ERBB2 mRNA Expression and Response to Ado-Trastuzumab Emtansine (T-DM1) in HER2-Positive Breast Cancer
Abstract
:1. Introduction
2. Results
2.1. ERBB2 mRNA in Advanced HER2+ BC Treated with T-DM1
2.2. Identification of an Optimized ERBB2 mRNA Cutoff
2.3. Validation of ERBB2 mRNA Expression in Early-Stage HER2+ BC Treated with Neoadjuvant T-DM1
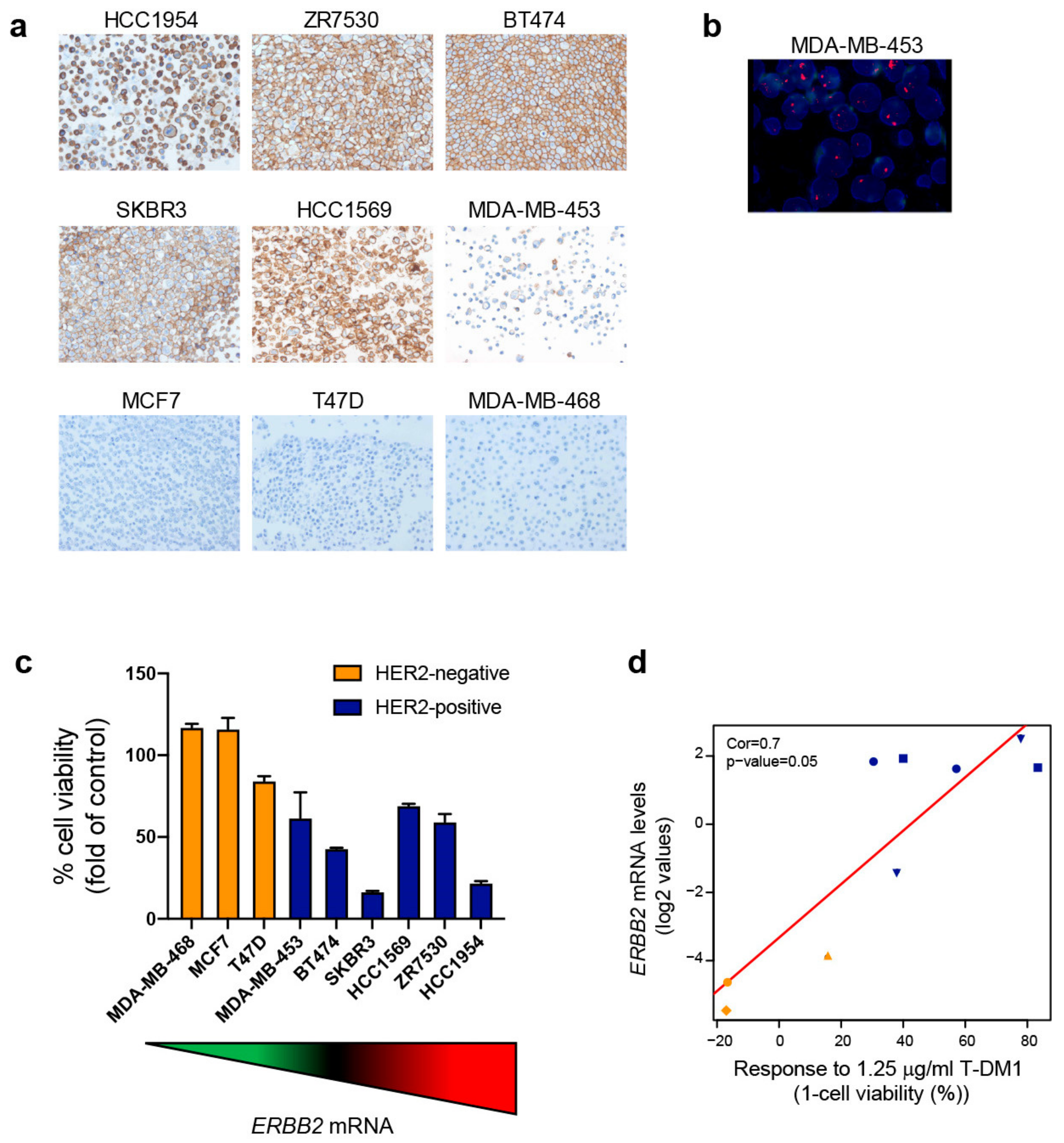
2.4. Exploring ERBB2 mRNA Expression and In Vitro Response to T-DM1
2.5. ERBB2 mRNA Expression in BC across the HER2 IHC-Based Groups
2.6. ERBB2 mRNA Expression across Cancer Types
2.7. ERBB2 mRNA Expression in HER2+ Gastric Cancer Treated with T-DM1
3. Discussion
4. Materials and Methods
4.1. Patient Datasets and Tumor Samples
4.2. In Vitro Cell Lines and T-DM1
4.3. HER2 Immunohistochemistry and Fluorescent In Situ Hybridization
4.4. In Vitro Cell Viability Assay
4.5. RNA Extraction
4.6. Gene Expression Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Verma, S.; Miles, D.; Gianni, L.; Krop, I.E.; Welslau, M.; Baselga, J.; Pegram, M.; Oh, D.-Y.; Diéras, V.; Guardino, E.; et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. New Engl. J. Med. 2012, 367, 1783–1791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krop, I.E.; Kim, S.-B.; González-Martín, A.; Lorusso, P.M.; Ferrero, J.-M.; Smitt, M.; Yu, R.; Leung, A.C.F.; Wildiers, H. Trastuzumab emtansine versus treatment of physician’s choice for pretreated HER2-positive advanced breast cancer (TH3RESA): A randomised, open-label, phase 3 trial. Lancet Oncol. 2014, 15, 689–699. [Google Scholar] [CrossRef]
- Von Minckwitz, G.; Huang, C.-S.; Mano, M.S.; Loibl, S.; Mamounas, E.P.; Untch, M.; Wolmark, N.; Rastogi, P.; Schneeweiss, A.; Redondo, A.; et al. Trastuzumab Emtansine for Residual Invasive HER2-Positive Breast Cancer. New Engl. J. Med. 2019, 380, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Wolff, A.C.; Hammond, M.E.H.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.; Bilous, M.; Fitzgibbons, P.; et al. Recommendations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Update. J. Clin. Oncol. 2013, 31, 3997–4013. [Google Scholar] [CrossRef]
- Slamon, D.J.; Leyland-Jones, B.; Shak, S.; Fuchs, H.; Paton, V.; Bajamonde, A.; Fleming, T.; Eiermann, W.; Wolter, J.; Pegram, M.D.; et al. Use of Chemotherapy plus a Monoclonal Antibody against HER2 for Metastatic Breast Cancer That Overexpresses HER2. New Engl. J. Med. 2001, 344, 783–792. [Google Scholar] [CrossRef]
- Lee, H.J.; Na Seo, A.; Kim, E.J.; Jang, M.H.; Suh, K.J.; Ryu, H.S.; Kim, Y.J.; Kim, J.H.; Im, S.-A.; Gong, G.; et al. HER2 Heterogeneity Affects Trastuzumab Responses and Survival in Patients With HER2-Positive Metastatic Breast Cancer. Am. J. Clin. Pathol. 2014, 142, 755–766. [Google Scholar] [CrossRef] [Green Version]
- Filho, O.M.; Viale, G.; Trippa, L.; Li, T.; Yardley, D.A.; Mayer, I.A.; Abramson, V.G.; Arteaga, C.L.; Spring, L.; Waks, A.G.; et al. HER2 heterogeneity as a predictor of response to neoadjuvant T-DM1 plus pertuzumab: Results from a prospective clinical trial. J. Clin. Oncol. 2019, 37, 502. [Google Scholar] [CrossRef]
- Hou, Y.; Nitta, H.; Wei, L.; Banks, P.M.; Portier, B.; Parwani, A.V.; Li, Z. HER2 intratumoral heterogeneity is independently associated with incomplete response to anti-HER2 neoadjuvant chemotherapy in HER2-positive breast carcinoma. Breast Cancer Res. Treat. 2017, 166, 447–457. [Google Scholar] [CrossRef]
- Modi, S.; Saura, C.; Yamashita, T.; Park, Y.H.; Kim, S.-B.; Tamura, K.; Andre, F.; Iwata, H.; Ito, Y.; Tsurutani, J.; et al. Trastuzumab Deruxtecan in Previously Treated HER2-Positive Breast Cancer. New Engl. J. Med. 2019, 382, 610–621. [Google Scholar] [CrossRef]
- Shitara, K.; Iwata, H.; Takahashi, S.; Tamura, K.; Park, H.; Modi, S.; Tsurutani, J.; Kadowaki, S.; Yamaguchi, K.; Iwasa, S.; et al. Trastuzumab deruxtecan (DS-8201a) in patients with advanced HER2-positive gastric cancer: a dose-expansion, phase 1 study. Lancet Oncol. 2019, 20, 827–836. [Google Scholar] [CrossRef]
- Banerji, U.; Van Herpen, C.M.L.; Saura, C.; Thistlethwaite, F.; Lord, S.; Moreno, V.; MacPherson, I.R.; Boni, V.; Rolfo, C.; E De Vries, E.G.; et al. Trastuzumab duocarmazine in locally advanced and metastatic solid tumours and HER2-expressing breast cancer: a phase 1 dose-escalation and dose-expansion study. Lancet Oncol. 2019, 20, 1124–1135. [Google Scholar] [CrossRef] [Green Version]
- Modi, S.; Park, H.; Murthy, R.K.; Iwata, H.; Tamura, K.; Tsurutani, J.; Moreno-Aspitia, A.; Doi, T.; Sagara, Y.; Redfern, C.; et al. Antitumor Activity and Safety of Trastuzumab Deruxtecan in Patients With HER2-Low-Expressing Advanced Breast Cancer: Results From a Phase Ib Study. J. Clin. Oncol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Gluz, O.; Christgen, M.; Kates, R.E.; Braun, M.; Küemmel, S.; Schumacher, C.; Potenberg, J.; Kraemer, S.; Kleine-Tebbe, A.; et al. De-Escalation Strategies in Human Epidermal Growth Factor Receptor 2 (HER2)–Positive Early Breast Cancer (BC): Final Analysis of the West German Study Group Adjuvant Dynamic Marker-Adjusted Personalized Therapy Trial Optimizing Risk Assessment and Therapy Response Prediction in Early BC HER2- and Hormone Receptor–Positive Phase II Randomized Trial—Efficacy, Safety, and Predictive Markers for 12 Weeks of Neoadjuvant Trastuzumab Emtansine With or Without Endocrine Therapy (ET) Versus Trastuzumab Plus ET. J. Clin. Oncol. 2017, 35, 3046–3054. [Google Scholar] [CrossRef] [PubMed]
- Llombart, A.; Cortés, J.; Paré, L.; Galván, P.; Bermejo, B.; Martínez, N.; Vidal, M.; Pernas, S.; López, R.; Muñoz, M.; et al. HER2-enriched subtype as a predictor of pathological complete response following trastuzumab and lapatinib without chemotherapy in early-stage HER2-positive breast cancer (PAMELA): an open-label, single-group, multicentre, phase 2 trial. Lancet Oncol. 2017, 18, 545–554. [Google Scholar] [CrossRef]
- Lambert, J.M.; Chari, R.V. Ado-trastuzumab Emtansine (T-DM1): An Antibody–Drug Conjugate (ADC) for HER2-Positive Breast Cancer. J. Med. Chem. 2014, 57, 6949–6964. [Google Scholar] [CrossRef]
- Phillips, G.D.L.; Li, G.; Dugger, D.L.; Crocker, L.M.; Parsons, K.L.; Mai, E.; Lambert, J.M.; Chari, R.V.; Lutz, R.J.; Wong, W.L.T.; et al. Targeting HER2-Positive Breast Cancer with Trastuzumab-DM1, an Antibody-Cytotoxic Drug Conjugate. Cancer Res. 2008, 68, 9280–9290. [Google Scholar] [CrossRef] [Green Version]
- Thuss-Patience, P.C.; A Shah, M.; Ohtsu, A.; Van Cutsem, E.; Ajani, J.A.; Castro, H.; Mansoor, W.; Chung, H.C.; Bodoky, G.; Shitara, K.; et al. Trastuzumab emtansine versus taxane use for previously treated HER2-positive locally advanced or metastatic gastric or gastro-oesophageal junction adenocarcinoma (GATSBY): an international randomised, open-label, adaptive, phase 2/3 study. Lancet Oncol. 2017, 18, 640–653. [Google Scholar] [CrossRef]
- Yazaki, S.; Hashimoto, J.; Ogita, S.; Nakano, E.; Yamauchi, T. Lower response to T-DM1 in metastatic breast cancer patients with HER2 IHC score of 2 and FISH positive compared with IHC score of 3. Ann. Oncol. 2017, 28, v102–v103. [Google Scholar] [CrossRef] [Green Version]
- Geyer, C.; Huang, C.-S.; Mano, M.; Loibl, S.; Mamounas, E.; Untch, M.; Wolmark, N.; Rastogi, P.; Fischer, H.; Redondo, A.; et al. Abstract GS1-10: Phase III study of trastuzumab emtansine (T-DM1) vs trastuzumab as adjuvant therapy in patients with HER2-positive early breast cancer with residual invasive disease after neoadjuvant chemotherapy and HER2-targeted therapy including trastuzumab: Primary results from KATHERINE. Gen. Sess. Abstr. 2019, 79, GS1–GS10. [Google Scholar] [CrossRef]
- Emens, L.; Esteva, F.; Beresford, M.; Saura, C.; De Laurentiis, M.; Kim, S.-B.; Im, S.-A.; Patre, M.; Wang, Y.; Mani, A.; et al. Abstract PD3-01: Results from KATE2, a randomized phase 2 study of atezolizumab (atezo)+trastuzumab emtansine (T-DM1) vs placebo (pbo)+T-DM1 in previously treated HER2+ advanced breast cancer (BC). Poster Discuss. Abstr. 2019, 79. [Google Scholar] [CrossRef]
- Shah, M.A.; Kang, Y.-K.; Thuss-Patience, P.C.; Ohtsu, A.; Ajani, J.A.; Van Cutsem, E.; Hoersch, S.; Harle-Yge, M.-L.; De Haas, S.L. Biomarker analysis of the GATSBY study of trastuzumab emtansine versus a taxane in previously treated HER2-positive advanced gastric/gastroesophageal junction cancer. Gastric Cancer 2019, 22, 803–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, S.; Stahel, R.A.; Bubendorf, L.; Bonomi, P.; Villegas, A.; Kowalski, D.M.; Baik, C.S.; Isla, D.; Carpeño, J.D.C.; Garrido, P.; et al. Trastuzumab Emtansine (T-DM1) in Patients with Previously Treated HER2-Overexpressing Metastatic Non–Small Cell Lung Cancer: Efficacy, Safety, and Biomarkers. Clin. Cancer Res. 2018, 25, 64–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baselga, J.; Phillips, G.D.L.; Verma, S.; Ro, J.; Huober, J.; Guardino, A.E.; Samant, M.K.; Olsen, S.; De Haas, S.L.; Pegram, M.D. Relationship between Tumor Biomarkers and Efficacy in EMILIA, a Phase III Study of Trastuzumab Emtansine in HER2-Positive Metastatic Breast Cancer. Clin. Cancer Res. 2016, 22, 3755–3763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.-B.; Wildiers, H.; Krop, I.E.; Smitt, M.; Yu, R.; De Haas, S.L.; Gonzalez-Martin, A. Relationship between tumor biomarkers and efficacy in TH3RESA, a phase III study of trastuzumab emtansine (T-DM1) versus treatment of physician’s choice in previously treated HER2-positive advanced breast cancer. Int. J. Cancer 2016, 139, 2336–2342. [Google Scholar] [CrossRef]
- A Perez, E.; Hurvitz, S.A.; Amler, L.C.; E Mundt, K.; Ng, V.W.; Guardino, E.; Gianni, L. Relationship between HER2 expression and efficacy with first-line trastuzumab emtansine compared with trastuzumab plus docetaxel in TDM4450g: a randomized phase II study of patients with previously untreated HER2-positive metastatic breast cancer. Breast Cancer Res. 2014, 16, R50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krop, I.E.; Lorusso, P.; Miller, K.D.; Modi, S.; Yardley, D.; Rodriguez, G.; Guardino, E.; Lu, M.; Zheng, M.; Girish, S.; et al. A Phase II Study of Trastuzumab Emtansine in Patients With Human Epidermal Growth Factor Receptor 2–Positive Metastatic Breast Cancer Who Were Previously Treated With Trastuzumab, Lapatinib, an Anthracycline, a Taxane, and Capecitabine. J. Clin. Oncol. 2012, 30, 3234–3241. [Google Scholar] [CrossRef]
- Burris, H.A.; Rugo, H.S.; Vukelja, S.J.; Vogel, C.L.; Borson, R.A.; Limentani, S.; Tan-Chiu, E.; Krop, I.E.; Michaelson, R.A.; Girish, S.; et al. Phase II Study of the Antibody Drug Conjugate Trastuzumab-DM1 for the Treatment of Human Epidermal Growth Factor Receptor 2 (HER2) –Positive Breast Cancer After Prior HER2-Directed Therapy. J. Clin. Oncol. 2011, 29, 398–405. [Google Scholar] [CrossRef]
- Denkert, C.; Lambertini, C.; Fasching, P.A.; Pogue-Geile, K.L.; Mano, M.S.; Untch, M.; Wolmark, N.; Huang, C.-S.; Loibl, S.; Mamounas, E.P.; et al. Biomarker data from KATHERINE: A phase III study of adjuvant trastuzumab emtansine (T-DM1) versus trastuzumab (H) in patients with residual invasive disease after neoadjuvant therapy for HER2-positive breast cancer. J. Clin. Oncol. 2020, 38, 502. [Google Scholar] [CrossRef]
- Doi, T.; Shitara, K.; Naito, Y.; Shimomura, A.; Fujiwara, Y.; Yonemori, K.; Shimizu, C.; Shimoi, T.; Kuboki, Y.; Matsubara, N.; et al. Safety, pharmacokinetics, and antitumour activity of trastuzumab deruxtecan (DS-8201), a HER2-targeting antibody–drug conjugate, in patients with advanced breast and gastric or gastro-oesophageal tumours: a phase 1 dose-escalation study. Lancet Oncol. 2017, 18, 1512–1522. [Google Scholar] [CrossRef]
- Cejalvo, J.M.; De Dueñas, E.M.; Galvan, P.; García-Recio, S.; Gasión, O.B.; Paré, L.; Antolin, S.; Martinello, R.; Blancas, I.; Adamo, B.; et al. Intrinsic Subtypes and Gene Expression Profiles in Primary and Metastatic Breast Cancer. Cancer Res. 2017, 77, 2213–2221. [Google Scholar] [CrossRef] [Green Version]




| Characteristics | n = 77 |
|---|---|
| Median age at BC diagnosis, years (range) | 49 (27–88) |
| Median age at start of T-DM1, years (range) | 51 (35–93) |
| Histology: Ductal | 71 (92%) |
| Lobular/other | 5 (6%) |
| NA | 1 (1%) |
| Histologic Grade: G1 | 2 (3%) |
| G2 | 10 (13%) |
| G3 | 28 (36%) |
| NA | 37 (48%) |
| Hormone-receptor: positive | 46 (60%) |
| negative | 31 (40%) |
| HER2 IHC status: IHC 0 | 5 (6%) |
| IHC 1+ | 3 (4%) |
| HC 2+ | 16 (21%) |
| IHC 3+ | 50 (65%) |
| NA * | 3 (4%) |
| HER2 ISH status in HER2 IHC 2+ cases: Amplified | 15 (94%) |
| Not evaluable * | 1 (6%) |
| HER2 ISH status in HER2 IHC 0/1+ cases: Amplified | 3 (38%) |
| Non-amplified * | 1 (12%) |
| Not available * | 4 (50%) |
| Previous (neo)adjuvant treatment | 44 (57%) |
| Median number previous lines HER2-targeted | 1 (0–4) |
| treatment for metastatic disease (range) | |
| Previously received: | |
| Pertuzumab-trastuzumab | 31 (40%) |
| Trastuzumab | 41 (53%) |
| Lapatinib | 14 (24%) |
| Visceral metastases at start of T-DM1 | 66 (86%) |
| Brain metastases at start of T-DM1 | 28 (36%) |
| Concomitant endocrine treatment during T-DM1 | 17 (22%) |
| Clinicopathological Variable | Univariate | Multivariable | |||
|---|---|---|---|---|---|
| Odds Ratio (95%CI) | p | Odds Ratio (95%CI) | p | ||
| Hormone-receptor status | negative | ref | 0.038 | ref | 0.152 |
| positive | 0.37 (0.14–0.95) | 0.39 (0.11–1.41) | |||
| De-novo metastatic disease | no | ref | 0.990 | ||
| yes | 0.99 (0.40–2.49) | ||||
| Visceral disease | no | ref | 0.577 | ||
| yes | 0.69 (0.19–2.50) | ||||
| Brain involvement | no | ref | 0.966 | ||
| yes | 0.98 (0.39–2.49) | ||||
| HER2 IHC | ≤2+ | ref | 0.002 | ref | 0.257 |
| 3+ | 1.84 (1.26–2.69) | 1.32 (0.82–2.13) | |||
| ERBB2 (continuous) | 1.73 (1.25–2.39) | 0.001 | 1.95 (1.22–3.12) | 0.006 | |
| Prior lines HER2-targeted therapy | 0–1 | ref | 0.009 | ref | 0.002 |
| ≥2 | 0.06 (0.01-0.50) | 0.02 (0.002-0.23) | |||
| Target Lesion | Screening | Pre Cycle 3 | Pre Cycle 5 | Pre Cycle 7 | Pre Cycle 9 |
|---|---|---|---|---|---|
| Right upper lobe lung metastasis | 14 mm | 8 mm | 8 mm | 8 mm | 10 mm |
| Left upper lobe lung metastasis | 10 mm | 10 mm | 10 mm | 10 mm | 12 mm |
| Mesentheric adenopathy | 17 mm | 11 mm | 11 mm | 11 mm | 18 mm |
| Retroperitoneal adenopathy | 18 mm | 9 mm | 9 mm | 9 mm | 17 mm |
| Total | 59 mm | 38 mm | 38 mm | 38 mm | 57 mm |
| Response | NA | 39% reduction | 39% reduction | 39% reduction | 36% increase |
| NA | PR | Maintained PR | Maintained PR | PD |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Griguolo, G.; Brasó-Maristany, F.; González-Farré, B.; Pascual, T.; Chic, N.; Saurí, T.; Kates, R.; Gluz, O.; Martínez, D.; Paré, L.; et al. ERBB2 mRNA Expression and Response to Ado-Trastuzumab Emtansine (T-DM1) in HER2-Positive Breast Cancer. Cancers 2020, 12, 1902. https://doi.org/10.3390/cancers12071902
Griguolo G, Brasó-Maristany F, González-Farré B, Pascual T, Chic N, Saurí T, Kates R, Gluz O, Martínez D, Paré L, et al. ERBB2 mRNA Expression and Response to Ado-Trastuzumab Emtansine (T-DM1) in HER2-Positive Breast Cancer. Cancers. 2020; 12(7):1902. https://doi.org/10.3390/cancers12071902
Chicago/Turabian StyleGriguolo, Gaia, Fara Brasó-Maristany, Blanca González-Farré, Tomás Pascual, Núria Chic, Tamara Saurí, Ronald Kates, Oleg Gluz, Débora Martínez, Laia Paré, and et al. 2020. "ERBB2 mRNA Expression and Response to Ado-Trastuzumab Emtansine (T-DM1) in HER2-Positive Breast Cancer" Cancers 12, no. 7: 1902. https://doi.org/10.3390/cancers12071902
APA StyleGriguolo, G., Brasó-Maristany, F., González-Farré, B., Pascual, T., Chic, N., Saurí, T., Kates, R., Gluz, O., Martínez, D., Paré, L., Tsvetkova, V., Pesantez, D., Vidal, M., Adamo, B., Muñoz, M., Galván, P., Barberá, L., Cuatrecasas, M., Christgen, M., ... Prat, A. (2020). ERBB2 mRNA Expression and Response to Ado-Trastuzumab Emtansine (T-DM1) in HER2-Positive Breast Cancer. Cancers, 12(7), 1902. https://doi.org/10.3390/cancers12071902







