Role of Telomeres and Telomeric Proteins in Human Malignancies and Their Therapeutic Potential
Abstract
1. Introduction
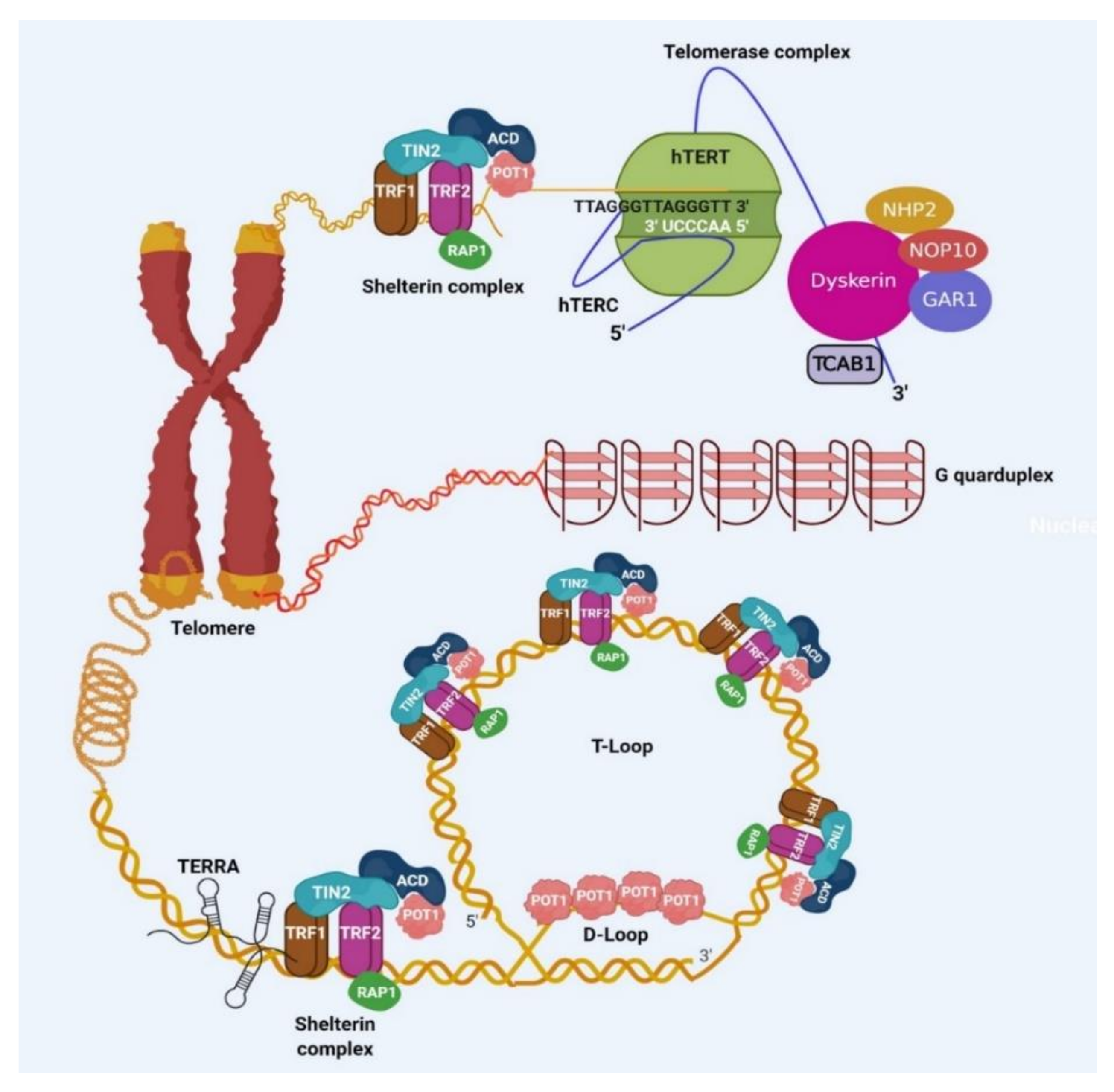
2. Importance of Shelterin Complex in Cancer
3. Importance of Telomerase in Cancer
3.1. TERT Promoter Mutations (TPMs)
3.2. TERT Gene Amplification
3.3. Rearrangement of TERT Locus
3.4. TERT Transcription through Telomere Position Effect-Over Long Distances (TPE-OLD)
3.5. Oncoviral DNA Insertions at TERT Locus
3.6. Alternative Splicing of TERT
3.7. Cohort Studies and Future Directions
4. Role of TERRA in Cancer
5. Targeting Telomeric Components in Cancer
5.1. TRF1 Inhibitors
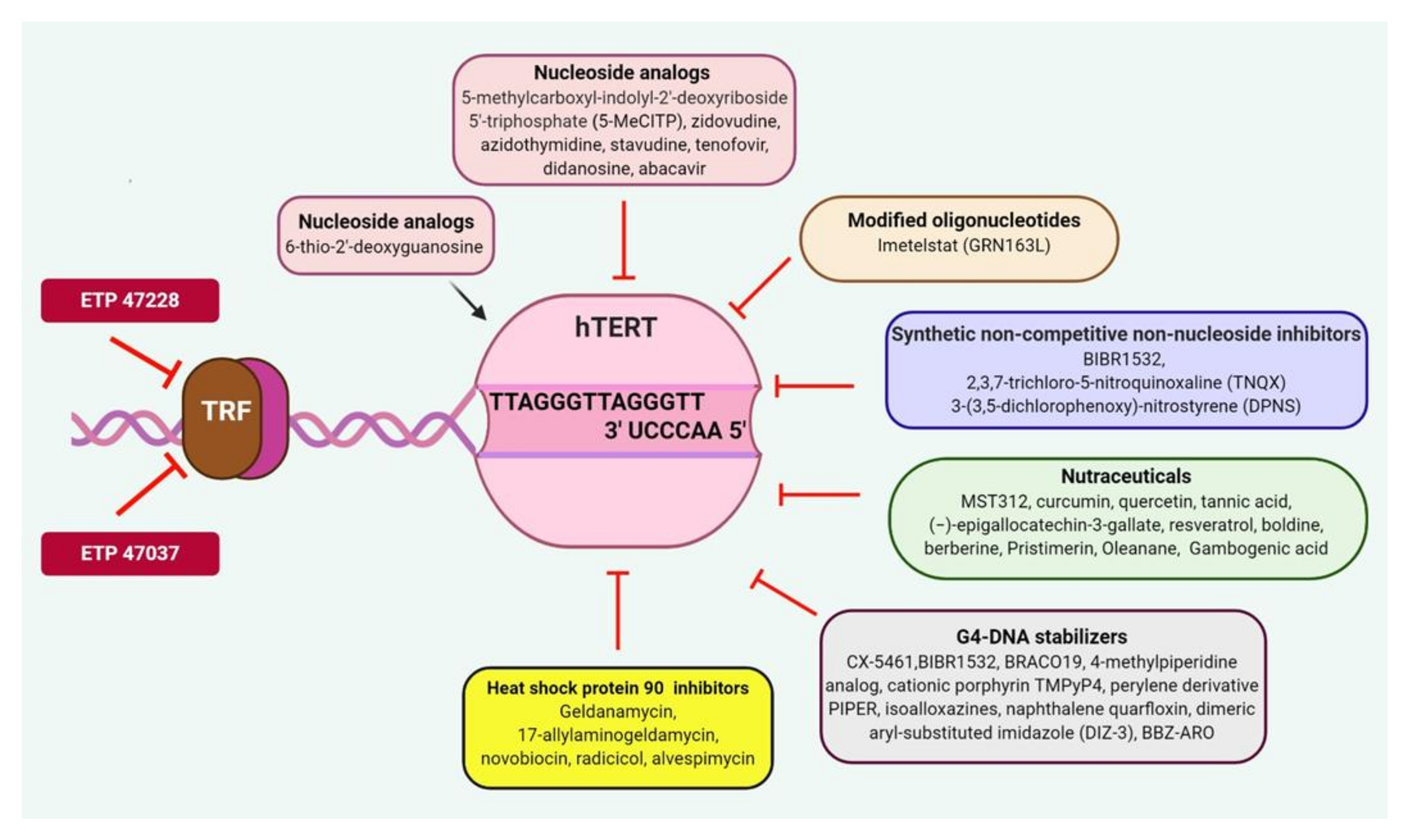
5.2. Telomerase Inhibitors
5.2.1. Nucleoside Analogs
5.2.2. Chemically Modified Oligonucleotides
5.2.3. Chemically Synthesized Mixed Type Noncompetitive Nonnucleoside Inhibitors
5.2.4. Natural Compounds and Derivatives
5.2.5. G4-DNA Stabilizers
5.2.6. Heat Shock Protein 90 (HSP90) Inhibitors
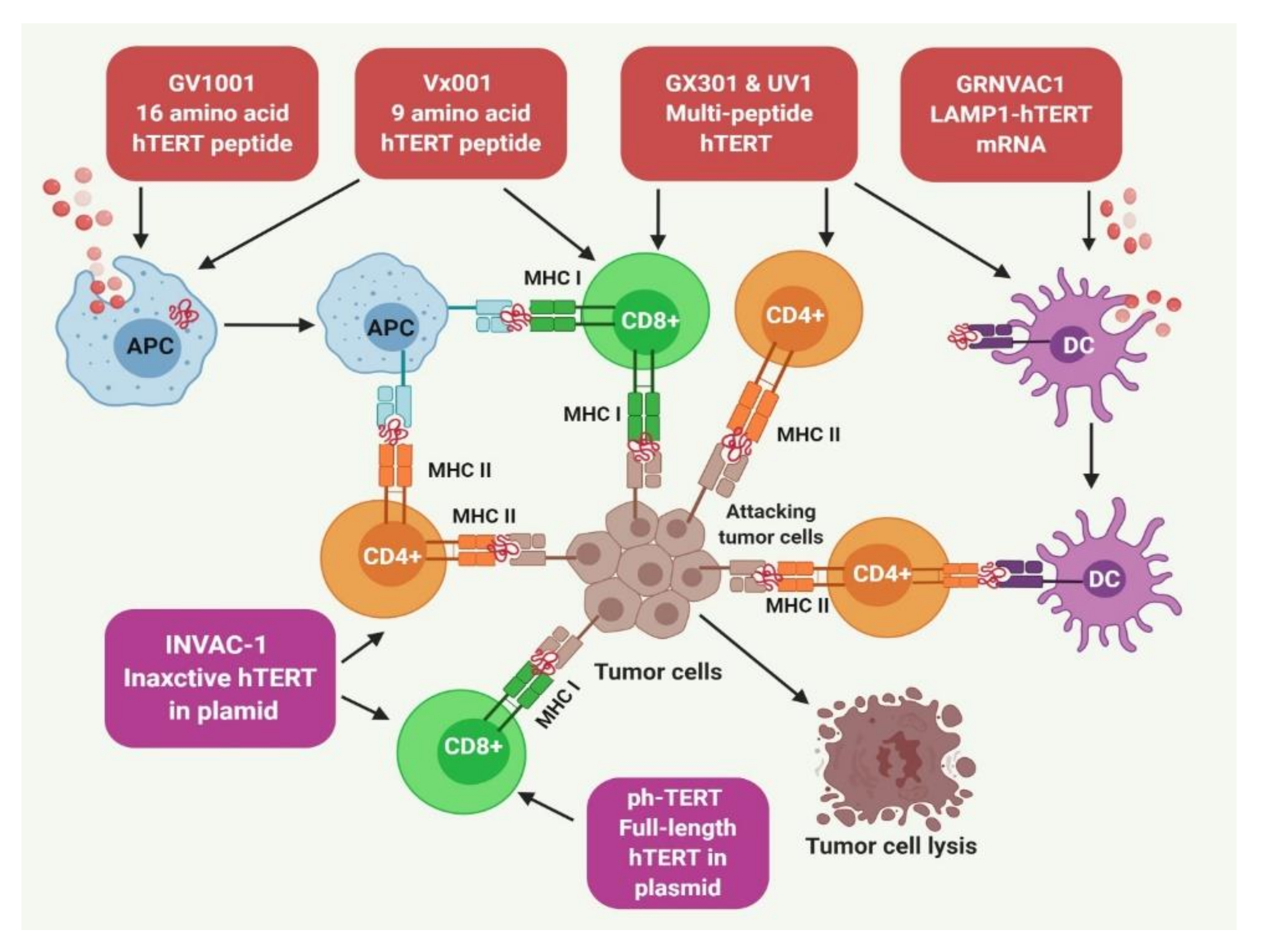
5.3. Human TERT Targeting Immunotherapy
5.3.1. Immunotherapy Using TERT-Derived Peptide Vaccines
5.3.2. TERT Targeting Dendritic Cells (DCs) for Immunotherapy
5.3.3. DNA Vaccines
5.3.4. Cell-Based Approaches
5.3.5. Gene-Modified T-Cell Therapy
5.3.6. TERT-Targeted Cancer Immunotherapy: Challenges and Future Perspectives
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Meyne, J.; Ratliff, R.L.; Moyzis, R.K. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc. Natl. Acad. Sci. USA 1989, 86, 7049–7053. [Google Scholar] [CrossRef] [PubMed]
- Wright, W.E.; Tesmer, V.M.; Huffman, K.E.; Levene, S.D.; Shay, J.W. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997, 11, 2801–2809. [Google Scholar] [CrossRef]
- Griffith, J.D.; Comeau, L.; Rosenfield, S.; Stansel, R.M.; Bianchi, A.; Moss, H.; De Lange, T. Mammalian telomeres end in a large duplex loop. Cell 1999, 97, 503–514. [Google Scholar] [CrossRef]
- Parkinson, G.N.; Lee, M.P.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef]
- Sundquist, W.I.; Klug, A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature 1989, 342, 825–829. [Google Scholar] [CrossRef] [PubMed]
- Saharia, A.; Guittat, L.; Crocker, S.; Lim, A.; Steffen, M.; Kulkarni, S.; Stewart, S.A. Flap endonuclease 1 contributes to telomere stability. Curr. Biol. 2008, 18, 496–500. [Google Scholar] [CrossRef]
- Vallur, A.C.; Maizels, N. Activities of human exonuclease 1 that promote cleavage of transcribed immunoglobulin switch regions. Proc. Natl. Acad. Sci. USA 2008, 105, 16508–16512. [Google Scholar] [CrossRef]
- De Lange, T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005, 19, 2100–2110. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.D. Origin of concatemeric T7 DNA. Nat. New Biol. 1972, 239, 197–201. [Google Scholar] [CrossRef]
- Weinrich, S.L.; Pruzan, R.; Ma, L.; Ouellette, M.; Tesmer, V.M.; Holt, S.E.; Bodnar, A.G.; Lichtsteiner, S.; Kim, N.W.; Trager, J.B.; et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 1997, 17, 498–502. [Google Scholar] [CrossRef]
- Schmidt, J.C.; Cech, T.R. Human telomerase: Biogenesis, trafficking, recruitment, and activation. Genes Dev. 2015, 29, 1095–1105. [Google Scholar] [CrossRef] [PubMed]
- Bodnar, A.G.; Ouellette, M.; Frolkis, M.; Holt, S.E.; Chiu, C.P.; Morin, G.B.; Harley, C.B.; Shay, J.W.; Lichtsteiner, S.; Wright, W.E. Extension of life-span by introduction of telomerase into normal human cells. Science 1998, 279, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Chiba, K.; Johnson, J.Z.; Vogan, J.M.; Wagner, T.; Boyle, J.M.; Hockemeyer, D. Cancer-associated TERT promoter mutations abrogate telomerase silencing. eLife 2015, 4, e07918. [Google Scholar] [CrossRef]
- Harley, C.B.; Futcher, A.B.; Greider, C.W. Telomeres shorten during ageing of human fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef]
- Lindsey, J.; McGill, N.I.; Lindsey, L.A.; Green, D.K.; Cooke, H.J. In vivo loss of telomeric repeats with age in humans. Mutat. Res. 1991, 256, 45–48. [Google Scholar] [CrossRef]
- Hastie, N.D.; Dempster, M.; Dunlop, M.G.; Thompson, A.M.; Green, D.K.; Allshire, R.C. Telomere reduction in human colorectal carcinoma and with ageing. Nature 1990, 346, 866–868. [Google Scholar] [CrossRef] [PubMed]
- Allsopp, R.C.; Vaziri, H.; Patterson, C.; Goldstein, S.; Younglai, E.V.; Futcher, A.B.; Greider, C.W.; Harley, C.B. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl. Acad. Sci. USA 1992, 89, 10114–10118. [Google Scholar] [CrossRef]
- Vaziri, H.; Schachter, F.; Uchida, I.; Wei, L.; Zhu, X.; Effros, R.; Cohen, D.; Harley, C.B. Loss of telomeric DNA during aging of normal and trisomy 21 human lymphocytes. Am. J. Hum. Genet. 1993, 52, 661–667. [Google Scholar]
- Kaul, Z.; Cesare, A.J.; Huschtscha, L.I.; Neumann, A.A.; Reddel, R.R. Five dysfunctional telomeres predict onset of senescence in human cells. EMBO Rep. 2011, 13, 52–59. [Google Scholar] [CrossRef]
- Wright, W.E.; Brasiskyte, D.; Piatyszek, M.A.; Shay, J.W. Experimental elongation of telomeres extends the lifespan of immortal x normal cell hybrids. EMBO J. 1996, 15, 1734–1741. [Google Scholar] [CrossRef]
- Wright, W.E.; Shay, J.W. The two-stage mechanism controlling cellular senescence and immortalization. Exp. Gerontol. 1992, 27, 383–389. [Google Scholar] [CrossRef]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Shay, J.W.; Bacchetti, S. A survey of telomerase activity in human cancer. Eur. J. Cancer 1997, 33, 787–791. [Google Scholar] [CrossRef]
- Takai, K.K.; Kibe, T.; Donigian, J.R.; Frescas, D.; De Lange, T. Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol. Cell 2011, 44, 647–659. [Google Scholar] [CrossRef] [PubMed]
- Arat, N.O.; Griffith, J.D. Human Rap1 interacts directly with telomeric DNA and regulates TRF2 localization at the telomere. J. Biol. Chem. 2012, 287, 41583–41594. [Google Scholar] [CrossRef]
- Zhang, Y.; Chiu, S.; Liang, X.; Gao, F.; Zhang, Z.; Liao, S.; Liang, Y.; Chai, Y.H.; Low, D.J.; Tse, H.F.; et al. Rap1-mediated nuclear factor-kappaB (NF-kappaB) activity regulates the paracrine capacity of mesenchymal stem cells in heart repair following infarction. Cell Death Discov. 2015, 1, 15007. [Google Scholar] [CrossRef]
- Sfeir, A.; Kosiyatrakul, S.T.; Hockemeyer, D.; MacRae, S.L.; Karlseder, J.; Schildkraut, C.L.; De Lange, T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 2009, 138, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.; Kibe, T.; Kabir, S.; De Lange, T. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev. 2014, 28, 2477–2491. [Google Scholar] [CrossRef] [PubMed]
- Vannier, J.B.; Sandhu, S.; Petalcorin, M.I.; Wu, X.; Nabi, Z.; Ding, H.; Boulton, S.J. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science 2013, 342, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Benarroch-Popivker, D.; Pisano, S.; Mendez-Bermudez, A.; Lototska, L.; Kaur, P.; Bauwens, S.; Djerbi, N.; Latrick, C.M.; Fraisier, V.; Pei, B.; et al. TRF2-Mediated Control of Telomere DNA Topology as a Mechanism for Chromosome-End Protection. Mol. Cell 2016, 61, 274–286. [Google Scholar] [CrossRef]
- Denchi, E.L.; De Lange, T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 2007, 448, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Sarek, G.; Vannier, J.B.; Panier, S.; Petrini, J.H.J.; Boulton, S.J. TRF2 recruits RTEL1 to telomeres in S phase to promote t-loop unwinding. Mol. Cell 2015, 57, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Janouskova, E.; Necasova, I.; Pavlouskova, J.; Zimmermann, M.; Hluchy, M.; Marini, V.; Novakova, M.; Hofr, C. Human Rap1 modulates TRF2 attraction to telomeric DNA. Nucleic Acids Res. 2015, 43, 2691–2700. [Google Scholar] [CrossRef]
- Sfeir, A.; Kabir, S.; van Overbeek, M.; Celli, G.B.; De Lange, T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science 2010, 327, 1657–1661. [Google Scholar] [CrossRef] [PubMed]
- Palm, W.; Hockemeyer, D.; Kibe, T.; De Lange, T. Functional dissection of human and mouse POT1 proteins. Mol. Cell. Biol. 2009, 29, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.Z.; De Lange, T. TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex. Nat. Genet. 2004, 36, 618–623. [Google Scholar] [CrossRef]
- Zeng, Z.; Wang, W.; Yang, Y.; Chen, Y.; Yang, X.; Diehl, J.A.; Liu, X.; Lei, M. Structural basis of selective ubiquitination of TRF1 by SCFFbx4. Dev. Cell 2010, 18, 214–225. [Google Scholar] [CrossRef]
- Kim, S.H.; Beausejour, C.; Davalos, A.R.; Kaminker, P.; Heo, S.J.; Campisi, J. TIN2 mediates functions of TRF2 at human telomeres. J. Biol. Chem. 2004, 279, 43799–43804. [Google Scholar] [CrossRef]
- Frank, A.K.; Tran, D.C.; Qu, R.W.; Stohr, B.A.; Segal, D.J.; Xu, L. The shelterin TIN2 subunit mediates recruitment of telomerase to telomeres. PLoS Genet. 2015, 11, e1005410. [Google Scholar] [CrossRef]
- Zaug, A.J.; Podell, E.R.; Nandakumar, J.; Cech, T.R. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010, 24, 613–622. [Google Scholar] [CrossRef]
- Wang, F.; Podell, E.R.; Zaug, A.J.; Yang, Y.; Baciu, P.; Cech, T.R.; Lei, M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 2007, 445, 506–510. [Google Scholar] [CrossRef] [PubMed]
- Latrick, C.M.; Cech, T.R. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 2010, 29, 924–933. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Takai, H.; De Lange, T. Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell 2012, 150, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Ohki, R.; Ishikawa, F. Telomere-bound TRF1 and TRF2 stall the replication fork at telomeric repeats. Nucleic Acids Res. 2004, 32, 1627–1637. [Google Scholar] [CrossRef]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Schoeftner, S.; Blasco, M.A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008, 10, 228–236. [Google Scholar] [CrossRef]
- Beishline, K.; Vladimirova, O.; Tutton, S.; Wang, Z.; Deng, Z.; Lieberman, P.M. CTCF driven TERRA transcription facilitates completion of telomere DNA replication. Nat. Commun. 2017, 8, 2114. [Google Scholar] [CrossRef]
- Diman, A.; Boros, J.; Poulain, F.; Rodriguez, J.; Purnelle, M.; Episkopou, H.; Bertrand, L.; Francaux, M.; Deldicque, L.; Decottignies, A. Nuclear respiratory factor 1 and endurance exercise promote human telomere transcription. Sci. Adv. 2016, 2, e1600031. [Google Scholar] [CrossRef]
- Lopez de Silanes, I.; Grana, O.; De Bonis, M.L.; Dominguez, O.; Pisano, D.G.; Blasco, M.A. Identification of TERRA locus unveils a telomere protection role through association to nearly all chromosomes. Nat. Commun. 2014, 5, 4723. [Google Scholar] [CrossRef]
- Arora, R.; Lee, Y.; Wischnewski, H.; Brun, C.M.; Schwarz, T.; Azzalin, C.M. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014, 5, 5220. [Google Scholar] [CrossRef]
- Chu, H.P.; Cifuentes-Rojas, C.; Kesner, B.; Aeby, E.; Lee, H.G.; Wei, C.; Oh, H.J.; Boukhali, M.; Haas, W.; Lee, J.T. TERRA RNA Antagonizes ATRX and protects telomeres. Cell 2017, 170, 86–101.e16. [Google Scholar] [CrossRef] [PubMed]
- Davoli, T.; Denchi, E.L.; De Lange, T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell 2010, 141, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Davoli, T.; De Lange, T. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell 2012, 21, 765–776. [Google Scholar] [CrossRef]
- Maciejowski, J.; Li, Y.; Bosco, N.; Campbell, P.J.; De Lange, T. Chromothripsis and kataegis induced by telomere crisis. Cell 2015, 163, 1641–1654. [Google Scholar] [CrossRef]
- Oh, B.K.; Kim, Y.J.; Park, C.; Park, Y.N. Up-regulation of telomere-binding proteins, TRF1, TRF2, and TIN2 is related to telomere shortening during human multistep hepatocarcinogenesis. Am. J. Pathol. 2005, 166, 73–80. [Google Scholar] [CrossRef]
- Nakanishi, K.; Kawai, T.; Kumaki, F.; Hiroi, S.; Mukai, M.; Ikeda, E.; Koering, C.E.; Gilson, E. Expression of mRNAs for telomeric repeat binding factor (TRF)-1 and TRF2 in atypical adenomatous hyperplasia and adenocarcinoma of the lung. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2003, 9, 1105–1111. [Google Scholar]
- Miyachi, K.; Fujita, M.; Tanaka, N.; Sasaki, K.; Sunagawa, M. Correlation between telomerase activity and telomeric-repeat binding factors in gastric cancer. J. Exp. Clin. Cancer Res. CR 2002, 21, 269–275. [Google Scholar]
- Ohyashiki, J.H.; Hayashi, S.; Yahata, N.; Iwama, H.; Ando, K.; Tauchi, T.; Ohyashiki, K. Impaired telomere regulation mechanism by TRF1 (telomere-binding protein), but not TRF2 expression, in acute leukemia cells. Int. J. Oncol. 2001, 18, 593–598. [Google Scholar] [CrossRef]
- Bellon, M.; Datta, A.; Brown, M.; Pouliquen, J.F.; Couppie, P.; Kazanji, M.; Nicot, C. Increased expression of telomere length regulating factors TRF1, TRF2 and TIN2 in patients with adult T-cell leukemia. Int. J. Cancer 2006, 119, 2090–2097. [Google Scholar] [CrossRef]
- Pal, D.; Sharma, U.; Singh, S.K.; Kakkar, N.; Prasad, R. Over-expression of telomere binding factors (TRF1 & TRF2) in renal cell carcinoma and their inhibition by using SiRNA induce apoptosis, reduce cell proliferation and migration invitro. PLoS ONE 2015, 10, e0115651. [Google Scholar]
- Chen, W.; Wang, Y.; Li, F.; Lin, W.; Liang, Y.; Ma, Z. Expression of telomere repeat binding factor 1 and TRF2 in prostate cancer and correlation with clinical parameters. BioMed Res. Int. 2017, 2017, 9764752. [Google Scholar] [CrossRef] [PubMed]
- Matsutani, N.; Yokozaki, H.; Tahara, E.; Tahara, H.; Kuniyasu, H.; Haruma, K.; Chayama, K.; Yasui, W.; Tahara, E. Expression of telomeric repeat binding factor 1 and 2 and TRF1-interacting nuclear protein 2 in human gastric carcinomas. Int. J. Oncol. 2001, 19, 507–512. [Google Scholar] [CrossRef]
- Panero, J.; Stanganelli, C.; Arbelbide, J.; Fantl, D.B.; Kohan, D.; Garcia Rivello, H.; Rabinovich, G.A.; Slavutsky, I. Expression profile of shelterin components in plasma cell disorders. Clinical significance of POT1 overexpression. Blood Cells Mol. Dis. 2014, 52, 134–139. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, J.; Long, Y.; Lu, X. Expression of telomere binding proteins in gastric cancer and correlation with clinicopathological parameters. Asia-Pac. J. Clin. Oncol. 2011, 7, 339–345. [Google Scholar] [CrossRef]
- Tang, T.; Zhou, F.X.; Lei, H.; Yu, H.J.; Xie, C.H.; Zhou, Y.F.; Liu, S.Q. Increased expression of telomere-related proteins correlates with resistance to radiation in human laryngeal cancer cell lines. Oncol. Rep. 2009, 21, 1505–1509. [Google Scholar] [PubMed]
- Zizza, P.; Dinami, R.; Porru, M.; Cingolani, C.; Salvati, E.; Rizzo, A.; D’Angelo, C.; Petti, E.; Amoreo, C.A.; Mottolese, M.; et al. TRF2 positively regulates SULF2 expression increasing VEGF-A release and activity in tumor microenvironment. Nucleic Acids Res. 2019, 47, 3365–3382. [Google Scholar] [CrossRef] [PubMed]
- El Mai, M.; Wagner, K.D.; Michiels, J.F.; Ambrosetti, D.; Borderie, A.; Destree, S.; Renault, V.; Djerbi, N.; Giraud-Panis, M.J.; Gilson, E.; et al. The telomeric Protein TRF2 regulates angiogenesis by binding and activating the PDGFRbeta promoter. Cell Rep. 2014, 9, 1047–1060. [Google Scholar] [CrossRef] [PubMed]
- Akincilar, S.C.; Khattar, E.; Boon, P.L.; Unal, B.; Fullwood, M.J.; Tergaonkar, V. Long-range chromatin interactions drive mutant TERT promoter activation. Cancer Discov. 2016, 6, 1276–1291. [Google Scholar] [CrossRef] [PubMed]
- Biroccio, A.; Cherfils-Vicini, J.; Augereau, A.; Pinte, S.; Bauwens, S.; Ye, J.; Simonet, T.; Horard, B.; Jamet, K.; Cervera, L.; et al. TRF2 inhibits a cell-extrinsic pathway through which natural killer cells eliminate cancer cells. Nat. Cell Biol. 2013, 15, 818–828. [Google Scholar] [CrossRef]
- Bejarano, L.; Schuhmacher, A.J.; Mendez, M.; Megias, D.; Blanco-Aparicio, C.; Martinez, S.; Pastor, J.; Squatrito, M.; Blasco, M.A. Inhibition of TRF1 telomere protein impairs tumor initiation and progression in glioblastoma mouse models and patient-derived xenografts. Cancer Cell 2017, 32, 590–607.e4. [Google Scholar] [CrossRef]
- Garcia-Beccaria, M.; Martinez, P.; Mendez-Pertuz, M.; Martinez, S.; Blanco-Aparicio, C.; Canamero, M.; Mulero, F.; Ambrogio, C.; Flores, J.M.; Megias, D.; et al. Therapeutic inhibition of TRF1 impairs the growth of p53-deficient K-RasG12V-induced lung cancer by induction of telomeric DNA damage. EMBO Mol. Med. 2015, 7, 930–949. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Yagihashi, A.; Yamada, M.; Asanuma, K.; Moriai, R.; Kobayashi, D.; Tsuji, N.; Watanabe, N. Decreased gene expression for telomeric-repeat binding factors and TIN2 in malignant hematopoietic cells. Anticancer Res. 2002, 22, 1315–1320. [Google Scholar]
- Poncet, D.; Belleville, A.; t’kint de Roodenbeke, C.; Roborel de Climens, A.; Ben Simon, E.; Merle-Beral, H.; Callet-Bauchu, E.; Salles, G.; Sabatier, L.; Delic, J.; et al. Changes in the expression of telomere maintenance genes suggest global telomere dysfunction in B-chronic lymphocytic leukemia. Blood 2008, 111, 2388–2391. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Yagihashi, A.; Nasu, S.; Izawa, Y.; Nakamura, M.; Kobayashi, D.; Tsuji, N.; Watanabe, N. Gene expression for suppressors of telomerase activity (telomeric-repeat binding factors) in breast cancer. Jpn. J. Cancer Res. Gann 2002, 93, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.K.; Shen, Q.; Wang, J.C.; Zheng, X.Z.; Hou, L.; Zhang, B. Correlation of telomere length and the expression of its regulating proteins in mesenchymal sarcomas. Beijing Da Xue Xue Bao Yi Xue Ban J. Peking Univ. Health Sci. 2008, 40, 363–368. [Google Scholar]
- Yamada, M.; Tsuji, N.; Nakamura, M.; Moriai, R.; Kobayashi, D.; Yagihashi, A.; Watanabe, N. Down-regulation of TRF1, TRF2 and TIN2 genes is important to maintain telomeric DNA for gastric cancers. Anticancer Res. 2002, 22, 3303–3307. [Google Scholar]
- Ferrandon, S.; Saultier, P.; Carras, J.; Battiston-Montagne, P.; Alphonse, G.; Beuve, M.; Malleval, C.; Honnorat, J.; Slatter, T.; Hung, N.; et al. Telomere profiling: Toward glioblastoma personalized medicine. Mol. Neurobiol. 2013, 47, 64–76. [Google Scholar] [CrossRef]
- Pal, D.; Singh, S.K.; Kakkar, N.; Prasad, R. Expression of telomere binding proteins (RAP1 and POT1) in renal cell carcinoma and their correlation with clinicopathological parameters. Indian J. Clin. Biochem. 2017, 32, 301–305. [Google Scholar] [CrossRef]
- Speedy, H.E.; Kinnersley, B.; Chubb, D.; Broderick, P.; Law, P.J.; Litchfield, K.; Jayne, S.; Dyer, M.J.S.; Dearden, C.; Follows, G.A.; et al. Germ line mutations in shelterin complex genes are associated with familial chronic lymphocytic leukemia. Blood 2016, 128, 2319–2326. [Google Scholar] [CrossRef]
- Robles-Espinoza, C.D.; Harland, M.; Ramsay, A.J.; Aoude, L.G.; Quesada, V.; Ding, Z.; Pooley, K.A.; Pritchard, A.L.; Tiffen, J.C.; Petljak, M.; et al. POT1 loss-of-function variants predispose to familial melanoma. Nat. Genet. 2014, 46, 478–481. [Google Scholar] [CrossRef]
- Shi, J.; Yang, X.R.; Ballew, B.; Rotunno, M.; Calista, D.; Fargnoli, M.C.; Ghiorzo, P.; Bressac-de Paillerets, B.; Nagore, E.; Avril, M.F.; et al. Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat. Genet. 2014, 46, 482–486. [Google Scholar] [CrossRef] [PubMed]
- Calvete, O.; Martinez, P.; Garcia-Pavia, P.; Benitez-Buelga, C.; Paumard-Hernandez, B.; Fernandez, V.; Dominguez, F.; Salas, C.; Romero-Laorden, N.; Garcia-Donas, J.; et al. A mutation in the POT1 gene is responsible for cardiac angiosarcoma in TP53-negative Li-Fraumeni-like families. Nat. Commun. 2015, 6, 8383. [Google Scholar] [CrossRef]
- Chubb, D.; Broderick, P.; Dobbins, S.E.; Frampton, M.; Kinnersley, B.; Penegar, S.; Price, A.; Ma, Y.P.; Sherborne, A.L.; Palles, C.; et al. Rare disruptive mutations and their contribution to the heritable risk of colorectal cancer. Nat. Commun. 2016, 7, 11883. [Google Scholar] [CrossRef] [PubMed]
- McMaster, M.L.; Sun, C.; Landi, M.T.; Savage, S.A.; Rotunno, M.; Yang, X.R.; Jones, K.; Vogt, A.; Hutchinson, A.; Zhu, B.; et al. Germline mutations in protection of telomeres 1 in two families with Hodgkin lymphoma. Br. J. Haematol. 2018, 181, 372–377. [Google Scholar] [CrossRef] [PubMed]
- Shen, E.; Xiu, J.; Lopez, G.Y.; Bentley, R.; Jalali, A.; Heimberger, A.B.; Bainbridge, M.N.; Bondy, M.L.; Walsh, K.M. POT1 mutation spectrum in tumour types commonly diagnosed among POT1-associated hereditary cancer syndrome families. J. Med. Genet. 2020. [Google Scholar] [CrossRef] [PubMed]
- Bainbridge, M.N.; Armstrong, G.N.; Gramatges, M.M.; Bertuch, A.A.; Jhangiani, S.N.; Doddapaneni, H.; Lewis, L.; Tombrello, J.; Tsavachidis, S.; Liu, Y.; et al. Germline mutations in shelterin complex genes are associated with familial glioma. J. Natl. Cancer Inst. 2015, 107, 384. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jima, D.; Moffitt, A.B.; Liu, Q.; Czader, M.; Hsi, E.D.; Fedoriw, Y.; Dunphy, C.H.; Richards, K.L.; Gill, J.I.; et al. The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood 2014, 123, 2988–2996. [Google Scholar] [CrossRef]
- Gong, Y.; Stock, A.J.; Liu, Y. The enigma of excessively long telomeres in cancer: Lessons learned from rare human POT1 variants. Curr. Opin. Genet. Dev. 2020, 60, 48–55. [Google Scholar] [CrossRef]
- Ramsay, A.J.; Quesada, V.; Foronda, M.; Conde, L.; Martinez-Trillos, A.; Villamor, N.; Rodriguez, D.; Kwarciak, A.; Garabaya, C.; Gallardo, M.; et al. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat. Genet. 2013, 45, 526–530. [Google Scholar] [CrossRef]
- Pinzaru, A.M.; Hom, R.A.; Beal, A.; Phillips, A.F.; Ni, E.; Cardozo, T.; Nair, N.; Choi, J.; Wuttke, D.S.; Sfeir, A.; et al. Telomere replication stress induced by POT1 inactivation accelerates tumorigenesis. Cell Rep. 2016, 15, 2170–2184. [Google Scholar] [CrossRef]
- Aoude, L.G.; Pritchard, A.L.; Robles-Espinoza, C.D.; Wadt, K.; Harland, M.; Choi, J.; Gartside, M.; Quesada, V.; Johansson, P.; Palmer, J.M.; et al. Nonsense mutations in the shelterin complex genes ACD and TERF2IP in familial melanoma. J. Natl. Cancer Inst. 2015, 107. [Google Scholar] [CrossRef] [PubMed]
- Munoz, P.; Blanco, R.; Flores, J.M.; Blasco, M.A. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat. Genet. 2005, 37, 1063–1071. [Google Scholar] [CrossRef]
- Munoz, P.; Blanco, R.; De Carcer, G.; Schoeftner, S.; Benetti, R.; Flores, J.M.; Malumbres, M.; Blasco, M.A. TRF1 controls telomere length and mitotic fidelity in epithelial homeostasis. Mol. Cell. Biol. 2009, 29, 1608–1625. [Google Scholar] [CrossRef] [PubMed]
- Khattar, E.; Maung, K.Z.Y.; Chew, C.L.; Ghosh, A.; Mok, M.M.H.; Lee, P.; Zhang, J.; Chor, W.H.J.; Cildir, G.; Wang, C.Q.; et al. Rap1 regulates hematopoietic stem cell survival and affects oncogenesis and response to chemotherapy. Nat. Commun. 2019, 10, 5349. [Google Scholar] [CrossRef] [PubMed]
- Saternus, K.S.; Thrun, C. Traumatology of the alar ligaments. Aktuelle Traumatol. 1987, 17, 214–218. [Google Scholar] [PubMed]
- Martinez, P.; Thanasoula, M.; Carlos, A.R.; Gomez-Lopez, G.; Tejera, A.M.; Schoeftner, S.; Dominguez, O.; Pisano, D.G.; Tarsounas, M.; Blasco, M.A. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat. Cell Biol. 2010, 12, 768–780. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Xiong, Y.; Kim, H.; He, Q.; Li, Y.; Chen, R.; Songyang, Z. Human telomeric proteins occupy selective interstitial sites. Cell Res. 2011, 21, 1013–1027. [Google Scholar] [CrossRef]
- Yeung, F.; Ramirez, C.M.; Mateos-Gomez, P.A.; Pinzaru, A.; Ceccarini, G.; Kabir, S.; Fernandez-Hernando, C.; Sfeir, A. Nontelomeric role for Rap1 in regulating metabolism and protecting against obesity. Cell Rep. 2013, 3, 1847–1856. [Google Scholar] [CrossRef]
- Teo, H.; Ghosh, S.; Luesch, H.; Ghosh, A.; Wong, E.T.; Malik, N.; Orth, A.; De Jesus, P.; Perry, A.S.; Oliver, J.D.; et al. Telomere-independent Rap1 is an IKK adaptor and regulates NF-kappaB-dependent gene expression. Nat. Cell Biol. 2010, 12, 758–767. [Google Scholar] [CrossRef]
- Hiyama, E.; Hiyama, K. Telomerase as tumor marker. Cancer Lett. 2003, 194, 221–233. [Google Scholar] [CrossRef]
- Hahn, W.C.; Stewart, S.A.; Brooks, M.W.; York, S.G.; Eaton, E.; Kurachi, A.; Beijersbergen, R.L.; Knoll, J.H.; Meyerson, M.; Weinberg, R.A. Inhibition of telomerase limits the growth of human cancer cells. Nat. Med. 1999, 5, 1164–1170. [Google Scholar] [CrossRef] [PubMed]
- Slusher, A.L.; Kim, J.J.; Ludlow, A.T. The role of alternative RNA splicing in the regulation of hTERT, telomerase, and telomeres: Implications for cancer therapeutics. Cancers 2020, 12, 1514. [Google Scholar] [CrossRef]
- Hrdlickova, R.; Nehyba, J.; Bose, H.R., Jr. Alternatively spliced telomerase reverse transcriptase variants lacking telomerase activity stimulate cell proliferation. Mol. Cell. Biol. 2012, 32, 4283–4296. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Ludlow, A.T.; Min, J.; Robin, J.D.; Stadler, G.; Mender, I.; Lai, T.P.; Zhang, N.; Wright, W.E.; Shay, J.W. Regulation of the human telomerase gene TERT by telomere position effect-over long distances (TPE-OLD): Implications for aging and cancer. PLoS Biol. 2016, 14, e2000016. [Google Scholar] [CrossRef] [PubMed]
- Barthel, F.P.; Wei, W.; Tang, M.; Martinez-Ledesma, E.; Hu, X.; Amin, S.B.; Akdemir, K.C.; Seth, S.; Song, X.; Wang, Q.; et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet. 2017, 49, 349–357. [Google Scholar] [CrossRef]
- Akincilar, S.C.; Low, K.C.; Liu, C.Y.; Yan, T.D.; Oji, A.; Ikawa, M.; Li, S.; Tergaonkar, V. Quantitative assessment of telomerase components in cancer cell lines. FEBS Lett. 2015, 589, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Puar, Y.R.; Shanmugam, M.K.; Fan, L.; Arfuso, F.; Sethi, G.; Tergaonkar, V. Evidence for the involvement of the master transcription factor NF-kappaB in cancer initiation and progression. Biomedicines 2018, 6, 82. [Google Scholar] [CrossRef]
- Xu, X.; Li, Y.; Bharath, S.R.; Ozturk, M.B.; Bowler, M.W.; Loo, B.Z.L.; Tergaonkar, V.; Song, H. Structural basis for reactivating the mutant TERT promoter by cooperative binding of p52 and ETS1. Nat. Commun. 2018, 9, 3183. [Google Scholar] [CrossRef]
- Shin, E.M.; Hay, H.S.; Lee, M.H.; Goh, J.N.; Tan, T.Z.; Sen, Y.P.; Lim, S.W.; Yousef, E.M.; Ong, H.T.; Thike, A.A.; et al. DEAD-box helicase DP103 defines metastatic potential of human breast cancers. J. Clin. Investig. 2014, 124, 3807–3824. [Google Scholar] [CrossRef] [PubMed]
- Kirtonia, A.; Sethi, G.; Garg, M. The multifaceted role of reactive oxygen species in tumorigenesis. Cell. Mol. Life Sci. 2020. [CrossRef]
- Koh, C.M.; Khattar, E.; Leow, S.C.; Liu, C.Y.; Muller, J.; Ang, W.X.; Li, Y.; Franzoso, G.; Li, S.; Guccione, E.; et al. Telomerase regulates MYC-driven oncogenesis independent of its reverse transcriptase activity. J. Clin. Investig. 2015, 125, 2109–2122. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, O.G.; Assfalg, R.; Koch, S.; Schelling, A.; Meena, J.K.; Kraus, J.; Lechel, A.; Katz, S.F.; Benes, V.; Scharffetter-Kochanek, K.; et al. Telomerase stimulates ribosomal DNA transcription under hyperproliferative conditions. Nat. Commun. 2014, 5, 4599. [Google Scholar] [CrossRef]
- Khattar, E.; Kumar, P.; Liu, C.Y.; Akincilar, S.C.; Raju, A.; Lakshmanan, M.; Maury, J.J.; Qiang, Y.; Li, S.; Tan, E.Y.; et al. Telomerase reverse transcriptase promotes cancer cell proliferation by augmenting tRNA expression. J. Clin. Investig. 2016, 126, 4045–4060. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Ding, D.; Hao, W.; Yang, F.; Wu, X.; Wang, M.; Xu, X.; Ju, Z.; Liu, J.P.; Song, Z.; et al. hTERT promotes tumor angiogenesis by activating VEGF via interactions with the Sp1 transcription factor. Nucleic Acids Res. 2016, 44, 8693–8703. [Google Scholar] [CrossRef] [PubMed]
- Low, K.C.; Tergaonkar, V. Telomerase: Central regulator of all of the hallmarks of cancer. Trends Biochem. Sci. 2013, 38, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.W.; Hodis, E.; Xu, M.J.; Kryukov, G.V.; Chin, L.; Garraway, L.A. Highly recurrent TERT promoter mutations in human melanoma. Science 2013, 339, 957–959. [Google Scholar] [CrossRef]
- Horn, S.; Figl, A.; Rachakonda, P.S.; Fischer, C.; Sucker, A.; Gast, A.; Kadel, S.; Moll, I.; Nagore, E.; Hemminki, K.; et al. TERT promoter mutations in familial and sporadic melanoma. Science 2013, 339, 959–961. [Google Scholar] [CrossRef]
- Killela, P.J.; Reitman, Z.J.; Jiao, Y.; Bettegowda, C.; Agrawal, N.; Diaz, L.A., Jr.; Friedman, A.H.; Friedman, H.; Gallia, G.L.; Giovanella, B.C.; et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Natl. Acad. Sci. USA 2013, 110, 6021–6026. [Google Scholar] [CrossRef]
- Vinagre, J.; Almeida, A.; Populo, H.; Batista, R.; Lyra, J.; Pinto, V.; Coelho, R.; Celestino, R.; Prazeres, H.; Lima, L.; et al. Frequency of TERT promoter mutations in human cancers. Nat. Commun. 2013, 4, 2185. [Google Scholar] [CrossRef]
- Shain, A.H.; Yeh, I.; Kovalyshyn, I.; Sriharan, A.; Talevich, E.; Gagnon, A.; Dummer, R.; North, J.; Pincus, L.; Ruben, B.; et al. The genetic evolution of melanoma from precursor lesions. N. Engl. J. Med. 2015, 373, 1926–1936. [Google Scholar] [CrossRef]
- Kinde, I.; Munari, E.; Faraj, S.F.; Hruban, R.H.; Schoenberg, M.; Bivalacqua, T.; Allaf, M.; Springer, S.; Wang, Y.; Diaz, L.A., Jr.; et al. TERT promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in urine. Cancer Res. 2013, 73, 7162–7167. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Liu, T.; Sofiadis, A.; Juhlin, C.C.; Zedenius, J.; Hoog, A.; Larsson, C.; Xu, D. TERT promoter mutation as an early genetic event activating telomerase in follicular thyroid adenoma (FTA) and atypical FTA. Cancer 2014, 120, 2965–2979. [Google Scholar] [CrossRef] [PubMed]
- Heidenreich, B.; Nagore, E.; Rachakonda, P.S.; Garcia-Casado, Z.; Requena, C.; Traves, V.; Becker, J.; Soufir, N.; Hemminki, K.; Kumar, R. Telomerase reverse transcriptase promoter mutations in primary cutaneous melanoma. Nat. Commun. 2014, 5, 3401. [Google Scholar] [CrossRef] [PubMed]
- Hosler, G.A.; Davoli, T.; Mender, I.; Litzner, B.; Choi, J.; Kapur, P.; Shay, J.W.; Wang, R.C. A primary melanoma and its asynchronous metastasis highlight the role of BRAF, CDKN2A, and TERT. J. Cutan. Pathol. 2015, 42, 108–117. [Google Scholar] [CrossRef]
- Scott, G.A.; Laughlin, T.S.; Rothberg, P.G. Mutations of the TERT promoter are common in basal cell carcinoma and squamous cell carcinoma. Mod. Pathol. Off. J. United States Can. Acad. Pathol. Inc 2014, 27, 516–523. [Google Scholar] [CrossRef]
- Arita, H.; Narita, Y.; Fukushima, S.; Tateishi, K.; Matsushita, Y.; Yoshida, A.; Miyakita, Y.; Ohno, M.; Collins, V.P.; Kawahara, N.; et al. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol. 2013, 126, 267–276. [Google Scholar] [CrossRef]
- Stern, J.L.; Theodorescu, D.; Vogelstein, B.; Papadopoulos, N.; Cech, T.R. Mutation of the TERT promoter, switch to active chromatin, and monoallelic TERT expression in multiple cancers. Genes Dev. 2015, 29, 2219–2224. [Google Scholar] [CrossRef]
- Heidenreich, B.; Rachakonda, P.S.; Hemminki, K.; Kumar, R. TERT promoter mutations in cancer development. Curr. Opin. Genet. Dev. 2014, 24, 30–37. [Google Scholar] [CrossRef]
- Ekedahl, H.; Lauss, M.; Olsson, H.; Griewank, K.G.; Schadendorf, D.; Ingvar, C.; Jonsson, G. High TERT promoter mutation frequency in non-acral cutaneous metastatic melanoma. Pigment Cell Melanoma Res. 2016, 29, 598–600. [Google Scholar] [CrossRef]
- Bell, R.J.; Rube, H.T.; Kreig, A.; Mancini, A.; Fouse, S.D.; Nagarajan, R.P.; Choi, S.; Hong, C.; He, D.; Pekmezci, M.; et al. Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science 2015, 348, 1036–1039. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, Q.L.; Sun, W.; Chandrasekharan, P.; Cheng, H.S.; Ying, Z.; Lakshmanan, M.; Raju, A.; Tenen, D.G.; Cheng, S.Y.; et al. Non-canonical NF-kappaB signalling and ETS1/2 cooperatively drive C250T mutant TERT promoter activation. Nat. Cell Biol. 2015, 17, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, T.J.; Turajlic, S.; Rowan, A.; Nicol, D.; Farmery, J.H.R.; O’Brien, T.; Martincorena, I.; Tarpey, P.; Angelopoulos, N.; Yates, L.R.; et al. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx renal. Cell 2018, 173, 611–623.e17. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson, N.J.; Ny, L.; Nilsson, J.A.; Larsson, E. Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat. Genet. 2014, 46, 1258–1263. [Google Scholar] [CrossRef]
- Borah, S.; Xi, L.; Zaug, A.J.; Powell, N.M.; Dancik, G.M.; Cohen, S.B.; Costello, J.C.; Theodorescu, D.; Cech, T.R. Cancer. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science 2015, 347, 1006–1010. [Google Scholar]
- Huang, D.S.; Wang, Z.; He, X.J.; Diplas, B.H.; Yang, R.; Killela, P.J.; Meng, Q.; Ye, Z.Y.; Wang, W.; Jiang, X.T.; et al. Recurrent TERT promoter mutations identified in a large-scale study of multiple tumour types are associated with increased TERT expression and telomerase activation. Eur. J. Cancer 2015, 51, 969–976. [Google Scholar]
- Heidenreich, B.; Rachakonda, P.S.; Hosen, I.; Volz, F.; Hemminki, K.; Weyerbrock, A.; Kumar, R. TERT promoter mutations and telomere length in adult malignant gliomas and recurrences. Oncotarget 2015, 6, 10617–10633. [Google Scholar] [CrossRef]
- Liu, T.; Wang, N.; Cao, J.; Sofiadis, A.; Dinets, A.; Zedenius, J.; Larsson, C.; Xu, D. The age- and shorter telomere-dependent TERT promoter mutation in follicular thyroid cell-derived carcinomas. Oncogene 2014, 33, 4978–4984. [Google Scholar] [CrossRef]
- Ceccarelli, M.; Barthel, F.P.; Malta, T.M.; Sabedot, T.S.; Salama, S.R.; Murray, B.A.; Morozova, O.; Newton, Y.; Radenbaugh, A.; Pagnotta, S.M.; et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 2016, 164, 550–563. [Google Scholar] [CrossRef]
- Hayward, N.K.; Wilmott, J.S.; Waddell, N.; Johansson, P.A.; Field, M.A.; Nones, K.; Patch, A.M.; Kakavand, H.; Alexandrov, L.B.; Burke, H.; et al. Whole-genome landscapes of major melanoma subtypes. Nature 2017, 545, 175–180. [Google Scholar] [CrossRef]
- Nagore, E.; Heidenreich, B.; Rachakonda, S.; Garcia-Casado, Z.; Requena, C.; Soriano, V.; Frank, C.; Traves, V.; Quecedo, E.; Sanjuan-Gimenez, J.; et al. TERT promoter mutations in melanoma survival. Int. J. Cancer 2016, 139, 75–84. [Google Scholar] [CrossRef]
- Jeong, D.E.; Woo, S.R.; Nam, H.; Nam, D.H.; Lee, J.H.; Joo, K.M. Preclinical and clinical implications of TERT promoter mutation in glioblastoma multiforme. Oncol. Lett. 2017, 14, 8213–8219. [Google Scholar] [CrossRef] [PubMed]
- Xing, M.; Liu, R.; Liu, X.; Murugan, A.K.; Zhu, G.; Zeiger, M.A.; Pai, S.; Bishop, J. BRAF V600E and TERT promoter mutations cooperatively identify the most aggressive papillary thyroid cancer with highest recurrence. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2014, 32, 2718–2726. [Google Scholar] [CrossRef] [PubMed]
- Rachakonda, P.S.; Hosen, I.; De Verdier, P.J.; Fallah, M.; Heidenreich, B.; Ryk, C.; Wiklund, N.P.; Steineck, G.; Schadendorf, D.; Hemminki, K.; et al. TERT promoter mutations in bladder cancer affect patient survival and disease recurrence through modification by a common polymorphism. Proc. Natl. Acad. Sci. USA 2013, 110, 17426–17431. [Google Scholar] [CrossRef]
- Bagci, O.; Kurtgoz, S. Amplification of cellular oncogenes in solid tumors. N. Am. J. Med. Sci. 2015, 7, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Zheng, C.; Hou, M.; Lindvall, C.; Li, K.J.; Erlandsson, F.; Bjorkholm, M.; Gruber, A.; Blennow, E.; Xu, D. Deletion of the telomerase reverse transcriptase gene and haploinsufficiency of telomere maintenance in Cri du chat syndrome. Am. J. Hum. Genet. 2003, 72, 940–948. [Google Scholar] [CrossRef]
- Zhang, A.; Zheng, C.; Lindvall, C.; Hou, M.; Ekedahl, J.; Lewensohn, R.; Yan, Z.; Yang, X.; Henriksson, M.; Blennow, E.; et al. Frequent amplification of the telomerase reverse transcriptase gene in human tumors. Cancer Res. 2000, 60, 6230–6235. [Google Scholar]
- Xie, H.; Liu, T.; Wang, N.; Bjornhagen, V.; Hoog, A.; Larsson, C.; Lui, W.O.; Xu, D. TERT promoter mutations and gene amplification: Promoting TERT expression in Merkel cell carcinoma. Oncotarget 2014, 5, 10048–10057. [Google Scholar] [CrossRef]
- Wang, N.; Kjellin, H.; Sofiadis, A.; Fotouhi, O.; Juhlin, C.C.; Backdahl, M.; Zedenius, J.; Xu, D.; Lehtio, J.; Larsson, C. Genetic and epigenetic background and protein expression profiles in relation to telomerase activation in medullary thyroid carcinoma. Oncotarget 2016, 7, 21332–21346. [Google Scholar] [CrossRef]
- Piscuoglio, S.; Ng, C.K.; Murray, M.; Burke, K.A.; Edelweiss, M.; Geyer, F.C.; Macedo, G.S.; Inagaki, A.; Papanastasiou, A.D.; Martelotto, L.G.; et al. Massively parallel sequencing of phyllodes tumours of the breast reveals actionable mutations, and TERT promoter hotspot mutations and TERT gene amplification as likely drivers of progression. J. Pathol. 2016, 238, 508–518. [Google Scholar] [CrossRef]
- Hoadley, K.A.; Yau, C.; Wolf, D.M.; Cherniack, A.D.; Tamborero, D.; Ng, S.; Leiserson, M.D.M.; Niu, B.; McLellan, M.D.; Uzunangelov, V.; et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 2014, 158, 929–944. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, S.; Popova, E.Y.; Grigoryev, S.A.; Zhu, J. Rearrangement of upstream sequences of the hTERT gene during cellular immortalization. Genes Chromosomes Cancer 2009, 48, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Nagel, I.; Szczepanowski, M.; Martin-Subero, J.I.; Harder, L.; Akasaka, T.; Ammerpohl, O.; Callet-Bauchu, E.; Gascoyne, R.D.; Gesk, S.; Horsman, D.; et al. Deregulation of the telomerase reverse transcriptase (TERT) gene by chromosomal translocations in B-cell malignancies. Blood 2010, 116, 1317–1320. [Google Scholar] [CrossRef] [PubMed]
- Schilling, G.; Penas, E.M.; Janjetovic, S.; Oliveira-Ferrer, L.; Braig, M.; Behrmann, P.; Bokemeyer, C.; Dierlamm, J. Molecular characterization of chromosomal band 5p15.33: A recurrent breakpoint region in mantle cell lymphoma involving the TERT-CLPTM1L locus. Leuk. Res. 2013, 37, 280–286. [Google Scholar] [CrossRef]
- Valentijn, L.J.; Koster, J.; Zwijnenburg, D.A.; Hasselt, N.E.; van Sluis, P.; Volckmann, R.; van Noesel, M.M.; George, R.E.; Tytgat, G.A.; Molenaar, J.J.; et al. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat. Genet. 2015, 47, 1411–1414. [Google Scholar] [CrossRef] [PubMed]
- Peifer, M.; Hertwig, F.; Roels, F.; Dreidax, D.; Gartlgruber, M.; Menon, R.; Kramer, A.; Roncaioli, J.L.; Sand, F.; Heuckmann, J.M.; et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature 2015, 526, 700–704. [Google Scholar] [CrossRef]
- Ackermann, S.; Cartolano, M.; Hero, B.; Welte, A.; Kahlert, Y.; Roderwieser, A.; Bartenhagen, C.; Walter, E.; Gecht, J.; Kerschke, L.; et al. A mechanistic classification of clinical phenotypes in neuroblastoma. Science 2018, 362, 1165–1170. [Google Scholar] [CrossRef]
- Law, M.J.; Lower, K.M.; Voon, H.P.; Hughes, J.R.; Garrick, D.; Viprakasit, V.; Mitson, M.; De Gobbi, M.; Marra, M.; Morris, A.; et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell 2010, 143, 367–378. [Google Scholar] [CrossRef]
- Xue, Y.; Gibbons, R.; Yan, Z.; Yang, D.; McDowell, T.L.; Sechi, S.; Qin, J.; Zhou, S.; Higgs, D.; Wang, W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl. Acad. Sci. USA 2003, 100, 10635–10640. [Google Scholar] [CrossRef]
- Heaphy, C.M.; De Wilde, R.F.; Jiao, Y.; Klein, A.P.; Edil, B.H.; Shi, C.; Bettegowda, C.; Rodriguez, F.J.; Eberhart, C.G.; Hebbar, S.; et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science 2011, 333, 425. [Google Scholar] [CrossRef]
- Baur, J.A.; Zou, Y.; Shay, J.W.; Wright, W.E. Telomere position effect in human cells. Science 2001, 292, 2075–2077. [Google Scholar] [CrossRef]
- Bellon, M.; Nicot, C. Regulation of telomerase and telomeres: Human tumor viruses take control. J. Natl. Cancer Inst. 2008, 100, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Horikawa, I.; Barrett, J.C. cis-Activation of the human telomerase gene (hTERT) by the hepatitis B virus genome. J. Natl. Cancer Inst. 2001, 93, 1171–1173. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Kost, J.; Sulovari, A.; Wong, N.; Liang, W.S.; Cao, J.; Li, D. A virome-wide clonal integration analysis platform for discovering cancer viral etiology. Genome Res. 2019, 29, 819–830. [Google Scholar] [CrossRef]
- Sung, W.K.; Zheng, H.; Li, S.; Chen, R.; Liu, X.; Li, Y.; Lee, N.P.; Lee, W.H.; Ariyaratne, P.N.; Tennakoon, C.; et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat. Genet. 2012, 44, 765–769. [Google Scholar] [CrossRef] [PubMed]
- Nault, J.C.; Datta, S.; Imbeaud, S.; Franconi, A.; Mallet, M.; Couchy, G.; Letouze, E.; Pilati, C.; Verret, B.; Blanc, J.F.; et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat. Genet. 2015, 47, 1187–1193. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.P.; Wang, C.I.; Pickering, C.R.; Huang, Y.; Tsai, C.N.; Tsang, N.M.; Kao, H.K.; Cheng, M.H.; Myers, J.N. Prevalence of promoter mutations in the TERT gene in oral cavity squamous cell carcinoma. Head Neck 2017, 39, 1131–1137. [Google Scholar] [CrossRef] [PubMed]
- Ludlow, A.T.; Slusher, A.L.; Sayed, M.E. Insights into telomerase/hTERT alternative splicing regulation using bioinformatics and network analysis in cancer. Cancers 2019, 11, 666. [Google Scholar] [CrossRef]
- Yi, X.; White, D.M.; Aisner, D.L.; Baur, J.A.; Wright, W.E.; Shay, J.W. An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia 2000, 2, 433–440. [Google Scholar] [CrossRef]
- Saeboe-Larssen, S.; Fossberg, E.; Gaudernack, G. Characterization of novel alternative splicing sites in human telomerase reverse transcriptase (hTERT): Analysis of expression and mutual correlation in mRNA isoforms from normal and tumour tissues. BMC Mol. Biol. 2006, 7, 26. [Google Scholar] [CrossRef]
- Ulaner, G.A.; Hu, J.F.; Vu, T.H.; Giudice, L.C.; Hoffman, A.R. Tissue-specific alternate splicing of human telomerase reverse transcriptase (hTERT) influences telomere lengths during human development. Int. J. Cancer 2001, 91, 644–649. [Google Scholar] [CrossRef]
- Ludlow, A.T.; Wong, M.S.; Robin, J.D.; Batten, K.; Yuan, L.; Lai, T.P.; Dahlson, N.; Zhang, L.; Mender, I.; Tedone, E.; et al. NOVA1 regulates hTERT splicing and cell growth in non-small cell lung cancer. Nat. Commun. 2018, 9, 3112. [Google Scholar] [CrossRef] [PubMed]
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar]
- Sieverling, L.; Hong, C.; Koser, S.D.; Ginsbach, P.; Kleinheinz, K.; Hutter, B.; Braun, D.M.; Cortes-Ciriano, I.; Xi, R.; Kabbe, R.; et al. Genomic footprints of activated telomere maintenance mechanisms in cancer. Nat. Commun. 2020, 11, 733. [Google Scholar] [CrossRef] [PubMed]
- Bryan, T.M.; Englezou, A.; Gupta, J.; Bacchetti, S.; Reddel, R.R. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995, 14, 4240–4248. [Google Scholar] [CrossRef]
- Bryan, T.M.; Englezou, A.; Dalla-Pozza, L.; Dunham, M.A.; Reddel, R.R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 1997, 3, 1271–1274. [Google Scholar] [CrossRef]
- Henson, J.D.; Reddel, R.R. Assaying and investigating alternative lengthening of telomeres activity in human cells and cancers. FEBS Lett. 2010, 584, 3800–3811. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Azzalin, C.M. Telomere elongation chooses TERRA ALTernatives. RNA Biol. 2015, 12, 938–941. [Google Scholar] [CrossRef]
- Redon, S.; Reichenbach, P.; Lingner, J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010, 38, 5797–5806. [Google Scholar] [CrossRef]
- Bae, S.U.; Park, W.J.; Jeong, W.K.; Baek, S.K.; Lee, H.W.; Lee, J.H. Prognostic impact of telomeric repeat-containing RNA expression on long-term oncologic outcomes in colorectal cancer. Medicine 2019, 98, e14932. [Google Scholar] [CrossRef] [PubMed]
- Karlseder, J.; Broccoli, D.; Dai, Y.; Hardy, S.; De Lange, T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science 1999, 283, 1321–1325. [Google Scholar] [CrossRef]
- Smogorzewska, A.; De Lange, T. Different telomere damage signaling pathways in human and mouse cells. EMBO J. 2002, 21, 4338–4348. [Google Scholar] [CrossRef] [PubMed]
- Martinez, P.; Thanasoula, M.; Munoz, P.; Liao, C.; Tejera, A.; McNees, C.; Flores, J.M.; Fernandez-Capetillo, O.; Tarsounas, M.; Blasco, M.A. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009, 23, 2060–2075. [Google Scholar] [CrossRef] [PubMed]
- Strahl, C.; Blackburn, E.H. Effects of reverse transcriptase inhibitors on telomere length and telomerase activity in two immortalized human cell lines. Mol. Cell. Biol. 1996, 16, 53–65. [Google Scholar] [CrossRef]
- Leeansyah, E.; Cameron, P.U.; Solomon, A.; Tennakoon, S.; Velayudham, P.; Gouillou, M.; Spelman, T.; Hearps, A.; Fairley, C.; Smit de, V.; et al. Inhibition of telomerase activity by human immunodeficiency virus (HIV) nucleos(t)ide reverse transcriptase inhibitors: A potential factor contributing to HIV-associated accelerated aging. J. Infect. Dis. 2013, 207, 1157–1165. [Google Scholar] [CrossRef]
- Mender, I.; Gryaznov, S.; Dikmen, Z.G.; Wright, W.E.; Shay, J.W. Induction of telomere dysfunction mediated by the telomerase substrate precursor 6-thio-2′-deoxyguanosine. Cancer Discov. 2015, 5, 82–95. [Google Scholar] [CrossRef] [PubMed]
- Mender, I.; LaRanger, R.; Luitel, K.; Peyton, M.; Girard, L.; Lai, T.P.; Batten, K.; Cornelius, C.; Dalvi, M.P.; Ramirez, M.; et al. Telomerase-mediated strategy for overcoming non-small cell lung cancer targeted therapy and chemotherapy resistance. Neoplasia 2018, 20, 826–837. [Google Scholar] [CrossRef]
- Sengupta, S.; Sobo, M.; Lee, K.; Senthil Kumar, S.; White, A.R.; Mender, I.; Fuller, C.; Chow, L.M.L.; Fouladi, M.; Shay, J.W.; et al. Induced telomere damage to treat telomerase expressing therapy-resistant pediatric brain tumors. Mol. Cancer Ther. 2018, 17, 1504–1514. [Google Scholar] [CrossRef]
- Hernandez-Sanchez, W.; Huang, W.; Plucinsky, B.; Garcia-Vazquez, N.; Robinson, N.J.; Schiemann, W.P.; Berdis, A.J.; Skordalakes, E.; Taylor, D.J. A non-natural nucleotide uses a specific pocket to selectively inhibit telomerase activity. PLoS Biol. 2019, 17, e3000204. [Google Scholar] [CrossRef]
- Akiyama, M.; Hideshima, T.; Shammas, M.A.; Hayashi, T.; Hamasaki, M.; Tai, Y.T.; Richardson, P.; Gryaznov, S.; Munshi, N.C.; Anderson, K.C. Effects of oligonucleotide N3′-->P5′ thio-phosphoramidate (GRN163) targeting telomerase RNA in human multiple myeloma cells. Cancer Res. 2003, 63, 6187–6194. [Google Scholar]
- Frink, R.E.; Peyton, M.; Schiller, J.H.; Gazdar, A.F.; Shay, J.W.; Minna, J.D. Telomerase inhibitor imetelstat has preclinical activity across the spectrum of non-small cell lung cancer oncogenotypes in a telomere length dependent manner. Oncotarget 2016, 7, 31639–31651. [Google Scholar] [CrossRef]
- Wu, X.; Smavadati, S.; Nordfjall, K.; Karlsson, K.; Qvarnstrom, F.; Simonsson, M.; Bergqvist, M.; Gryaznov, S.; Ekman, S.; Paulsson-Karlsson, Y. Telomerase antagonist imetelstat inhibits esophageal cancer cell growth and increases radiation-induced DNA breaks. Biochim. Biophys. Acta 2012, 1823, 2130–2135. [Google Scholar] [CrossRef] [PubMed]
- Joseph, I.; Tressler, R.; Bassett, E.; Harley, C.; Buseman, C.M.; Pattamatta, P.; Wright, W.E.; Shay, J.W.; Go, N.F. The telomerase inhibitor imetelstat depletes cancer stem cells in breast and pancreatic cancer cell lines. Cancer Res. 2010, 70, 9494–9504. [Google Scholar] [CrossRef] [PubMed]
- Marian, C.O.; Cho, S.K.; McEllin, B.M.; Maher, E.A.; Hatanpaa, K.J.; Madden, C.J.; Mickey, B.E.; Wright, W.E.; Shay, J.W.; Bachoo, R.M. The telomerase antagonist, imetelstat, efficiently targets glioblastoma tumor-initiating cells leading to decreased proliferation and tumor growth. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Roth, A.; Vercauteren, S.; Sutherland, H.J.; Lansdorp, P.M. Telomerase is limiting the growth of acute myeloid leukemia cells. Leukemia 2003, 17, 2410–2417. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, A.; Tauchi, T.; Sashida, G.; Sumi, M.; Abe, K.; Yamamoto, K.; Ohyashiki, J.H.; Ohyashiki, K. Telomerase inhibition enhances apoptosis in human acute leukemia cells: Possibility of antitelomerase therapy. Leukemia 2003, 17, 560–567. [Google Scholar] [CrossRef]
- Koziel, J.E.; Herbert, B.S. The telomerase inhibitor imetelstat alone, and in combination with trastuzumab, decreases the cancer stem cell population and self-renewal of HER2+ breast cancer cells. Breast Cancer Res. Treat. 2015, 149, 607–618. [Google Scholar] [CrossRef]
- Damm, K.; Hemmann, U.; Garin-Chesa, P.; Hauel, N.; Kauffmann, I.; Priepke, H.; Niestroj, C.; Daiber, C.; Enenkel, B.; Guilliard, B.; et al. A highly selective telomerase inhibitor limiting human cancer cell proliferation. EMBO J. 2001, 20, 6958–6968. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, J.H.; Lee, G.E.; Kim, S.W.; Chung, I.K. Identification of a quinoxaline derivative that is a potent telomerase inhibitor leading to cellular senescence of human cancer cells. Biochem. J. 2003, 373 Pt 2, 523–529. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, J.H.; Lee, G.E.; Lee, J.E.; Chung, I.K. Potent inhibition of human telomerase by nitrostyrene derivatives. Mol. Pharmacol. 2003, 63, 1117–1124. [Google Scholar] [CrossRef]
- Ganesan, K.; Xu, B. Telomerase inhibitors from natural products and their anticancer potential. Int. J. Mol. Sci. 2017, 19, 13. [Google Scholar] [CrossRef]
- Seimiya, H.; Oh-hara, T.; Suzuki, T.; Naasani, I.; Shimazaki, T.; Tsuchiya, K.; Tsuruo, T. Telomere shortening and growth inhibition of human cancer cells by novel synthetic telomerase inhibitors MST-312, MST-295, and MST-1991. Mol. Cancer Ther. 2002, 1, 657–665. [Google Scholar] [PubMed]
- Fatemi, A.; Safa, M.; Kazemi, A. MST-312 induces G2/M cell cycle arrest and apoptosis in APL cells through inhibition of telomerase activity and suppression of NF-kappaB pathway. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2015, 36, 8425–8437. [Google Scholar] [CrossRef]
- Gurung, R.L.; Lim, S.N.; Low, G.K.; Hande, M.P. MST-312 alters telomere dynamics, gene expression profiles and growth in human breast cancer cells. J. Nutr. Nutr. 2014, 7, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Sumi, M.; Tauchi, T.; Sashida, G.; Nakajima, A.; Gotoh, A.; Shin-Ya, K.; Ohyashiki, J.H.; Ohyashiki, K. A G-quadruplex-interactive agent, telomestatin (SOT-095), induces telomere shortening with apoptosis and enhances chemosensitivity in acute myeloid leukemia. Int. J. Oncol. 2004, 24, 1481–1487. [Google Scholar] [PubMed]
- Phatak, P.; Cookson, J.C.; Dai, F.; Smith, V.; Gartenhaus, R.B.; Stevens, M.F.; Burger, A.M. Telomere uncapping by the G-quadruplex ligand RHPS4 inhibits clonogenic tumour cell growth in vitro and in vivo consistent with a cancer stem cell targeting mechanism. Br. J. Cancer 2007, 96, 1223–1233. [Google Scholar] [CrossRef]
- Huang, F.C.; Chang, C.C.; Wang, J.M.; Chang, T.C.; Lin, J.J. Induction of senescence in cancer cells by the G-quadruplex stabilizer, BMVC4, is independent of its telomerase inhibitory activity. Br. J. Pharmacol. 2012, 167, 393–406. [Google Scholar] [CrossRef]
- Kim, M.Y.; Gleason-Guzman, M.; Izbicka, E.; Nishioka, D.; Hurley, L.H. The different biological effects of telomestatin and TMPyP4 can be attributed to their selectivity for interaction with intramolecular or intermolecular G-quadruplex structures. Cancer Res. 2003, 63, 3247–3256. [Google Scholar]
- Gowan, S.M.; Heald, R.; Stevens, M.F.; Kelland, L.R. Potent inhibition of telomerase by small-molecule pentacyclic acridines capable of interacting with G-quadruplexes. Mol. Pharmacol. 2001, 60, 981–988. [Google Scholar] [CrossRef]
- Sun, D.; Thompson, B.; Cathers, B.E.; Salazar, M.; Kerwin, S.M.; Trent, J.O.; Jenkins, T.C.; Neidle, S.; Hurley, L.H. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997, 40, 2113–2116. [Google Scholar] [CrossRef]
- Islam, M.K.; Jackson, P.J.; Rahman, K.M.; Thurston, D.E. Recent advances in targeting the telomeric G-quadruplex DNA sequence with small molecules as a strategy for anticancer therapies. Future Med. Chem. 2016, 8, 1259–1290. [Google Scholar] [CrossRef]
- Hu, M.H.; Lin, X.T.; Liu, B.; Tan, J.H. Dimeric aryl-substituted imidazoles may inhibit ALT cancer by targeting the multimeric G-quadruplex in telomere. Eur. J. Med. Chem. 2020, 186, 111891. [Google Scholar] [CrossRef] [PubMed]
- Yadav, K.; Meka, P.N.R.; Sadhu, S.; Guggilapu, S.D.; Kovvuri, J.; Kamal, A.; Srinivas, R.; Devayani, P.; Babu, B.N.; Nagesh, N. Telomerase Inhibition and Human Telomeric G-Quadruplex DNA Stabilization by a beta-Carboline-Benzimidazole Derivative at Low Concentrations. Biochemistry 2017, 56, 4392–4404. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Gao, S.; Cao, T.; Liu, J.; Gao, X.; Hao, J.; Lv, C.; Huang, H.; Xu, J.; Yao, T. Targeting human telomeric G-quadruplex DNA and inhibition of telomerase activity with [(dmb)2Ru(obip)Ru(dmb)2](4+). PLoS ONE 2013, 8, e84419. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, Y. Pediatric neurology. Nihon rinsho. Jpn. J. Clin. Med. 1987, 45, 2161–2170. [Google Scholar]
- Drygin, D.; Lin, A.; Bliesath, J.; Ho, C.B.; O’Brien, S.E.; Proffitt, C.; Omori, M.; Haddach, M.; Schwaebe, M.K.; Siddiqui-Jain, A.; et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011, 71, 1418–1430. [Google Scholar] [CrossRef]
- Che, T.; Chen, S.B.; Tu, J.L.; Wang, B.; Wang, Y.Q.; Zhang, Y.; Wang, J.; Wang, Z.Q.; Zhang, Z.P.; Ou, T.M.; et al. Discovery of novel schizocommunin derivatives as telomeric G-quadruplex ligands that trigger telomere dysfunction and the deoxyribonucleic acid (DNA) damage response. J. Med. Chem. 2018, 61, 3436–3453. [Google Scholar] [CrossRef]
- Sur, S.; Tiwari, V.; Sinha, D.; Kamran, M.Z.; Dubey, K.D.; Suresh Kumar, G.; Tandon, V. naphthalenediimide-linked bisbenzimidazole derivatives as telomeric G-quadruplex-stabilizing ligands with improved anticancer activity. ACS Omega 2017, 2, 966–980. [Google Scholar] [CrossRef]
- Street, S.T.G.; Chin, D.N.; Hollingworth, G.J.; Berry, M.; Morales, J.C.; Galan, M.C. Divalent Naphthalene Diimide Ligands Display High Selectivity for the Human Telomeric G-quadruplex in K(+) Buffer. Chemistry 2017, 23, 6953–6958. [Google Scholar] [CrossRef]
- Gao, C.; Zhang, W.; He, S.; Li, S.; Liu, F.; Jiang, Y. Synthesis and antiproliferative activity of 2,7-diamino l0-(3,5-dimethoxy)benzyl-9(10H)-acridone derivatives as potent telomeric G-quadruplex DNA ligands. Bioorganic Chem. 2015, 60, 30–36. [Google Scholar] [CrossRef]
- Qin, Q.P.; Qin, J.L.; Chen, M.; Li, Y.L.; Meng, T.; Zhou, J.; Liang, H.; Chen, Z.F. Chiral platinum (II)-4-(2,3-dihydroxypropyl)- formamide oxo-aporphine (FOA) complexes promote tumor cells apoptosis by directly targeting G-quadruplex DNA in vitro and in vivo. Oncotarget 2017, 8, 61982–61997. [Google Scholar] [CrossRef]
- Chen, Z.F.; Qin, Q.P.; Qin, J.L.; Liu, Y.C.; Huang, K.B.; Li, Y.L.; Meng, T.; Zhang, G.H.; Peng, Y.; Luo, X.J.; et al. Stabilization of G-quadruplex DNA, inhibition of telomerase activity, and tumor cell apoptosis by organoplatinum(II) complexes with oxoisoaporphine. J. Med. Chem. 2015, 58, 2159–2179. [Google Scholar] [CrossRef]
- Mancini, J.; Rousseau, P.; Castor, K.J.; Sleiman, H.F.; Autexier, C. Platinum(II) phenanthroimidazole G-quadruplex ligand induces selective telomere shortening in A549 cancer cells. Biochimie 2016, 121, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Suzuki, Y.; Ito, K.; Komiyama, M. Telomeric repeat-containing RNA structure in living cells. Proc. Natl. Acad. Sci. USA 2010, 107, 14579–14584. [Google Scholar] [CrossRef] [PubMed]
- Tauchi, T.; Shin-Ya, K.; Sashida, G.; Sumi, M.; Nakajima, A.; Shimamoto, T.; Ohyashiki, J.H.; Ohyashiki, K. Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: Involvement of ATM-dependent DNA damage response pathways. Oncogene 2003, 22, 5338–5347. [Google Scholar] [CrossRef] [PubMed]
- Holt, S.E.; Aisner, D.L.; Baur, J.; Tesmer, V.M.; Dy, M.; Ouellette, M.; Trager, J.B.; Morin, G.B.; Toft, D.O.; Shay, J.W.; et al. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 1999, 13, 817–826. [Google Scholar] [CrossRef] [PubMed]
- Villa, R.; Folini, M.; Porta, C.D.; Valentini, A.; Pennati, M.; Daidone, M.G.; Zaffaroni, N. Inhibition of telomerase activity by geldanamycin and 17-allylamino, 17-demethoxygeldanamycin in human melanoma cells. Carcinogenesis 2003, 24, 851–859. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mizukoshi, E.; Kaneko, S. Telomerase-targeted cancer immunotherapy. Int. J. Mol. Sci. 2019, 20, 1823. [Google Scholar] [CrossRef]
- Mizukoshi, E.; Nakagawa, H.; Kitahara, M.; Yamashita, T.; Arai, K.; Sunagozaka, H.; Fushimi, K.; Kobayashi, E.; Kishi, H.; Muraguchi, A.; et al. Immunological features of T cells induced by human telomerase reverse transcriptase-derived peptides in patients with hepatocellular carcinoma. Cancer Lett. 2015, 364, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Vonderheide, R.H. Telomerase as a universal tumor-associated antigen for cancer immunotherapy. Oncogene 2002, 21, 674–679. [Google Scholar] [CrossRef]
- Vonderheide, R.H. Prospects and challenges of building a cancer vaccine targeting telomerase. Biochimie 2008, 90, 173–180. [Google Scholar] [CrossRef]
- Jafri, M.A.; Ansari, S.A.; Alqahtani, M.H.; Shay, J.W. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016, 8, 69. [Google Scholar] [CrossRef] [PubMed]
- Brunsvig, P.F.; Aamdal, S.; Gjertsen, M.K.; Kvalheim, G.; Markowski-Grimsrud, C.J.; Sve, I.; Dyrhaug, M.; Trachsel, S.; Moller, M.; Eriksen, J.A.; et al. Telomerase peptide vaccination: A phase I/II study in patients with non-small cell lung cancer. Cancer Immunol. Immunother. 2006, 55, 1553–1564. [Google Scholar] [CrossRef]
- Kim, B.K.; Kim, B.R.; Lee, H.J.; Lee, S.A.; Kim, B.J.; Kim, H.; Won, Y.S.; Shon, W.J.; Lee, N.R.; Inn, K.S.; et al. Tumor-suppressive effect of a telomerase-derived peptide by inhibiting hypoxia-induced HIF-1alpha-VEGF signaling axis. Biomaterials 2014, 35, 2924–2933. [Google Scholar] [CrossRef]
- Kim, H.; Seo, E.H.; Lee, S.H.; Kim, B.J. The telomerase-derived anticancer peptide vaccine GV1001 as an extracellular heat shock protein-mediated cell-penetrating peptide. Int. J. Mol. Sci. 2016, 17, 2054. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.H.; Jung, A.R.; Kim, G.E.; Kim, M.Y.; Sung, J.W.; Shin, D.; Cho, H.J.; Ha, U.S.; Hong, S.H.; Kim, S.W.; et al. GV1001 inhibits cell viability and induces apoptosis in castration-resistant prostate cancer cells through the AKT/NF-kappaB/VEGF pathway. J. Cancer 2019, 10, 6269–6277. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Lee, S.G.; Yang, W.M.; Arfuso, F.; Um, J.Y.; Kumar, A.P.; Bian, J.; Sethi, G.; Ahn, K.S. Formononetin-induced oxidative stress abrogates the activation of STAT3/5 signaling axis and suppresses the tumor growth in multiple myeloma preclinical model. Cancer Lett. 2018, 431, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.E.; Jung, A.R.; Kim, M.Y.; Lee, J.B.; Im, J.H.; Lee, K.W.; Park, Y.H.; Lee, J.Y. GV1001 Induces Apoptosis by Reducing Angiogenesis in Renal Cell Carcinoma Cells Both In Vitro and In Vivo. Urology 2018, 113, 129–137. [Google Scholar] [CrossRef]
- Staff, C.; Mozaffari, F.; Frodin, J.E.; Mellstedt, H.; Liljefors, M. Telomerase (GV1001) vaccination together with gemcitabine in advanced pancreatic cancer patients. Int. J. Oncol. 2014, 45, 1293–1303. [Google Scholar] [CrossRef]
- Middleton, G.; Silcocks, P.; Cox, T.; Valle, J.; Wadsley, J.; Propper, D.; Coxon, F.; Ross, P.; Madhusudan, S.; Roques, T.; et al. Gemcitabine and capecitabine with or without telomerase peptide vaccine GV1001 in patients with locally advanced or metastatic pancreatic cancer (TeloVac): An open-label, randomised, phase 3 trial. Lancet Oncol. 2014, 15, 829–840. [Google Scholar] [CrossRef]
- Kyte, J.A.; Gaudernack, G.; Dueland, S.; Trachsel, S.; Julsrud, L.; Aamdal, S. Telomerase peptide vaccination combined with temozolomide: A clinical trial in stage IV melanoma patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011, 17, 4568–4580. [Google Scholar] [CrossRef]
- Inderberg-Suso, E.M.; Trachsel, S.; Lislerud, K.; Rasmussen, A.M.; Gaudernack, G. Widespread CD4+ T-cell reactivity to novel hTERT epitopes following vaccination of cancer patients with a single hTERT peptide GV1001. Oncoimmunology 2012, 1, 670–686. [Google Scholar] [CrossRef] [PubMed]
- Greten, T.F.; Forner, A.; Korangy, F.; N’Kontchou, G.; Barget, N.; Ayuso, C.; Ormandy, L.A.; Manns, M.P.; Beaugrand, M.; Bruix, J. A phase II open label trial evaluating safety and efficacy of a telomerase peptide vaccination in patients with advanced hepatocellular carcinoma. BMC Cancer 2010, 10, 209. [Google Scholar] [CrossRef] [PubMed]
- Hunger, R.E.; Kernland Lang, K.; Markowski, C.J.; Trachsel, S.; Moller, M.; Eriksen, J.A.; Rasmussen, A.M.; Braathen, L.R.; Gaudernack, G. Vaccination of patients with cutaneous melanoma with telomerase-specific peptides. Cancer Immunol. Immunother. 2011, 60, 1553–1564. [Google Scholar] [CrossRef] [PubMed]
- Fenoglio, D.; Traverso, P.; Parodi, A.; Tomasello, L.; Negrini, S.; Kalli, F.; Battaglia, F.; Ferrera, F.; Sciallero, S.; Murdaca, G.; et al. A multi-peptide, dual-adjuvant telomerase vaccine (GX301) is highly immunogenic in patients with prostate and renal cancer. Cancer Immunol. Immunother. 2013, 62, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Fenoglio, D.; Parodi, A.; Lavieri, R.; Kalli, F.; Ferrera, F.; Tagliamacco, A.; Guastalla, A.; Lamperti, M.G.; Giacomini, M.; Filaci, G. Immunogenicity of GX301 cancer vaccine: Four (telomerase peptides) are better than one. Hum. Vaccines Immunother. 2015, 11, 838–850. [Google Scholar] [CrossRef] [PubMed]
- Lilleby, W.; Gaudernack, G.; Brunsvig, P.F.; Vlatkovic, L.; Schulz, M.; Mills, K.; Hole, K.H.; Inderberg, E.M. Phase I/IIa clinical trial of a novel hTERT peptide vaccine in men with metastatic hormone-naive prostate cancer. Cancer Immunol. Immunother. 2017, 66, 891–901. [Google Scholar] [CrossRef]
- Ruden, M.; Puri, N. Novel anticancer therapeutics targeting telomerase. Cancer Treat. Rev. 2013, 39, 444–456. [Google Scholar] [CrossRef]
- Menez-Jamet, J.; Gallou, C.; Rougeot, A.; Kosmatopoulos, K. Optimized tumor cryptic peptides: The basis for universal neo-antigen-like tumor vaccines. Ann. Transl. Med. 2016, 4, 266. [Google Scholar] [CrossRef]
- Kotsakis, A.; Vetsika, E.K.; Christou, S.; Hatzidaki, D.; Vardakis, N.; Aggouraki, D.; Konsolakis, G.; Georgoulias, V.; Christophyllakis, C.; Cordopatis, P.; et al. Clinical outcome of patients with various advanced cancer types vaccinated with an optimized cryptic human telomerase reverse transcriptase (TERT) peptide: Results of an expanded phase II study. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2012, 23, 442–449. [Google Scholar] [CrossRef]
- Bolonaki, I.; Kotsakis, A.; Papadimitraki, E.; Aggouraki, D.; Konsolakis, G.; Vagia, A.; Christophylakis, C.; Nikoloudi, I.; Magganas, E.; Galanis, A.; et al. Vaccination of patients with advanced non-small-cell lung cancer with an optimized cryptic human telomerase reverse transcriptase peptide. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2007, 25, 2727–2734. [Google Scholar] [CrossRef]
- Mavroudis, D.; Bolonakis, I.; Cornet, S.; Myllaki, G.; Kanellou, P.; Kotsakis, A.; Galanis, A.; Nikoloudi, I.; Spyropoulou, M.; Menez, J.; et al. A phase I study of the optimized cryptic peptide TERT(572y) in patients with advanced malignancies. Oncology 2006, 70, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Georgoulias, V.; Douillard, J.Y.; Khayat, D.; Manegold, C.; Rosell, R.; Rossi, A.; Menez-Jamet, J.; Iche, M.; Kosmatopoulos, K.; Gridelli, C. A multicenter randomized phase IIb efficacy study of Vx-001, a peptide-based cancer vaccine as maintenance treatment in advanced non-small-cell lung cancer: Treatment rationale and protocol dynamics. Clin. Lung Cancer 2013, 14, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Ono, I.; Komamine, H.; Kato, T.; Tanaka, T.; Tanaka, R. A family with Thomsen’s disease. Naika Intern. Med. 1971, 28, 789–793. [Google Scholar]
- Vetsika, E.K.; Konsolakis, G.; Aggouraki, D.; Kotsakis, A.; Papadimitraki, E.; Christou, S.; Menez-Jamet, J.; Kosmatopoulos, K.; Georgoulias, V.; Mavroudis, D. Immunological responses in cancer patients after vaccination with the therapeutic telomerase-specific vaccine Vx-001. Cancer Immunol. Immunother. 2012, 61, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Kotsakis, A.; Papadimitraki, E.; Vetsika, E.K.; Aggouraki, D.; Dermitzaki, E.K.; Hatzidaki, D.; Kentepozidis, N.; Mavroudis, D.; Georgoulias, V. A phase II trial evaluating the clinical and immunologic response of HLA-A2(+) non-small cell lung cancer patients vaccinated with an hTERT cryptic peptide. Lung Cancer 2014, 86, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Dannull, J.; Yang, B.K.; Dahm, P.; Coleman, D.; Yancey, D.; Sichi, S.; Niedzwiecki, D.; Boczkowski, D.; Gilboa, E.; et al. Telomerase mRNA-transfected dendritic cells stimulate antigen-specific CD8+ and CD4+ T cell responses in patients with metastatic prostate cancer. J. Immunol. 2005, 174, 3798–3807. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.K.; Heiser, A.; Boczkowski, D.; Majumdar, A.; Naoe, M.; Lebkowski, J.S.; Vieweg, J.; Gilboa, E. Induction of cytotoxic T cell responses and tumor immunity against unrelated tumors using telomerase reverse transcriptase RNA transfected dendritic cells. Nat. Med. 2000, 6, 1011–1017. [Google Scholar] [CrossRef]
- Aloysius, M.M.; Mc Kechnie, A.J.; Robins, R.A.; Verma, C.; Eremin, J.M.; Farzaneh, F.; Habib, N.A.; Bhalla, J.; Hardwick, N.R.; Satthaporn, S.; et al. Generation in vivo of peptide-specific cytotoxic T cells and presence of regulatory T cells during vaccination with hTERT (class I and II) peptide-pulsed DCs. J. Transl. Med. 2009, 7, 18. [Google Scholar] [CrossRef]
- Khoury, H.J.; Collins, R.H., Jr.; Blum, W.; Stiff, P.S.; Elias, L.; Lebkowski, J.S.; Reddy, A.; Nishimoto, K.P.; Sen, D.; Wirth, E.D., III; et al. Immune responses and long-term disease recurrence status after telomerase-based dendritic cell immunotherapy in patients with acute myeloid leukemia. Cancer 2017, 123, 3061–3072. [Google Scholar] [CrossRef]
- Brower, V. Telomerase-based therapies emerging slowly. J. Natl. Cancer Inst. 2010, 102, 520–521. [Google Scholar] [CrossRef]
- Sioud, M.; Nyakas, M.; Saeboe-Larssen, S.; Mobergslien, A.; Aamdal, S.; Kvalheim, G. Diversification of antitumour immunity in a patient with metastatic melanoma treated with ipilimumab and an IDO-silenced dendritic cell vaccine. Case Rep. Med. 2016, 2016, 9639585. [Google Scholar] [CrossRef] [PubMed]
- Frolkis, M.; Fischer, M.B.; Wang, Z.; Lebkowski, J.S.; Chiu, C.P.; Majumdar, A.S. Dendritic cells reconstituted with human telomerase gene induce potent cytotoxic T-cell response against different types of tumors. Cancer Gene Ther. 2003, 10, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Onfray, F.; Pereda, C.; Reyes, D.; Lopez, M.N. TAPCells, the Chilean dendritic cell vaccine against melanoma and prostate cancer. Biol. Res. 2013, 46, 431–440. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mehrotra, S.; Britten, C.D.; Chin, S.; Garrett-Mayer, E.; Cloud, C.A.; Li, M.; Scurti, G.; Salem, M.L.; Nelson, M.H.; Thomas, M.B.; et al. Vaccination with poly(IC:LC) and peptide-pulsed autologous dendritic cells in patients with pancreatic cancer. J. Hematol. Oncol. 2017, 10, 82. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.Y.; Wu, Y.; Luo, Y.; Su, J.M.; Li, Q.; Zhang, X.W.; Liu, J.Y.; He, Q.M.; Yang, L.; Tian, L.; et al. Mannan-modified adenovirus as a vaccine to induce antitumor immunity. Gene Ther. 2007, 14, 657–663. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, J.; Wang, Y.; Wu, Y.; Ding, Z.Y.; Luo, X.M.; Zhong, W.N.; Liu, J.; Xia, X.Y.; Deng, G.H.; Deng, Y.T.; et al. Mannan-modified adenovirus encoding VEGFR-2 as a vaccine to induce anti-tumor immunity. J. Cancer Res. Clin. Oncol. 2014, 140, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Borch, T.H.; Engell-Noerregaard, L.; Zeeberg Iversen, T.; Ellebaek, E.; Met, O.; Hansen, M.; Andersen, M.H.; Thor Straten, P.; Svane, I.M. mRNA-transfected dendritic cell vaccine in combination with metronomic cyclophosphamide as treatment for patients with advanced malignant melanoma. Oncoimmunology 2016, 5, e1207842. [Google Scholar] [CrossRef]
- Yan, J.; Pankhong, P.; Shin, T.H.; Obeng-Adjei, N.; Morrow, M.P.; Walters, J.N.; Khan, A.S.; Sardesai, N.Y.; Weiner, D.B. Highly optimized DNA vaccine targeting human telomerase reverse transcriptase stimulates potent antitumor immunity. Cancer Immunol. Res. 2013, 1, 179–189. [Google Scholar] [CrossRef]
- Rana, R.; Jagadish, N.; Garg, M.; Mishra, D.; Dahiya, N.; Chaurasiya, D.; Suri, A. Immunogenicity study of recombinant human sperm-associated antigen 9 in bonnet macaque (Macaca radiata). Hum. Reprod. 2006, 21, 2894–2900. [Google Scholar] [CrossRef][Green Version]
- Yang, B.; Jeang, J.; Yang, A.; Wu, T.C.; Hung, C.F. DNA vaccine for cancer immunotherapy. Hum. Vaccines Immunother. 2014, 10, 3153–3164. [Google Scholar] [CrossRef]
- Thalmensi, J.; Pliquet, E.; Liard, C.; Escande, M.; Bestetti, T.; Julithe, M.; Kostrzak, A.; Pailhes-Jimenez, A.S.; Bourges, E.; Loustau, M.; et al. Anticancer DNA vaccine based on human telomerase reverse transcriptase generates a strong and specific T cell immune response. Oncoimmunology 2016, 5, e1083670. [Google Scholar] [CrossRef]
- Teixeira, L.; Medioni, J.; Garibal, J.; Adotevi, O.; Doucet, L.; Durey, M.D.; Ghrieb, Z.; Kiladjian, J.J.; Brizard, M.; Laheurte, C.; et al. A first-in-human Phase I study of INVAC-1, an optimized human telomerase DNA vaccine in patients with advanced solid tumors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Jagadish, N.; Rana, R.; Mishra, D.; Garg, M.; Selvi, R.; Suri, A. Characterization of immune response in mice to plasmid DNA encoding human sperm associated antigen 9 (SPAG9). Vaccine 2006, 24, 3695–3703. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Sang, Y.; Fang, C.; Shao, B.; Yang, L.; Yao, K.; Zhao, X.; Gou, J.; Wei, Y.; Yi, T.; et al. Immunotherapy of tumors with human telomerase reverse transcriptase immortalized human umbilical vein endothelial cells. Int. J. Oncol. 2015, 47, 1901–1911. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.A.; Dudley, M.E.; Wunderlich, J.R.; Hughes, M.S.; Yang, J.C.; Sherry, R.M.; Royal, R.E.; Topalian, S.L.; Kammula, U.S.; Restifo, N.P.; et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science 2006, 314, 126–129. [Google Scholar] [CrossRef]
- Johnson, L.A.; Morgan, R.A.; Dudley, M.E.; Cassard, L.; Yang, J.C.; Hughes, M.S.; Kammula, U.S.; Royal, R.E.; Sherry, R.M.; Wunderlich, J.R.; et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood 2009, 114, 535–546. [Google Scholar] [CrossRef]
- Jackson, H.J.; Rafiq, S.; Brentjens, R.J. Driving CAR T-cells forward. Nat. Rev. Clin. Oncol. 2016, 13, 370–383. [Google Scholar] [CrossRef]
- Neelapu, S.S.; Locke, F.L.; Bartlett, N.L.; Lekakis, L.J.; Miklos, D.B.; Jacobson, C.A.; Braunschweig, I.; Oluwole, O.O.; Siddiqi, T.; Lin, Y.; et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 2017, 377, 2531–2544. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Riviere, I.; Gonen, M.; Wang, X.; Senechal, B.; Curran, K.J.; Sauter, C.; Wang, Y.; Santomasso, B.; Mead, E.; et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N. Engl. J. Med. 2018, 378, 449–459. [Google Scholar] [CrossRef]
- Kyte, J.A.; Gaudernack, G.; Faane, A.; Lislerud, K.; Inderberg, E.M.; Brunsvig, P.; Aamdal, S.; Kvalheim, G.; Walchli, S.; Pule, M. T-helper cell receptors from long-term survivors after telomerase cancer vaccination for use in adoptive cell therapy. Oncoimmunology 2016, 5, e1249090. [Google Scholar] [CrossRef]
- Miyazaki, Y.; Fujiwara, H.; Asai, H.; Ochi, F.; Ochi, T.; Azuma, T.; Ishida, T.; Okamoto, S.; Mineno, J.; Kuzushima, K.; et al. Development of a novel redirected T-cell-based adoptive immunotherapy targeting human telomerase reverse transcriptase for adult T-cell leukemia. Blood 2013, 121, 4894–4901. [Google Scholar] [CrossRef] [PubMed]
- Ohta, R.; Demachi-Okamura, A.; Akatsuka, Y.; Fujiwara, H.; Kuzushima, K. Improving TCR affinity on 293T cells. J. Immunol. Methods 2019, 466, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Hwang, S.S.; Liesa, M.; Gan, B.; Sahin, E.; Jaskelioff, M.; Ding, Z.; Ying, H.; Boutin, A.T.; Zhang, H.; et al. Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell 2012, 148, 651–663. [Google Scholar] [CrossRef]
- Sharma, P.; Allison, J.P. The future of immune checkpoint therapy. Science 2015, 348, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Marquez-Rodas, I.; Cerezuela, P.; Soria, A.; Berrocal, A.; Riso, A.; Gonzalez-Cao, M.; Martin-Algarra, S. Immune checkpoint inhibitors: Therapeutic advances in melanoma. Ann. Transl. Med. 2015, 3, 267. [Google Scholar] [PubMed]
- Larkin, J.; Hodi, F.S.; Wolchok, J.D. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 2015, 373, 1270–1271. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Ciuleanu, T.E.; Pluzanski, A.; Lee, J.S.; Otterson, G.A.; Audigier-Valette, C.; Minenza, E.; Linardou, H.; Burgers, S.; Salman, P.; et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N. Engl. J. Med. 2018, 378, 2093–2104. [Google Scholar] [CrossRef]
- Galon, J.; Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019, 18, 197–218. [Google Scholar] [CrossRef]
| Shelterin Subunit | Disease Reported with Upregulation | Ref. | Disease Reported with Downregulation | Ref. | Disease Reported with the Genetic Mutation | Ref. |
|---|---|---|---|---|---|---|
| TRF1 | Hepatocellular carcinoma Lung cancer Gastric carcinoma Acute lymphoblastic leukemia T-cell leukemia Renal cell carcinoma Prostate cancer Glioblastoma | [55] [56] [57] [58] [59] [60] [61] [70] | Gastric cancer Acute myeloid leukemia B-chronic lymphocytic leukemia Breast cancer Mesenchymal sarcomas | [24] [72] [73] [74] [75] | ||
| TRF2 | Hepatocellular carcinoma Lung cancer Gastric carcinoma T-cell leukemia Renal cell carcinoma Prostate cancer Skin carcinoma | [55] [56] [57,62] [59] [60] [61] [91] | Acute myeloid leukemia Breast cancer Gastric cancer | [72] [74] [76] | Chronic lymphocytic leukemia | [78] |
| RAP1 | Gastric carcinoma Multiple myeloma Renal cell carcinoma | [62] [63] [77] | B-chronic lymphocytic leukemia | [73] | Melanoma | [91] |
| TIN2 | Hepatocellular carcinoma T-cell leukemia Multiple myeloma | [55] [59] [63] | Acute myeloid leukemia Gastric cancer | [72,76] | ||
| POT1 | Multiple myeloma Gastric cancer Laryngeal cancer | [63] [64] [65] | B-chronic lymphocytic leukemia Mesenchymal sarcomas | [73,75] | Angiosarcoma Glioma Mantle cell lymphoma Chronic lymphocytic leukemia Melanoma | [82,85] [85,86] [87] [79,89,90] [80,81,85,91] |
| ACD | Multiple myeloma Laryngeal cancer B-chronic lymphocytic leukemia | [63] [65] [73] | Chronic lymphocytic leukemia Melanoma | [79] [91] |
| Different Types or Classes of Telomerase Inhibitors | Name of Drugs or Agents | Mechanism of Their Action | Identification Methods | Outcomes and Safety Profile | References |
|---|---|---|---|---|---|
| Nucleoside analogs | AZT, stavudine, tenofovir, didanosine and abacavir 6-thio-dG, 5-MeCITP | Incorporate into telomeric DNA to prevents the addition of dNTP and telomerase activity resulting into impairment of telomere except for 6-thio-dG | TRAP method as well as direct telomerase assay were used for validation | Lower efficacy in preclinical cancer models as well as associated with toxicity and nonspecific Less cytotoxic than AZT | [183,184,185,186,187,188] |
| Modified oligonucleotide | Imetelstat (GRN163L) 13-mer oligonucleotide sequence with thio- phosphoramidate and palmitoyl lipid group | Robustly binds to the human telomerase RNA (hTR) template to hamper its recruitment to telomeric DNA leading telomerase inhibition and shortening of telomeric ends | TRAP method was used for validation | Suppress cellular and tumor growth Limited toxicity in phase I/II clinical trials | [189,190,191,192,193,194,195,196] |
| Synthetic mixed noncompetitive nonnucleoside inhibitor | BIBR1532 TNQX (2,3,7-trichloro-5-nitroquinoxaline), DPNS (3,5-dichlorophenoxy-nitrostyrene) | Suppress telomerase dependent telomere lengthening | TRAP method was used for validation. | Suppress cellular growth and induce cell death High doses were associated with cytotoxicity | [197,198,199] |
| Nutraceuticals | MST-312, EGCG, curcumin, quercetin, tannic acid, rhodacyanine, genistein, resveratrol, gambogic acid boldine, gambogenic acid oleanane, berberine, pristimerin | Suppress telomerase activity and telomere shortening | Nutraceuticals and their derivatives were validated via TRAP assay | Reduced tumor growth in a preclinical model Lower stability and bioavailability | [200,201,202,203] |
| Isothiazolone derivatives | TMPI | Isothiazolone moiety may bind with the sulfhydryl of cysteines in the active site of the TERT to attenuate telomerase enzymatic activity | High-throughput using the TRAP method discovered isothiazolone derivatives including TMPI | No data for effects on cancer cell proliferation | [197,198,199] |
| G4-DNA stabilizers | CX-5461, BIBR1532, telomestatin, RHPS4, BRACO-19 and TMPyP4, fluorenones, 4-methylpiperidine analog, perylene derivative PIPER, isoalloxazines, quarfloxin naphthalene, TERRA, BBZ-ARO | G-quadruplex has displayed to suppress telomerase activity and telomeric elongation | TRAP method was used for the validation of G-quadruplex stabilizers in blocking the telomere elongation. | Limited stability, pharmacokinetics Bind nonspecifically to g-quadruplex in the promoter and other regions in the genome associated with off-target effects | [204,205,206,207,208,209,210,211,212,213,214,215,216,217,218,219,220,221] [222,223,224] |
| HSP90 inhibitors | Geldanamycin, 17-allylaminogeldamycin, novobiocin, radicicol, and alvespimycin | Hamper the assemble of telomerase | Small molecule inhibitors against HSP90 were verified using TRAP assay | Inhibit cellular growth and induce apoptosis of cancer cells | [225,226] |
| Clinical Trial Identifier | Phases | Human Malignancies/Conditions | Objective | Design | Results |
|---|---|---|---|---|---|
| NCT00594126 | I | Refractory or relapsed multiple myeloma | Evaluation of maximum tolerated dose (MTD) and safety profile | 3 + 3 cohort; dose escalation study | Dose limiting toxicity (DLT): anemia, thrombocytopenia, neutropenia, a PTT prolongation, fatigue, nausea, anorexia dizziness |
| NCT00732056I | I | Recurrent or metastatic breast cancer | Evaluation of MTD and safety profile Efficacy in combination with paclitaxel and bevacizumab | 3 + 3 cohort; dose escalation study | DLT including thrombocytopenia and neutropenia |
| NCT00310895 | I | Refractory or relapsed solid tumors | Evaluation of MTD and safety profile | 3 + 3 cohort; dose escalation study | DLT including thrombocytopenia and myelosuppression |
| NCT 00718601 | I | Multiple myeloma | Evaluation of MTD and safety profile. Efficacy in combination with bortezomib and dexamethasone | 3 + 3 cohort; dose escalation study | Results are unavailable |
| NCT00124189 | I | Refractory chronic lymphoproliferative disease | Evaluation of MTD, safety, tolerability, DLT | Sequential dose cohort, open label, escalation trial evaluating one infusion duration of 2 h; weekly intravenous infusion | Results are unavailable |
| NCT00510445 | I | Non-small cell lung cancer with metastasis | Evaluation of safety, DLT, MTD in combination with a standard paclitaxel/ carboplatin regimen | Dose cohorts with a minimum of three patients | Patients with imetelstat plus short autologous tumor lysate (TL) displayed longer median progression free survival (PFS) and overall survival (OS). On the other hand, imetelstat plus long TL had no improvement in median PFS or OS Adverse drug reactions (ADRs) includes neutropenia, and thrombocytopenia |
| NCT01265927 | I | HER2+ breast cancer | Evaluation of DLT in combination with trastuzumab | Open label, nonrandomized study | Results are unavailable |
| NCT01242930 | II | Multiple myeloma | Improved outcome in patients previously treated with imetelstat | Imetelstat 2 h intravenous Infusion on day 1 and day 8 of a 28-day cycle | Results are unavailable |
| NCT02426086 | II | Patients with myelofibrosis and previously treated with JAK inhibitors | Evaluation of safety and efficacy | Randomized, single-blind, multicenter | Recruiting patients |
| NCT01731951 | II | Primary or secondary Myelofibrosis | Efficacy | Open label, parallel, active, not recruiting | Complete or partial remission in 21% patients. Bone marrow fibrosis was reversed in a few patients. |
| NCT01243073 | II | Essential thrombocythemia | Evaluation of safety and efficacy | Open label, single group | Eighteen patients and all with positive hematologic response. Positive molecular response in most patients with JAK2 V617 F mutation. ADRs includes neutropenia, anemia |
| NCT02598661 | III | Myelodysplastic syndrome | Safety and efficacy | Randomized, double-blind | Recruiting patients |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes, S.G.; Dsouza, R.; Pandya, G.; Kirtonia, A.; Tergaonkar, V.; Lee, S.Y.; Garg, M.; Khattar, E. Role of Telomeres and Telomeric Proteins in Human Malignancies and Their Therapeutic Potential. Cancers 2020, 12, 1901. https://doi.org/10.3390/cancers12071901
Fernandes SG, Dsouza R, Pandya G, Kirtonia A, Tergaonkar V, Lee SY, Garg M, Khattar E. Role of Telomeres and Telomeric Proteins in Human Malignancies and Their Therapeutic Potential. Cancers. 2020; 12(7):1901. https://doi.org/10.3390/cancers12071901
Chicago/Turabian StyleFernandes, Stina George, Rebecca Dsouza, Gouri Pandya, Anuradha Kirtonia, Vinay Tergaonkar, Sook Y. Lee, Manoj Garg, and Ekta Khattar. 2020. "Role of Telomeres and Telomeric Proteins in Human Malignancies and Their Therapeutic Potential" Cancers 12, no. 7: 1901. https://doi.org/10.3390/cancers12071901
APA StyleFernandes, S. G., Dsouza, R., Pandya, G., Kirtonia, A., Tergaonkar, V., Lee, S. Y., Garg, M., & Khattar, E. (2020). Role of Telomeres and Telomeric Proteins in Human Malignancies and Their Therapeutic Potential. Cancers, 12(7), 1901. https://doi.org/10.3390/cancers12071901







