystemic phenotypes of the 8q24.3 copy-number variant. Am. J. Hum. Genet. 2013, 93, 798–811. [Google Scholar] [CrossRef]
- El Chehadeh, S.; Kerstjens-Frederikse, W.S.; Thevenon, J.; Kuentz, P.; Bruel, A.L.; Thauvin-Robinet, C.; Bensignor, C.; Dollfus, H.; Laugel, V.; Riviere, J.B.; et al. Dominant variants in the splicing factor PUF60 cause a recognizable syndrome with intellectual disability, heart defects and short stature. Eur. J. Hum. Genet. 2016, 25, 43–51. [Google Scholar] [CrossRef]
- Matsushita, K.; Tomonaga, T.; Shimada, H.; Shioya, A.; Higashi, M.; Matsubara, H.; Harigaya, K.; Nomura, F.; Libutti, D.; Levens, D.; et al. An essential role of alternative splicing of c-myc suppressor FUSE-binding protein-interacting repressor in carcinogenesis. Cancer Res. 2006, 66, 1409–1417. [Google Scholar] [CrossRef] [PubMed]
- Malz, M.; Bovet, M.; Samarin, J.; Rabenhorst, U.; Sticht, C.; Bissinger, M.; Roessler, S.; Bermejo, J.L.; Renner, M.; Calvisi, D.F.; et al. Overexpression of far upstream element (FUSE) binding protein (FBP)-interacting repressor (FIR) supports growth of hepatocellular carcinoma. Hepatology 2014, 60, 1241–1250. [Google Scholar] [CrossRef] [PubMed]
- Kralovicova, J.; Vorechovsky, I. Allele-dependent recognition of the 3′ splice site of INS intron 1. Hum. Genet. 2010, 128, 383–400. [Google Scholar] [CrossRef] [PubMed]
- Kralovicova, J.; Knut, M.; Cross, N.C.; Vorechovsky, I. Identification of U2AF(35)-dependent exons by RNA-Seq reveals a link between 3′ splice-site organization and activity of U2AF-related proteins. Nucleic Acids Res. 2015, 43, 3747–3763. [Google Scholar] [CrossRef] [PubMed]
- Kralovicova, J.; Vorechovsky, I. Alternative splicing of U2AF1 reveals a shared repression mechanism for duplicated exons. Nucleic Acids Res. 2017, 45, 417–434. [Google Scholar] [CrossRef] [PubMed]
- Kralovicova, J.; Houngninou-Molango, S.; Kramer, A.; Vorechovsky, I. Branch site haplotypes that control alternative splicing. Hum. Mol. Genet. 2004, 13, 3189–3202. [Google Scholar] [CrossRef]
- Sormanni, P.; Aprile, F.A.; Vendruscolo, M. The CamSol method of rational design of protein mutants with enhanced solubility. J. Mol. Biol. 2015, 427, 478–490. [Google Scholar] [CrossRef]
- Pires, D.E.; Ascher, D.B.; Blundell, T.L. mCSM: Predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 2014, 30, 335–342. [Google Scholar] [CrossRef]
- Vorechovsky, I.; Luo, L.; Dyer, M.J.; Catovsky, D.; Amlot, P.L.; Yaxley, J.C.; Foroni, L.; Hammarstrom, L.; Webster, A.D.; Yuille, M.A. Clustering of missense mutations in the ataxia-telangiectasia gene in a sporadic T-cell leukaemia. Nat. Genet. 1997, 17, 96–99. [Google Scholar] [CrossRef]
- Yang, F.; Petsalaki, E.; Rolland, T.; Hill, D.E.; Vidal, M.; Roth, F.P. Protein domain-level landscape of cancer-type specific somatic mutations. PLoS Comput. Biol. 2015, 11, e1004147. [Google Scholar] [CrossRef]
- Cartegni, L.; Chew, S.L.; Krainer, A.R. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 2002, 3, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Pagani, F.; Baralle, F.E. Genomic variants in exons and introns: Identifying the splicing spoilers. Nat. Rev. Genet. 2004, 5, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Raponi, M.; Kralovicova, J.; Copson, E.; Divina, P.; Eccles, D.; Johnson, P.M.; Baralle, D.; Vorechovsky, I. Prediction of single-nucleotide substitutions that result in exon skipping: Identification of a splicing silencer in BRCA1 exon 5. Hum. Mutat. 2011, 32, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Niesen, F.H.; Berglund, H.; Vedadi, M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007, 2, 2212–2221. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.A.; Salsi, E.; Chatrikhi, R.; Henderson, S.; Jenkins, J.L.; Green, M.R.; Ermolenko, D.N.; Kielkopf, C.L. An extended U2AF(65)-RNA-binding domain recognizes the 3′ splice site signal. Nat. Commun. 2016, 7, 10950. [Google Scholar] [CrossRef]
- Zarnack, K.; Konig, J.; Tajnik, M.; Martincorena, I.; Eustermann, S.; Stevant, I.; Reyes, A.; Anders, S.; Luscombe, N.M.; Ule, J. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 2013, 152, 453–466. [Google Scholar] [CrossRef]
- Cukier, C.D.; Hollingworth, D.; Martin, S.R.; Kelly, G.; Diaz-Moreno, I.; Ramos, A. Molecular basis of FIR-mediated c-myc transcriptional control. Nat. Struct. Mol. Biol. 2010, 17, 1058–1064. [Google Scholar] [CrossRef]
- Wang, Z.L.; Li, B.; Luo, Y.X.; Lin, Q.; Liu, S.R.; Zhang, X.Q.; Zhou, H.; Yang, J.H.; Qu, L.H. Comprehensive Genomic Characterization of RNA-Binding Proteins across Human Cancers. Cell Rep. 2018, 22, 286–298. [Google Scholar] [CrossRef]
- Cheng, L.; Wang, P.; Yang, S.; Yang, Y.; Zhang, Q.; Zhang, W.; Xiao, H.; Gao, H.; Zhang, Q. Identification of genes with a correlation between copy number and expression in gastric cancer. BMC Med. Genomics 2012, 5, 14. [Google Scholar] [CrossRef]
- Ramakrishna, M.; Williams, L.H.; Boyle, S.E.; Bearfoot, J.L.; Sridhar, A.; Speed, T.P.; Gorringe, K.L.; Campbell, I.G. Identification of candidate growth promoting genes in ovarian cancer through integrated copy number and expression analysis. PLoS ONE 2010, 5, e9983. [Google Scholar] [CrossRef]
- Muller, B.; Bovet, M.; Yin, Y.; Stichel, D.; Malz, M.; Gonzalez-Vallinas, M.; Middleton, A.; Ehemann, V.; Schmitt, J.; Muley, T.; et al. Concomitant expression of far upstream element (FUSE) binding protein (FBP) interacting repressor (FIR) and its splice variants induce migration and invasion of non-small cell lung cancer (NSCLC) cells. J. Pathol. 2015, 237, 390–401. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.T.; Ayrault, O.; Hunt, H.W.; Taherbhoy, A.M.; Duda, D.M.; Scott, D.C.; Borg, L.A.; Neale, G.; Murray, P.J.; Roussel, M.F.; et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol. Cell 2009, 33, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Morgan, M.A.; Sun, Y. Targeting Neddylation pathways to inactivate cullin-RING ligases for anticancer therapy. Antioxid. Redox. Signal 2014, 21, 2383–2400. [Google Scholar] [CrossRef]
- Zhou, W.; Xu, J.; Tan, M.; Li, H.; Li, H.; Wei, W.; Sun, Y. UBE2M is a Stress-Inducible Dual E2 for Neddylation and Ubiquitylation that Promotes Targeted Degradation of UBE2F. Mol. Cell 2018, 70, 1008–1024. [Google Scholar] [CrossRef]
- Zhou, W.; Xu, J.; Li, H.; Xu, M.; Chen, Z.J.; Wei, W.; Pan, Z.; Sun, Y. Neddylation E2 UBE2F Promotes the Survival of Lung Cancer Cells by Activating CRL5 to Degrade NOXA via the K11 Linkage. Clin. Cancer Res. 2017, 23, 1104–1116. [Google Scholar] [CrossRef]
- Dowhan, D.H.; Hong, E.P.; Auboeuf, D.; Dennis, A.P.; Wilson, M.M.; Berget, S.M.; O’Malley, B.W. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol. Cell 2005, 17, 429–439. [Google Scholar] [CrossRef]
- Li, Y.; Sun, N.; Lu, Z.; Sun, S.; Huang, J.; Chen, Z.; He, J. Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett. 2017, 393, 40–51. [Google Scholar] [CrossRef]
- Han, S.S.; Kim, W.J.; Hong, Y.; Hong, S.H.; Lee, S.J.; Ryu, D.R.; Lee, W.; Cho, Y.H.; Lee, S.; Ryu, Y.J.; et al. RNA sequencing identifies novel markers of non-small cell lung cancer. Lung Cancer 2014, 84, 229–235. [Google Scholar] [CrossRef]
- Baugh, E.H.; Ke, H.; Levine, A.J.; Bonneau, R.A.; Chan, C.S. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 2018, 25, 154–160. [Google Scholar] [CrossRef]
- Dixit, A.; Verkhivker, G.M. Structure-functional prediction and analysis of cancer mutation effects in protein kinases. Comp. Math. Methods Med. 2014, 2014, 653487. [Google Scholar] [CrossRef]
- Arendt, C.W.; Dawicki, W.; Ostergaard, H.L. Alternative splicing of transcripts encoding the alpha- and beta-subunits of mouse glucosidase II in T lymphocytes. Glycobiology 1999, 9, 277–283. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Satoh, T.; Toshimori, T.; Yan, G.; Yamaguchi, T.; Kato, K. Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control. Sci. Rep. 2016, 6, 20575. [Google Scholar] [CrossRef] [PubMed]
- Akimov, V.; Barrio-Hernandez, I.; Hansen, S.V.F.; Hallenborg, P.; Pedersen, A.K.; Bekker-Jensen, D.B.; Puglia, M.; Christensen, S.D.K.; Vanselow, J.T.; Nielsen, M.M.; et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 2018, 25, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Boussadia, Z.; Lamberti, J.; Mattei, F.; Pizzi, E.; Puglisi, R.; Zanetti, C.; Pasquini, L.; Fratini, F.; Fantozzi, L.; Felicetti, F.; et al. Acidic microenvironment plays a key role in human melanoma progression through a sustained exosome mediated transfer of clinically relevant metastatic molecules. J. Exp. Clin. Cancer Res. 2018, 37, 245. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Yan, W.; Zubia, E.; Zhou, Y.; Zhang, Y.; Duan, Q.; Narayan, M.; Xu, G. Quantitative proteomics and biochemical analyses reveal the role of endoplasmin in the regulation of the expression and secretion of A Disintegrin And Metalloproteinase 12. J. Proteom. 2018, 182, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Rauscher, B.; Heigwer, F.; Henkel, L.; Hielscher, T.; Voloshanenko, O.; Boutros, M. Toward an integrated map of genetic interactions in cancer cells. Mol. Syst. Biol. 2018, 14, e7656. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.L.; Ulanet, D.B.; Pirman, D.; Mahoney, C.E.; Coco, J.; Si, Y.; Chen, Y.; Huang, L.; Ren, J.; Choe, S.; et al. Differential Aspartate Usage Identifies a Subset of Cancer Cells Particularly Dependent on OGDH. Cell Rep. 2016, 17, 876–890. [Google Scholar] [CrossRef]
- Sickmier, E.A.; Frato, K.E.; Shen, H.; Paranawithana, S.R.; Green, M.R.; Kielkopf, C.L. Structural Basis for Polypyrimidine Tract Recognition by the Essential Pre-mRNA Splicing Factor U2AF65. Mol. Cell 2006, 23, 49–59. [Google Scholar] [CrossRef]
- Giannakis, M.; Hodis, E.; Jasmine Mu, X.; Yamauchi, M.; Rosenbluh, J.; Cibulskis, K.; Saksena, G.; Lawrence, M.S.; Qian, Z.R.; Nishihara, R.; et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat. Genet. 2014, 46, 1264–1266. [Google Scholar] [CrossRef]
- Barbieri, C.E.; Baca, S.C.; Lawrence, M.S.; Demichelis, F.; Blattner, M.; Theurillat, J.P.; White, T.A.; Stojanov, P.; Van Allen, E.; Stransky, N.; et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012, 44, 685–689. [Google Scholar] [CrossRef]
- Champion-Arnaud, P.; Reed, R. The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes Dev. 1994, 8, 1974–1983. [Google Scholar] [CrossRef] [PubMed]
- Golas, M.M.; Sander, B.; Will, C.L.; Luhrmann, R.; Stark, H. Major conformational change in the complex SF3b upon integration into the spliceosomal U11/U12 di-snRNP as revealed by electron cryomicroscopy. Mol. Cell 2005, 17, 869–883. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Ohta, A.; Terashima, K.; Sakamoto, H. Polycistronic expression and RNA-binding specificity of the C. elegans homologue of the spliceosome-associated protein SAP49. J. Biochem. 1997, 121, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Nam, S.W. SF3B4 as an early-stage diagnostic marker and driver of hepatocellular carcinoma. BMB Rep. 2018, 51, 57–58. [Google Scholar] [CrossRef] [PubMed]
- Marques, F.; Tenney, J.; Duran, I.; Martin, J.; Nevarez, L.; Pogue, R.; Krakow, D.; Cohn, D.H.; Li, B. Altered mRNA Splicing, Chondrocyte Gene Expression and Abnormal Skeletal Development due to SF3B4 Mutations in Rodriguez Acrofacial Dysostosis. PLoS Genet. 2016, 12, e1006307. [Google Scholar] [CrossRef]

| Oligoribonucleotide | Sequence (5′-3′) 1 |
|---|---|
| AdML 2 | *UUCGUGCUGACCUGUCCCUUUUUUUUCCACAGC |
| UBE2F-can3′ss-32 | *UUUUUUUGUUUUGUUUUGUGUUUUUUGAUAGA |
| UBE2F-cr3′ss-32 | *CUUUUUGUUUCCUUUUUUUUUUAAUUUAAAGG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kralovicova, J.; Borovska, I.; Kubickova, M.; Lukavsky, P.J.; Vorechovsky, I. Cancer-Associated Substitutions in RNA Recognition Motifs of PUF60 and U2AF65 Reveal Residues Required for Correct Folding and 3′ Splice-Site Selection. Cancers 2020, 12, 1865. https://doi.org/10.3390/cancers12071865
Kralovicova J, Borovska I, Kubickova M, Lukavsky PJ, Vorechovsky I. Cancer-Associated Substitutions in RNA Recognition Motifs of PUF60 and U2AF65 Reveal Residues Required for Correct Folding and 3′ Splice-Site Selection. Cancers. 2020; 12(7):1865. https://doi.org/10.3390/cancers12071865
Chicago/Turabian StyleKralovicova, Jana, Ivana Borovska, Monika Kubickova, Peter J. Lukavsky, and Igor Vorechovsky. 2020. "Cancer-Associated Substitutions in RNA Recognition Motifs of PUF60 and U2AF65 Reveal Residues Required for Correct Folding and 3′ Splice-Site Selection" Cancers 12, no. 7: 1865. https://doi.org/10.3390/cancers12071865
APA StyleKralovicova, J., Borovska, I., Kubickova, M., Lukavsky, P. J., & Vorechovsky, I. (2020). Cancer-Associated Substitutions in RNA Recognition Motifs of PUF60 and U2AF65 Reveal Residues Required for Correct Folding and 3′ Splice-Site Selection. Cancers, 12(7), 1865. https://doi.org/10.3390/cancers12071865






