The Activity of KIF14, Mieap, and EZR in a New Type of the Invasive Component, Torpedo-Like Structures, Predetermines the Metastatic Potential of Breast Cancer
Abstract
:1. Introduction
2. Results
2.1. Molecules Potentially Associated with Breast Cancer Invasion
2.2. Positive Expression of KIF14 and Mieap and Negative Expression of EZR Are Associated with Breast Cancer Metastasis
2.3. Patterns of Expression of KIF14, Mieap, and EZR in Breast Cancer
2.4. Transcriptomic Profile of KIF14-Positive Tumor Cells Located in Torpedo-Like Structures
2.5. Transcriptomic Profile of Mieap-Positive Tumor Cells Located in Torpedo-Like Structures
2.6. Transcriptomic Profile of EZR-Negative Tumor Cells Located in Torpedo-Like Structures
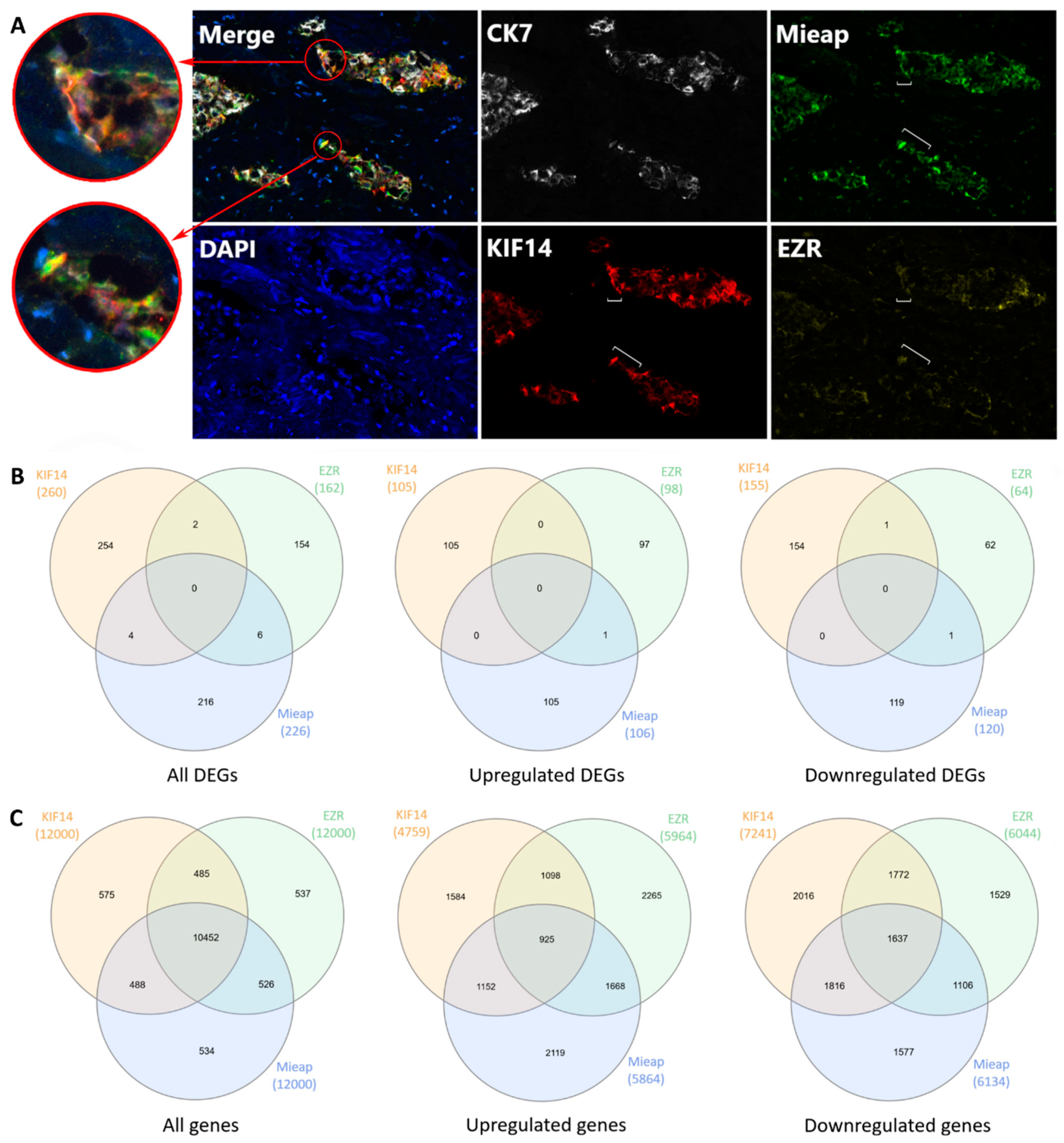
2.7. KIF14- and Mieap-Positive and EZR-Negative Cells are Co-Localized in Torpedo-Like Structures
2.8. Expression of Classic Markers of Invasion in KIF14-Positive, Mieap-Positive, and EZR-Negative Cells
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Gene Expression Microarrays
4.3. Immunohistochemistry
4.4. Multiplex Immunohistochemistry
4.5. RNA-Preserving Immunolabeling and Fluorescence-Guided Laser Microdissection
4.6. RNA Extraction, Library Preparation, Sequencing, and Bioinformatic Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedl, P.; Alexander, S. Cancer invasion and the microenvironment: Plasticity and reciprocity. Cell 2011, 147, 992–1009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedl, P.; Gilmour, D. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 2009, 10, 445–457. [Google Scholar] [CrossRef]
- Khalil, A.A.; Ilina, O.; Gritsenko, P.G.; Bult, P.; Span, P.N.; Friedl, P. Collective invasion in ductal and lobular breast cancer associates with distant metastasis. Clin. Exp. Metastasis 2017, 34, 421–429. [Google Scholar] [CrossRef] [Green Version]
- Pandya, P.; Orgaz, J.L.; Sanz-Moreno, V. Modes of invasion during tumour dissemination. Mol. Oncol. 2017, 11, 5–27. [Google Scholar] [CrossRef] [Green Version]
- Brabletz, T.; Kalluri, R.; Nieto, M.A.; Weinberg, R.A. EMT in cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef]
- Nieto, M.A.; Huang, R.Y.Y.J.; Jackson, R.A.A.; Thiery, J.P.P. Emt: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [Green Version]
- Grigore, A.; Jolly, M.; Jia, D.; Farach-Carson, M.; Levine, H. Tumor Budding: The Name is EMT. Partial EMT. J. Clin. Med. 2016, 5, 51. [Google Scholar] [CrossRef]
- Westcott, J.M.; Prechtl, A.M.; Maine, E.A.; Dang, T.T.; Esparza, M.A.; Sun, H.; Zhou, Y.; Xie, Y.; Pearson, G.W. An epigenetically distinct breast cancer cell subpopulation promotes collective invasion. J. Clin. Investig. 2015, 125, 1927–1943. [Google Scholar] [CrossRef]
- Gerashchenko, T.S.; Novikov, N.M.; Krakhmal, N.V.; Zolotaryova, S.Y.; Zavyalova, M.V.; Cherdyntseva, N.V.; Denisov, E.V.; Perelmuter, V.M. Markers of Cancer Cell Invasion: Are They Good Enough? J. Clin. Med. 2019, 8, 1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denisov, E.V.; Skryabin, N.A.; Gerashchenko, T.S.; Tashireva, L.A.; Wilhelm, J.; Buldakov, M.A.; Sleptcov, A.A.; Lebedev, I.N.; Vtorushin, S.V.; Zavyalova, M.V.; et al. Clinically relevant morphological structures in breast cancer represent transcriptionally distinct tumor cell populations with varied degrees of epithelial–mesenchymal transition and CD44+CD24- stemness. Oncotarget 2017, 8, 61163–61180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zavjalova, M.V.; Perelmuter, V.M.; Slonimskaya, E.M.; Vtorushin, S.V.; Garbukov, E.Y.; Gluschenko, S.A. Conjugation of lymphogenous metastatic spread and histologic pattern of infiltrative component of ductal breast cancer. Sib. J. Oncol. 2006, 1, 32–35. [Google Scholar]
- Gerashchenko, T.S.; Zavyalova, M.V.; Denisov, E.V.; Krakhmal, N.V.; Pautova, D.N.; Litviakov, N.V.; Vtorushin, S.V.; Cherdyntseva, N.V.; Perelmuter, V.M. Intratumoral morphological heterogeneity of breast cancer as an indicator of the metastatic potential and tumor chemosensitivity. Acta Naturae 2017, 9, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Tashireva, L.A.; Zavyalova, M.V.; Savelieva, O.E.; Gerashchenko, T.S.; Kaigorodova, E.V.; Denisov, E.V.; Perelmuter, V.M. Single Tumor Cells With Epithelial-Like Morphology Are Associated With Breast Cancer Metastasis. Front. Oncol. 2020, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef] [Green Version]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef] [Green Version]
- Miki, H.; Okada, Y.; Hirokawa, N. Analysis of the kinesin superfamily: Insights into structure and function. Trends Cell Biol. 2005, 15, 467–476. [Google Scholar] [CrossRef]
- Gruneberg, U.; Neef, R.; Li, X.; Chan, E.H.Y.; Chalamalasetty, R.B.; Nigg, E.A.; Barr, F.A. KIF14 and citron kinase act together to promote efficient cytokinesis. J. Cell Biol. 2006, 172, 363–372. [Google Scholar] [CrossRef] [Green Version]
- Corson, T.W.; Gallie, B.L. KIF14 mRNA expression is a predictor of grade and outcome in breast cancer. Int. J. Cancer 2006, 119, 1088–1094. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.M.; Thériault, B.L.; Uppalapati, M.; Chiu, C.W.N.; Gallie, B.L.; Sidhu, S.S.; Angers, S. KIF14 negatively regulates Rap 1 a-Radil signaling during breast cancer progression. J. Cell Biol. 2012, 199, 951–967. [Google Scholar] [CrossRef] [Green Version]
- Benjamin, D.I.; Cozzo, A.; Ji, X.; Roberts, L.S.; Louie, S.M.; Mulvihill, M.M.; Luo, K.; Nomura, D.K. Ether lipid generating enzyme AGPS alters the balance of structural and signaling lipids to fuel cancer pathogenicity. Proc. Natl. Acad. Sci. USA 2013, 110, 14912–14917. [Google Scholar] [CrossRef] [Green Version]
- Brooks, S.P.; Coccia, M.; Tang, H.R.; Kanuga, N.; Machesky, L.M.; Bailly, M.; Cheetham, M.E.; Hardcastle, A.J. The Nance-Horan syndrome protein encodes a functional WAVE homology domain (WHD) and is important for co-ordinating actin remodelling and maintaining cell morphology. Hum. Mol. Genet. 2010, 19, 2421–2432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, P.; Otten, A.B.C.; Cheng, B.; Ishii, M.A.; Zhang, W.; Huang, B.; Qu, K.; Sun, B.K. A genome-wide long noncoding RNA CRISPRi screen identifies PRANCR as a novel regulator of epidermal homeostasis. Genome Res. 2020, 30, 22–34. [Google Scholar] [CrossRef]
- Muppala, S.; Xiao, R.; Krukovets, I.; Verbovetsky, D.; Yendamuri, R.; Habib, N.; Raman, P.; Plow, E.; Stenina-Adognravi, O. Thrombospondin-4 mediates TGF-β-induced angiogenesis. Oncogene 2017, 36, 5189–5198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muppala, S.; Xiao, R.; Gajeton, J.; Krukovets, I.; Verbovetskiy, D.; Stenina-Adognravi, O. Thrombospondin-4 mediates hyperglycemia- and TGF-beta-induced inflammation in breast cancer. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kitamura, N.; Nakamura, Y.; Miyamoto, Y.; Miyamoto, T.; Kabu, K.; Yoshida, M.; Futamura, M.; Ichinose, S.; Arakawa, H. Mieap, a p53-inducible protein, controls mitochondrial quality by repairing or eliminating unhealthy mitochondria. PLoS ONE 2011, 6, e16060. [Google Scholar] [CrossRef]
- Kamino, H.; Nakamura, Y.; Tsuneki, M.; Sano, H.; Miyamoto, Y.; Kitamura, N.; Futamura, M.; Kanai, Y.; Taniguchi, H.; Shida, D.; et al. Mieap-regulated mitochondrial quality control is frequently inactivated in human colorectal cancer. Oncogenesis 2016, 5, e181. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, Y.; Arakawa, H. Discovery of Mieap-regulated mitochondrial quality control as a new function of tumor suppressor p53. Cancer Sci. 2017, 108, 809–817. [Google Scholar] [CrossRef] [Green Version]
- Bordi, M.; Nazio, F.; Campello, S. The Close Interconnection between Mitochondrial Dynamics and Mitophagy in Cancer. Front. Oncol. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vyas, S.; Zaganjor, E.; Haigis, M.C. Mitochondria and Cancer. Cell 2016, 166, 555–566. [Google Scholar] [CrossRef]
- Zhang, R.; Li, X.; Liu, Z.; Wang, Y.; Zhang, H.; Xu, H. EZH2 inhibitors-mediated epigenetic reactivation of FOSB inhibits triple-negative breast cancer progress. Cancer Cell Int. 2020, 20, 175. [Google Scholar] [CrossRef] [PubMed]
- Hoskin, V.; Szeto, A.; Ghaffari, A.; Greer, P.A.; Côté, G.P.; Elliott, B.E. Ezrin regulates focal adhesion and invadopodia dynamics by altering calpain activity to promote breast cancer cell invasion. Mol. Biol. Cell 2015, 26, 3464–3479. [Google Scholar] [CrossRef] [PubMed]
- Jais, M.H.; Md Zin, R.R.; Muhd Hanapi, N.A.; Md Ali, S.A. Ezrin is Significantly Overexpressed in Luminal A, Luminal B, and HER2 Subtype Breast Cancer. Appl. Immunohistochem. Mol. Morphol. 2017, 25, 44–48. [Google Scholar] [CrossRef]
- Yu, Z.; Sun, M.; Jin, F.; Xiao, Q.; He, M.; Wu, H.; Ren, J.; Zhao, L.; Zhao, H.; Yao, W.; et al. Combined expression of ezrin and E-cadherin is associated with lymph node metastasis and poor prognosis in breast cancer. Oncol. Rep. 2015, 34, 165–174. [Google Scholar] [CrossRef]
- Ghaffari, A.; Hoskin, V.; Turashvili, G.; Varma, S.; Mewburn, J.; Mullins, G.; Greer, P.A.; Kiefer, F.; Day, A.G.; Madarnas, Y.; et al. Intravital imaging reveals systemic ezrin inhibition impedes cancer cell migration and lymph node metastasis in breast cancer. Breast Cancer Res. 2019, 21, 1–11. [Google Scholar] [CrossRef]
- Kim, S.; Karin, M. Role of TLR2-dependent inflammation in metastatic progression. Ann. N. Y. Acad. Sci. 2011, 1217, 191–206. [Google Scholar] [CrossRef]
- Wang, W.; Xu, G.-L.; Jia, W.-D.; Ma, J.-L.; Li, J.-S.; Ge, Y.-S.; Ren, W.-H.; Yu, J.-H.; Liu, W.-B. Ligation of TLR2 by Versican: A Link Between Inflammation and Metastasis. Arch. Med. Res. 2009, 40, 321–323. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.F.; Gao, W.D.; Li, Q.L.; Zhou, P.H.; Xu, M.D.; Yao, L.Q. SLIT2 inhibits cell migration in colorectal cancer through the AKT-GSK3β signaling pathway. Int. J. Colorectal Dis. 2013, 28, 933–940. [Google Scholar] [CrossRef]
- Sharma, G.; Mirza, S.; Prasad, C.P.; Srivastava, A.; Gupta, S.D.; Ralhan, R. Promoter hypermethylation of p16INK4A, p14ARF, CyclinD2 and Slit2 in serum and tumor DNA from breast cancer patients. Life Sci. 2007, 80, 1873–1881. [Google Scholar] [CrossRef]
- Zhang, C.; Guo, H.; Li, B.; Sui, C.; Zhang, Y.; Xia, X.; Qin, Y.; Ye, L.; Xie, F.; Wang, H.; et al. Effects of Slit3 silencing on the invasive ability of lung carcinoma A549 cells. Oncol. Rep. 2015, 34, 952–960. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Kim, J.-H.; Baek, S.-J.; Kim, S.-Y.; Kim, Y.S. Specific expression and methylation of SLIT1, SLIT2, SLIT3, and miR-218 in gastric cancer subtypes. Int. J. Oncol. 2016, 48, 2497–2507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, J.; Li, H.; Li, Q.; Yang, Y. CD109 is a potential target for triple-negative breast cancer. Tumor Biol. 2014, 35, 12083–12090. [Google Scholar] [CrossRef]
- Junankar, S.; Baker, L.A.; Roden, D.L.; Nair, R.; Elsworth, B.; Gallego-Ortega, D.; Lacaze, P.; Cazet, A.; Nikolic, I.; Teo, W.S.; et al. ID4 controls mammary stem cells and marks breast cancers with a stem cell-like phenotype. Nat. Commun. 2015, 6, 6548. [Google Scholar] [CrossRef]
- Nguyen, K.; Yan, Y.; Yuan, B.; Dasgupta, A.; Sun, J.; Mu, H.; Do, K.-A.; Ueno, N.T.; Andreeff, M.; Battula, V.L. ST8SIA1 Regulates Tumor Growth and Metastasis in TNBC by Activating the FAK–AKT–mTOR Signaling Pathway. Mol. Cancer Ther. 2018, 17, 2689–2701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, F.; Drabsch, Y.; Dekker, T.J.A.; de Vinuesa, A.G.; Li, Y.; Hawinkels, L.J.A.C.; Sheppard, K.-A.; Goumans, M.-J.; Luwor, R.B.; de Vries, C.J.; et al. Nuclear receptor NR4A1 promotes breast cancer invasion and metastasis by activating TGF-β signalling. Nat. Commun. 2014, 5, 3388. [Google Scholar] [CrossRef] [Green Version]
- Koboldt, D.C.; Fulton, R.S.; McLellan, M.D.; Schmidt, H.; Kalicki-Veizer, J.; McMichael, J.F.; Fulton, L.L.; Dooling, D.J.; Ding, L.; Mardis, E.R.; et al. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Goliwas, K.F.; Wang, W.; Taufalele, P.V.; Bordeleau, F.; Reinhart-King, C.A. Energetic regulation of coordinated leader–follower dynamics during collective invasion of breast cancer cells. Proc. Natl. Acad. Sci. USA 2019, 116, 7867–7872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aoki, T.; Nishita, M.; Sonoda, J.; Ikeda, T.; Kakeji, Y.; Minami, Y. Intraflagellar transport 20 promotes collective cancer cell invasion by regulating polarized organization of Golgi-associated microtubules. Cancer Sci. 2019, 110, 1306–1316. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Cao, M.; Liu, Y.; He, Y.; Du, Y.; Zhang, G.; Gao, F. Inducible formation of leader cells driven by CD44 switching gives rise to collective invasion and metastases in luminal breast carcinomas. Oncogene 2019, 38, 7113–7132. [Google Scholar] [CrossRef]
- Commander, R.; Wei, C.; Sharma, A.; Mouw, J.K.; Burton, L.J.; Summerbell, E.; Mahboubi, D.; Peterson, R.J.; Konen, J.; Zhou, W.; et al. Subpopulation targeting of pyruvate dehydrogenase and GLUT1 decouples metabolic heterogeneity during collective cancer cell invasion. Nat. Commun. 2020, 11, 1533. [Google Scholar] [CrossRef] [Green Version]
- Minn, A.J.; Gupta, G.P.; Siegel, P.M.; Bos, P.D.; Shu, W.; Giri, D.D.; Viale, A.; Olshen, A.B.; Gerald, W.L.; Massagué, J. Genes that mediate breast cancer metastasis to lung. Nature 2005, 436, 518–524. [Google Scholar] [CrossRef]
- Minn, A.J.; Gupta, G.P.; Padua, D.; Bos, P.; Nguyen, D.X.; Nuyten, D.; Kreike, B.; Zhang, Y.; Wang, Y.; Ishwaran, H.; et al. Lung metastasis genes couple breast tumor size and metastatic spread. Proc. Natl. Acad. Sci. USA 2007, 104, 6740–6745. [Google Scholar] [CrossRef] [Green Version]
- Gupta, G.P.; Nguyen, D.X.; Chiang, A.C.; Bos, P.D.; Kim, J.Y.; Nadal, C.; Gomis, R.R.; Manova-Todorova, K.; Massagué, J. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature 2007, 446, 765–770. [Google Scholar] [CrossRef]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [Green Version]
- Zavyalova, M.V.; Denisov, E.V.; Tashireva, L.A.; Gerashchenko, T.S.; Litviakov, N.V.; Skryabin, N.A.; Vtorushin, S.V.; Telegina, N.S.; Slonimskaya, E.M.; Cherdyntseva, N.V.; et al. Phenotypic Drift as a Cause for Intratumoral Morphological Heterogeneity of Invasive Ductal Breast Carcinoma Not Otherwise Specified. Biores. Open Access 2013, 2, 148–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Maayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 2013, 14, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
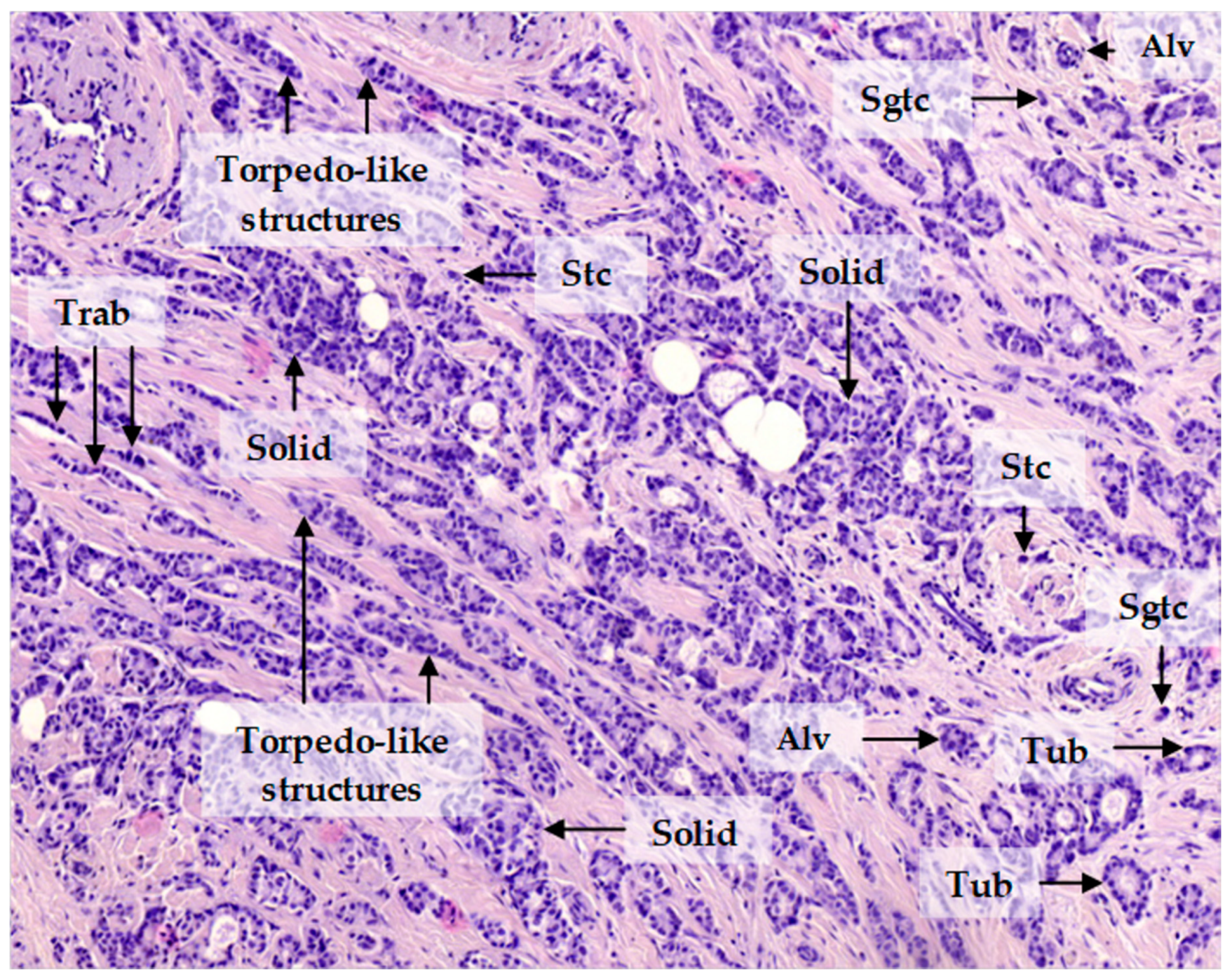

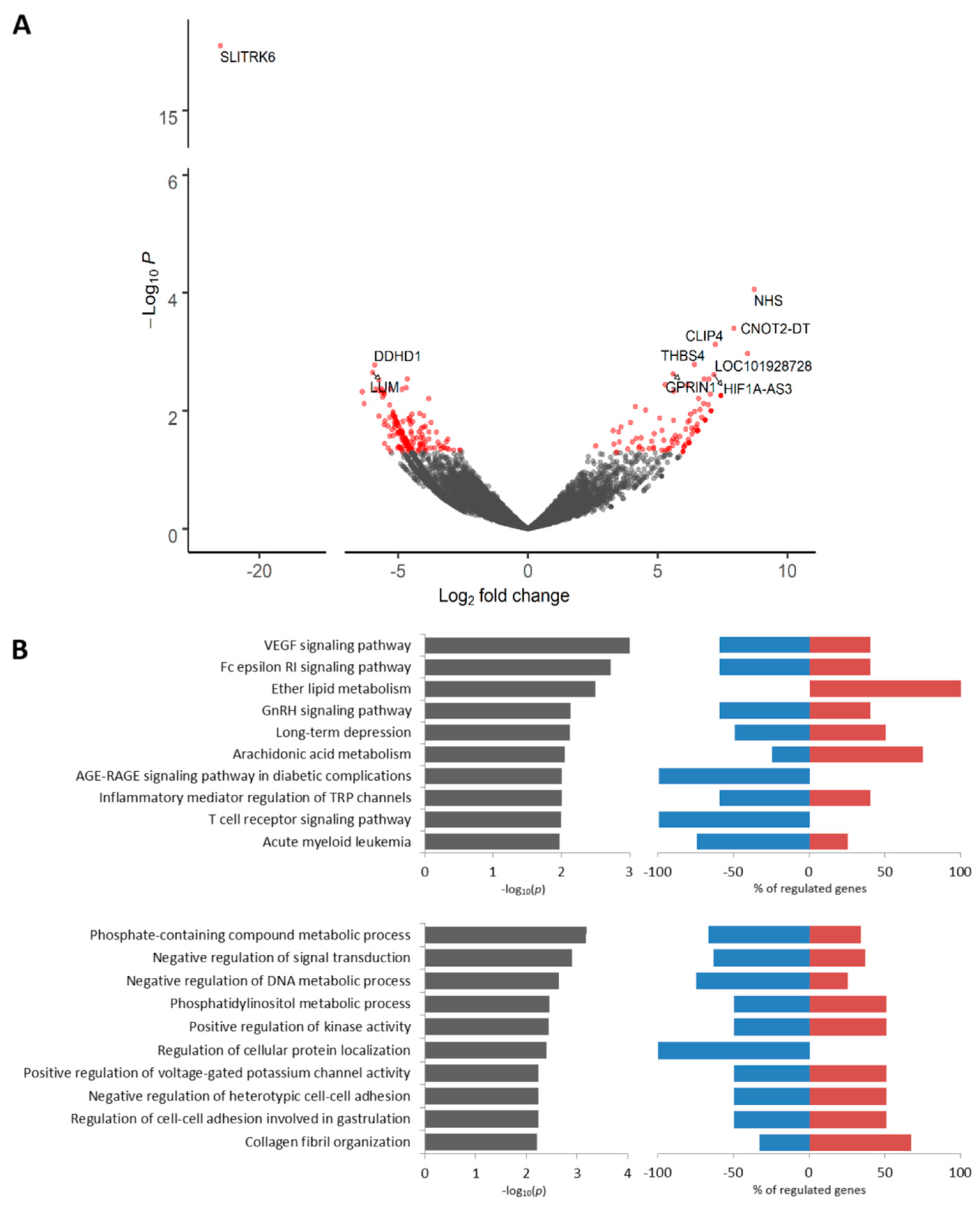
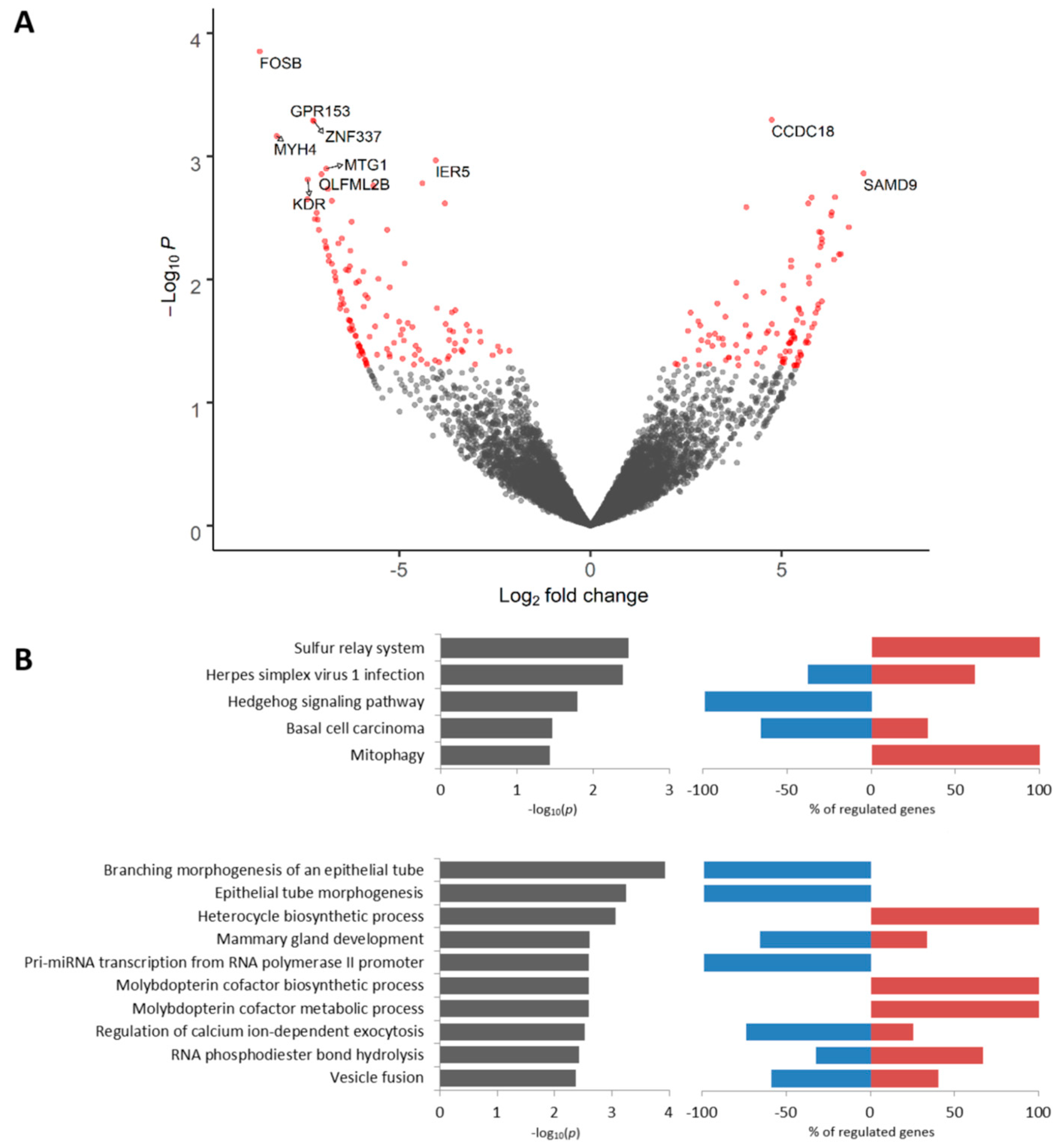
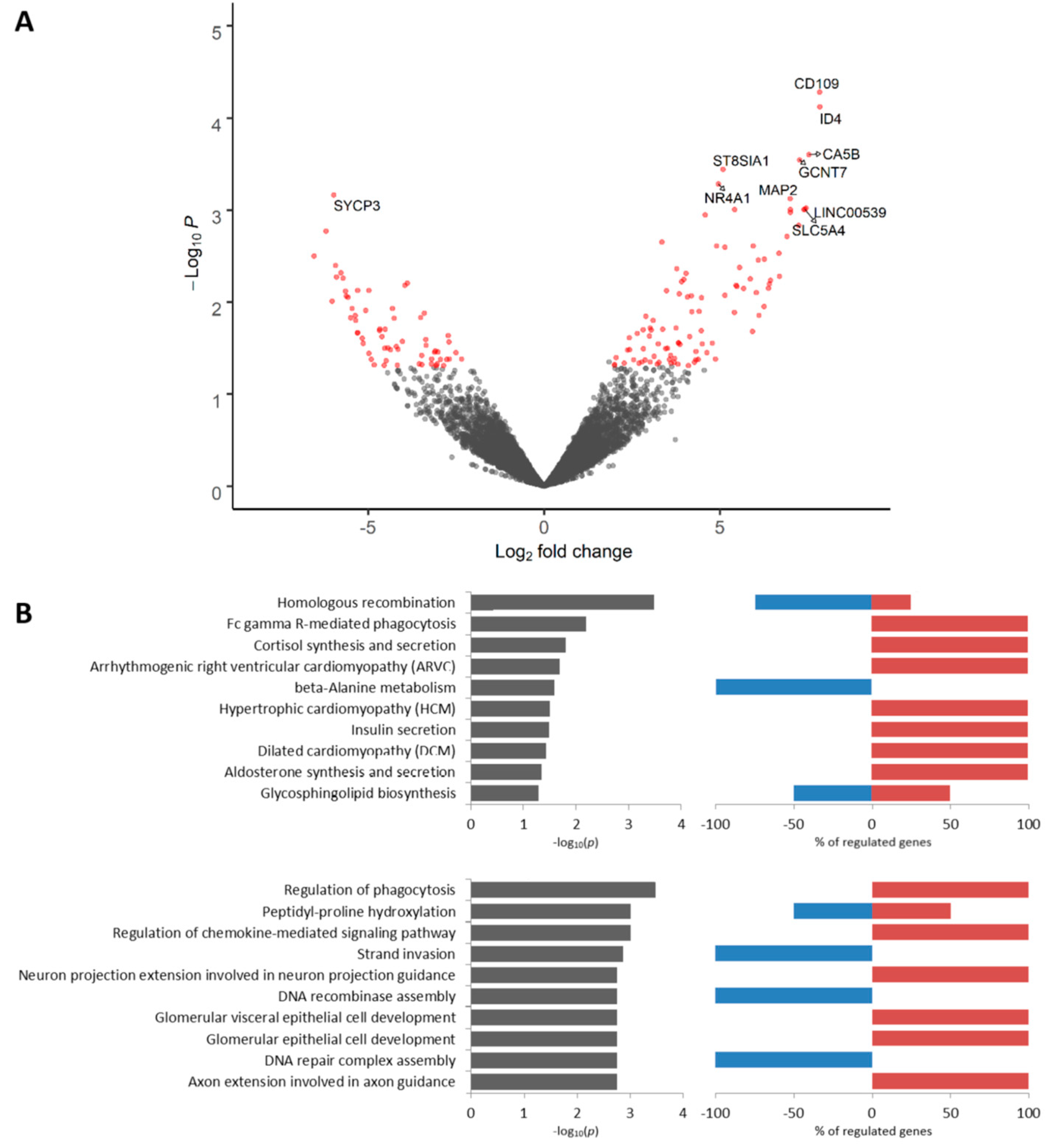
| Gene | Log-Fold Change | Function | Protein Expression Patterns in Breast Cancer ‡ | ||||
|---|---|---|---|---|---|---|---|
| Tub | Alv | Sol | Trab | Discr | |||
| HAX1 | 0.61 | 1.14 * | 1.42 † | 1.27 * | −0.21 | Regulator of cortical actin cytoskeleton | Positive expression at the periphery of solid structures |
| KIF14 | 1.34 | 2.56 | 3.87 * | 3.42 * | 2.73 | Microtubule motor protein | Positive expression at the tips of solid structures |
| SPATA18 (Mieap) | −1.68 * | −1.62 | −2.71 † | −3.02 † | −4.09 † | Regulation of mitochondrial quality and viability | Positive expression at the tips of solid structures |
| EZR | 1.73 * | 1.09 | 1.99 † | 1.91 † | ND | Connection of major cytoskeletal structures to the plasma membrane | Negative expression at the tips of solid structures |
| Distant Metastasis | Nuclear Expression of KIF14 | Cytoplasmic Expression of Mieap | Cytoplasmic Expression of EZR | |||
|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | Positive | Negative | |
| No | 7 (33.3) | 12 (85.7) | 6 (30.0) | 14 (87.5) | 13 (86.6) | 5 (26.3) |
| Yes | 14 (66.7) | 2 (14.3) | 14 (70.0) | 2 (12.5) | 2 (13.3) | 14 (73.7) |
| p-value * | 0.003 | 0.001 | 0.001 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gerashchenko, T.S.; Zolotaryova, S.Y.; Kiselev, A.M.; Tashireva, L.A.; Novikov, N.M.; Krakhmal, N.V.; Cherdyntseva, N.V.; Zavyalova, M.V.; Perelmuter, V.M.; Denisov, E.V. The Activity of KIF14, Mieap, and EZR in a New Type of the Invasive Component, Torpedo-Like Structures, Predetermines the Metastatic Potential of Breast Cancer. Cancers 2020, 12, 1909. https://doi.org/10.3390/cancers12071909
Gerashchenko TS, Zolotaryova SY, Kiselev AM, Tashireva LA, Novikov NM, Krakhmal NV, Cherdyntseva NV, Zavyalova MV, Perelmuter VM, Denisov EV. The Activity of KIF14, Mieap, and EZR in a New Type of the Invasive Component, Torpedo-Like Structures, Predetermines the Metastatic Potential of Breast Cancer. Cancers. 2020; 12(7):1909. https://doi.org/10.3390/cancers12071909
Chicago/Turabian StyleGerashchenko, Tatiana S., Sofia Y. Zolotaryova, Artem M. Kiselev, Liubov A. Tashireva, Nikita M. Novikov, Nadezhda V. Krakhmal, Nadezhda V. Cherdyntseva, Marina V. Zavyalova, Vladimir M. Perelmuter, and Evgeny V. Denisov. 2020. "The Activity of KIF14, Mieap, and EZR in a New Type of the Invasive Component, Torpedo-Like Structures, Predetermines the Metastatic Potential of Breast Cancer" Cancers 12, no. 7: 1909. https://doi.org/10.3390/cancers12071909
APA StyleGerashchenko, T. S., Zolotaryova, S. Y., Kiselev, A. M., Tashireva, L. A., Novikov, N. M., Krakhmal, N. V., Cherdyntseva, N. V., Zavyalova, M. V., Perelmuter, V. M., & Denisov, E. V. (2020). The Activity of KIF14, Mieap, and EZR in a New Type of the Invasive Component, Torpedo-Like Structures, Predetermines the Metastatic Potential of Breast Cancer. Cancers, 12(7), 1909. https://doi.org/10.3390/cancers12071909






