Integrin Crosstalk Contributes to the Complexity of Signalling and Unpredictable Cancer Cell Fates
Abstract
:1. Introduction
2. Regulation of Integrin Expression
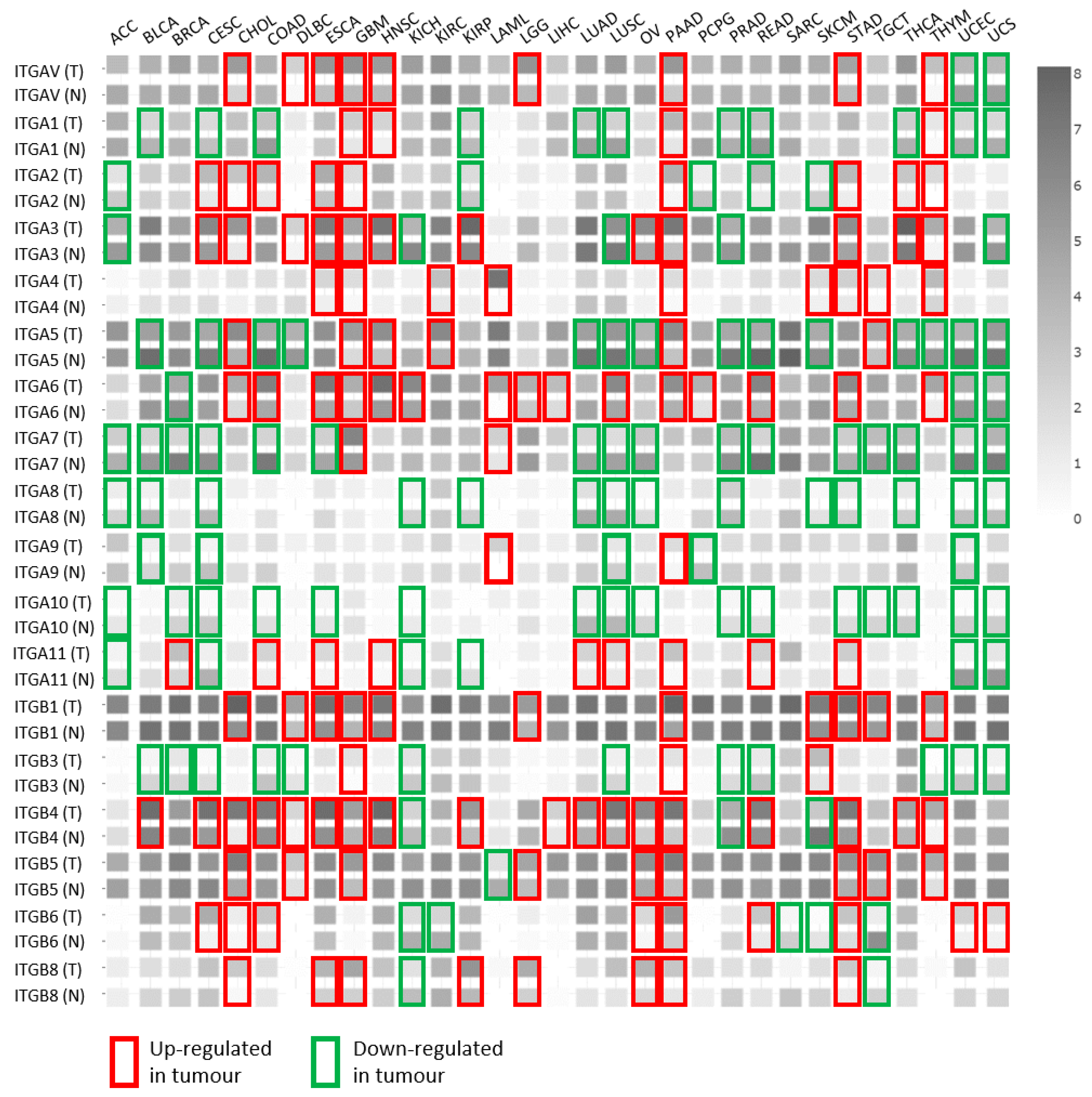
3. The Integrin Repertoire Is Changed or ”Switched” in Cancer Cells—an Indication of Integrin Crosstalk Events
4. Integrin Crosstalk
5. Molecular Mechanisms of Integrin Crosstalk
5.1. Changes in Stoichiometry of Integrin Subunits
5.2. Effects on Transcription
5.3. Effects on Integrin Maturation
5.4. Changes in Integrin Post-translational Modifications
5.5. Changes in Transport and Recycling
5.6. Competition for Intracellular Activators
6. Is Integrin Crosstalk One of the Reasons for Integrin Targeting Therapy Failure?
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ECM | cell-extracellular matrix |
| IACs | integrin adhesion complexes |
| NAs | nascent adhesions |
| FAs | focal adhesions |
| FBs | fibrillar adhesions |
| RAs | reticular adhesions |
| HDs | hemidesmosomes |
| ER | endoplasmic reticulum |
| MoAbs | monoclonal antibodies |
References
- Horton, E.R.; Humphries, J.D.; James, J.; Jones, M.C.; Askari, J.A.; Humphries, M.J. The integrin adhesome network at a glance. J. Cell Sci. 2016, 129, 4159–4163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamidi, H.; Ivaska, J. Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 2018, 18, 533–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, J.; Giancotti, F.G. Integrin Signaling in Cancer: Mechanotransduction, Stemness, Epithelial Plasticity, and Therapeutic Resistance. Cancer Cell 2019, 35, 347–367. [Google Scholar] [CrossRef]
- Green, H.; Brown, N.H. Integrin intracellular machinery in action. Exp. Cell Res. 2019, 378, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Humphries, J.D.; Chastney, M.R.; Askari, J.A.; Humphries, M.J. Signal transduction via integrin adhesion complexes. Curr. Opin. Cell Biol. 2019, 56, 14–21. [Google Scholar] [CrossRef] [Green Version]
- Michael, M.; Parsons, M. New perspectives on integrin-dependent adhesions. Curr. Opin. Cell Biol. 2020, 63, 31–37. [Google Scholar] [CrossRef]
- Seguin, L.; Desgrosellier, J.S.; Weis, S.M.; Cheresh, D.A. Integrins and cancer: Regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol. 2015, 25, 234–240. [Google Scholar] [CrossRef] [Green Version]
- Hamidi, H.; Pietilä, M.; Ivaska, J. The complexity of integrins in cancer and new scopes for therapeutic targeting. Br. J. Cancer 2016, 115, 1017–1023. [Google Scholar] [CrossRef] [Green Version]
- Raab-Westphal, S.; Marshall, J.; Goodman, S.L. Integrins as Therapeutic Targets: Successes and Cancers. Cancers 2017, 9, 110. [Google Scholar] [CrossRef]
- Vicente-Manzanares, M.; Sanchez-Madrid, F. Targeting the integrin interactome in human disease. Curr. Opin. Cell Biol. 2018, 55, 17–23. [Google Scholar] [CrossRef]
- Byron, A.; Frame, M. Adhesion protein networks reveal functions proximal and distal to cell-matrix contacts. Curr. Opin. Cell Biol. 2016, 39, 93–100. [Google Scholar] [CrossRef] [Green Version]
- Burridge, K. Focal adhesions: A personal perspective on a half century of progress. FEBS J. 2017, 284, 3355–3361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lock, J.G.; Jones, M.C.; Askari, J.; Gong, X.; Oddone, A.; Olofsson, H.; Göransson, S.; Lakadamyali, M.; Humphries, M.J.; Stromblad, S. Reticular adhesions are a distinct class of cell-matrix adhesions that mediate attachment during mitosis. Nature 2018, 20, 1290–1302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Zuidema, A.; Molder, L.T.; Nahidiazar, L.; Hoekman, L.; Schmidt, T.; Coppola, S.; Sonnenberg, A. Hemidesmosomes modulate force generation via focal adhesions. J. Cell Biol. 2020, 219. [Google Scholar] [CrossRef] [PubMed]
- Zaidel-Bar, R.; Itzkovitz, S.; Ma’Ayan, A.; Iyengar, R.; Geiger, B. Functional atlas of the integrin adhesome. Nature 2007, 9, 858–867. [Google Scholar] [CrossRef]
- Zaidel-Bar, R.; Geiger, B. The switchable integrin adhesome. J. Cell Sci. 2010, 123, 1385–1388. [Google Scholar] [CrossRef] [Green Version]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef] [Green Version]
- Aoudjit, F.; Vuori, K. Integrin Signaling in Cancer Cell Survival and Chemoresistance. Chemother. Res. Pract. 2012, 2012, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Winograd-Katz, S.E.; Fässler, R.; Geiger, B.; Legate, K. The integrin adhesome: From genes and proteins to human disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 273–288. [Google Scholar] [CrossRef]
- Horton, E.R.; Byron, A.; Askari, J.A.; Ng, D.H.J.; Millon-Frémillon, A.; Robertson, J.; Koper, E.J.; Paul, N.R.; Warwood, S.; Knight, D.; et al. Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nature 2015, 17, 1577–1587. [Google Scholar] [CrossRef] [Green Version]
- Walko, G.; Castañón, M.J.; Wiche, G. Molecular architecture and function of the hemidesmosome. Cell Tissue Res. 2014, 360, 363–378. [Google Scholar] [CrossRef] [Green Version]
- Costa, P.; Parsons, M. New Insights into the Dynamics of Cell Adhesions. Int. Rev. Cell Mol. Biol. 2010, 283, 57–91. [Google Scholar] [CrossRef] [PubMed]
- Albiges-Rizo, C.; Destaing, O.; Fourcade, B.; Planus, E.; Block, M.R. Actin machinery and mechanosensitivity in invadopodia, podosomes and focal adhesions. J. Cell Sci. 2009. [Google Scholar] [CrossRef] [Green Version]
- Zamir, E.; Katz, M.; Posen, Y.; Erez, N.; Yamada, K.M.; Katz, B.-Z.; Lin, S.; Lin, D.C.; Bershadsky, A.; Kam, Z.; et al. Dynamics and segregation of cell–matrix adhesions in cultured fibroblasts. Nature 2000, 2, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Zamir, E.; Katz, B.Z.; Aota, S.; Yamada, K.M.; Geiger, B.; Kam, Z. Molecular diversity of cell-matrix adhesions. J. Cell Sci. 1999, 112, 1655–1669. [Google Scholar]
- Lock, J.G.; Baschieri, F.; Jones, M.C.; Humphries, J.D.; Montagnac, G.; Stromblad, S.; Humphries, M.J. Clathrin-containing adhesion complexes. J. Cell Biol. 2019, 218, 2086–2095. [Google Scholar] [CrossRef]
- Schwartz, M.A.; Ginsberg, M.H. Networks and crosstalk: Integrin signalling spreads. Nat. Cell Biol. 2002. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.M.; Bhattacharya, R.; Dehart, G.W.; Jones, J.C. Transdominant regulation of integrin function: Mechanisms of crosstalk. Cell. Signal. 2010, 22, 578–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Díaz-González, F.; Forsyth, J.; Steiner, B.; Ginsberg, M.H. Trans-dominant inhibition of integrin function. Mol. Biol. Cell 1996, 7, 1939–1951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hight-Warburton, W.; Parsons, M. Regulation of cell migration by α4 and α9 integrins. Biochem. J. 2019, 476, 705–718. [Google Scholar] [CrossRef]
- Hynes, R.O. Integrins. Cell 2002, 110, 673–687. [Google Scholar] [CrossRef] [Green Version]
- Ho, M.K.; Springer, T.A. Biosynthesis and assembly of the alpha and beta subunits of Mac-1, a macrophage glycoprotein associated with complement receptor function. J. Biol. Chem. 1983, 258, 2766–2769. [Google Scholar]
- Lu, C.; Oxvig, C.; Springer, T.A. The Structure of the β-Propeller Domain and C-terminal Region of the Integrin αM Subunit. J. Biol. Chem. 1998, 273, 15138–15147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiwari, S.; Askari, J.A.; Humphries, M.J.; Bulleid, N.J. Divalent cations regulate the folding and activation status of integrins during their intracellular trafficking. J. Cell Sci. 2011, 124, 1672–1680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, B.-H.; Springer, T.A. Integrin structures and conformational signaling. Curr. Opin. Cell Biol. 2006, 18, 579–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, L.R.; Owens, T.W.; Naylor, M.J. Structural and mechanical functions of integrins. Biophys. Rev. 2013, 6, 203–213. [Google Scholar] [CrossRef] [Green Version]
- Campbell, I.D.; Humphries, M.J. Integrin Structure, Activation, and Interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a004994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Z.; Costell, M.; Fässler, R. Integrin activation by talin, kindlin and mechanical forces. Nature 2019, 21, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Ginsberg, M.H.; Partridge, A.; Shattil, S.J. Integrin regulation. Curr. Opin. Cell Biol. 2005, 17, 509–516. [Google Scholar] [CrossRef]
- Delcommenne, M.; Streuli, C.H. Control of Integrin Expression by Extracellular Matrix. J. Biol. Chem. 1995, 270, 26794–26801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, L.T.; Yamada, K.M. The Regulation of Expression of Integrin Receptors. Exp. Biol. Med. 1997, 214, 123–131. [Google Scholar] [CrossRef]
- Pechkovsky, D.; Scaffidi, A.K.; Hackett, T.L.; Ballard, J.; Shaheen, F.; Thompson, P.J.; Thannickal, V.J.; Knight, D.A. Transforming Growth Factor β1 Induces αvβ3 Integrin Expression in Human Lung Fibroblasts via a β3 Integrin-, c-Src-, and p38 MAPK-dependent Pathway. J. Biol. Chem. 2008, 283, 12898–12908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Harbeck, M.C.; Zhang, W.; Jacobson, J.R. MicroRNA regulation of integrins. Transl. Res. 2013, 162, 133–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.-J.; Park, M.-J.; Kim, B.-S.; Choi, H.-J.; Joo, B.; Lee, K.S.; Choi, J.-H.; Chung, T.-W.; Ha, K.-T. Transforming growth factor β1 enhances adhesion of endometrial cells to mesothelium by regulating integrin expression. BMB Rep. 2017, 50, 429–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, N.F.; Marshall, J. Integrin-Mediated TGFβ Activation Modulates the Tumour Microenvironment. Cancers 2019, 11, 1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koistinen, P.; Heino, J. The Selective Regulation of αVβ1 Integrin Expression Is Based on the Hierarchical Formation of αV-containing Heterodimers. J. Biol. Chem. 2002, 277, 24835–24841. [Google Scholar] [CrossRef] [Green Version]
- Heino, J.; Ignotz, R.A.; Hemler, M.E.; Crouse, C.; Massague, J. Regulation of cell adhesion receptors by transforming growth factor-β. Concomitant regulation of integrins that share a common β1 subunit. J. Biol. Chem. 1989, 264, 380–388. [Google Scholar]
- Moreno-Layseca, P.; Icha, J.; Hamidi, H.; Ivaska, J. Integrin trafficking in cells and tissues. Nature 2019, 21, 122–132. [Google Scholar] [CrossRef]
- O’Reilly, A.M.; Lee, H.-H.; Simon, M.A. Integrins control the positioning and proliferation of follicle stem cells in the Drosophila ovary. J. Cell Biol. 2008, 182, 801–815. [Google Scholar] [CrossRef] [Green Version]
- Bouvard, D.; Pouwels, J.; De Franceschi, N.; Ivaska, J. Integrin inactivators: Balancing cellular functions in vitro and in vivo. Nat. Rev. Mol. Cell Biol. 2013, 14, 430–442. [Google Scholar] [CrossRef]
- Ganguly, K.K.; Pal, S.; Moulik, S.; Chatterjee, A. Integrins and metastasis. Cell Adhes. Migr. 2013, 7, 251–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, N.R.; Jacquemet, G.; Caswell, P.T. Endocytic Trafficking of Integrins in Cell Migration. Curr. Biol. 2015, 25, R1092–R1105. [Google Scholar] [CrossRef]
- Ata, R.; Antonescu, C.N. Integrins and Cell Metabolism: An Intimate Relationship Impacting Cancer. Int. J. Mol. Sci. 2017, 18, 189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Pascalis, C.; Etienne-Manneville, S. Single and collective cell migration: The mechanics of adhesions. Mol. Biol. Cell 2017, 28, 1833–1846. [Google Scholar] [CrossRef]
- Longmate, W.; DiPersio, C.M. Beyond adhesion: Emerging roles for integrins in control of the tumor microenvironment. F1000Research 2017, 6, 1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alday-Parejo, B.; Stupp, R.; Rüegg, C.; Parejo, A. Are Integrins Still Practicable Targets for Anti-Cancer Therapy? Cancers 2019, 11, 978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madamanchi, A.; Zijlstra, A.; Zutter, M.M. Flipping the Switch: Integrin Switching Provides Metastatic Competence. Sci. Signal. 2014, 7, pe9. [Google Scholar] [CrossRef] [Green Version]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [Green Version]
- Jones, V.A.; Patel, P.M.; Gibson, F.T.; Cordova, A.; Amber, K.T. The Role of Collagen XVII in Cancer: Squamous Cell Carcinoma and Beyond. Front. Oncol. 2020, 10, 352. [Google Scholar] [CrossRef]
- Arun, A.S.; Tepper, C.G.; Lam, K.S. Identification of integrin drug targets for 17 solid tumor types. Oncotarget 2018, 9, 30146–30162. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, U.B.; Westphal, J.R.; Waas, E.T.; Becker, J.C.; Ruiter, D.J.; Van Muijen, G.N. Coexpression of Integrin αvβ3 and Matrix Metalloproteinase-2 (MMP-2) Coincides with MMP-2 Activation: Correlation with Melanoma Progression. J. Investig. Dermatol. 2000, 115, 625–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikkola, J.; Vihinen, P.; Vlaykova, T.; Hahka-Kemppinen, M.; Heino, J.; Pyrhönen, S. Integrin chains β1 and αv as prognostic factors in human metastatic melanoma. Melanoma Res. 2004, 14, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Ruffini, F.; D’Atri, S.; Lacal, P.M. Neuropilin-1 expression promotes invasiveness of melanoma cells through vascular endothelial growth factor receptor-2-dependent and -independent mechanisms. Int. J. Oncol. 2013, 43, 297–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stojanović, N.; Dekanić, A.; Paradžik, M.; Majhen, A.; Ferenčak, K.; Ruščić, J.; Bardak, I.; Supina, C.; Tomicic, M.T.; Christmann, M.; et al. Differential Effects of Integrin αv Knockdown and Cilengitide on Sensitization of Triple-Negative Breast Cancer and Melanoma Cells to Microtubule Poisons. Mol. Pharmacol. 2018, 94, 1334–1351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paradžik, M.; Humphries, J.D.; Stojanović, N.; Nestić, D.; Majhen, D.; Dekanić, A.; Samaržija, I.; Sedda, D.; Weber, I.; Humphries, M.J.; et al. KANK2 Links αVβ5 Focal Adhesions to Microtubules and Regulates Sensitivity to Microtubule Poisons and Cell Migration. Front. Cell Dev. Biol. 2020, 8, 125. [Google Scholar] [CrossRef] [PubMed]
- Logue, J.S.; Morrison, D.K. Complexity in the signaling network: Insights from the use of targeted inhibitors in cancer therapy. Genes Dev. 2012, 26, 641–650. [Google Scholar] [CrossRef] [Green Version]
- Trusolino, L.; Bertotti, A. Compensatory Pathways in Oncogenic Kinase Signaling and Resistance to Targeted Therapies: Six Degrees of Separation: Figure 1. Cancer Discov. 2012, 2, 876–880. [Google Scholar] [CrossRef] [Green Version]
- Von Manstein, V.; Yang, C.M.; Richter, D.; Delis, N.; Vafaizadeh, V.; Groner, B. Resistance of Cancer Cells to Targeted Therapies Through the Activation of Compensating Signaling Loops. Curr. Signal Transduct. Ther. 2013, 8, 193–202. [Google Scholar] [CrossRef]
- Parlato, R.; Mastroberardino, P.G. Editorial: Neuronal Self-Defense: Compensatory Mechanisms in Neurodegenerative Disorders. Front. Cell. Neurosci. 2016, 9. [Google Scholar] [CrossRef] [Green Version]
- Nish, S.; Medzhitov, R. Host Defense Pathways: Role of Redundancy and Compensation in Infectious Disease Phenotypes. Immunity 2011, 34, 629–636. [Google Scholar] [CrossRef] [Green Version]
- Blystone, S.D.; Graham, I.L.; Lindberg, F.P.; Brown, E.J. Integrin αvβ3 differentially regulates adhesive and phagocytic functions of the fibronectin receptor α5β1. J. Cell Biol. 1994, 127, 1129–1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popov, C.; Radic, T.; Haasters, F.; Prall, W.C.; Aszodi, A.; Gullberg, D.; Schieker, M.; Docheva, D. Integrins α2β1 and α11β1 regulate the survival of mesenchymal stem cells on collagen I. Cell Death Dis. 2011, 2, e186. [Google Scholar] [CrossRef] [PubMed]
- Borza, C.M.; Pedchenko, V.; Hellmark, T.; Hudson, B.G.; Zent, R.; Pozzi, A.; Borza, D.-B. Integrin α3β1, a Novel Receptor for 3(IV) Noncollagenous Domain and a Trans-dominant Inhibitor for Integrin αvβ3. J. Biol. Chem. 2006, 281, 20932–20939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, J.R.; Hyduk, S.J.; Cybulsky, M.I. α4β1 Integrin/VCAM-1 Interaction Activates αLβ2 Integrin-Mediated Adhesion to ICAM-1 in Human T Cells. J. Immunol. 2000, 164, 746–753. [Google Scholar] [CrossRef] [Green Version]
- Rose, D.M.; Liu, S.; Woodside, D.G.; Han, J.; Schlaepfer, D.D.; Ginsberg, M.H. Paxillin Binding to the α4 Integrin Subunit Stimulates LFA-1 (Integrin αLβ2)-Dependent T Cell Migration by Augmenting the Activation of Focal Adhesion Kinase/Proline-Rich Tyrosine Kinase-2. J. Immunol. 2003, 170, 5912–5918. [Google Scholar] [CrossRef] [Green Version]
- Cantor, J.M.; Rose, D.M.; Slepak, M.; Ginsberg, M.H. Fine-tuning Tumor Immunity with Integrin Trans-regulation. Cancer Immunol. Res. 2015, 3, 661–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Lu, H.; Dazin, P.; Kapila, Y.L. Functional differences between integrin α4 and integrins α5/αv in modulating the motility of human oral squamous carcinoma cells in response to the V region and heparin-binding domain of fibronectin. Exp. Cell Res. 2004, 295, 48–58. [Google Scholar] [CrossRef]
- Kim, S.; Harris, M.; Varner, J. Regulation of Integrin αvβ3-mediated Endothelial Cell Migration and Angiogenesis by Integrin α5β1and Protein Kinase A. J. Biol. Chem. 2000, 275, 33920–33928. [Google Scholar] [CrossRef] [Green Version]
- Diaz, C.; Neubauer, S.; Rechenmacher, F.; Kessler, H.; Missirlis, D. Recruitment of ανβ3 integrin to α5β1 integrin-induced clusters enables focal adhesion maturation and cell spreading. J. Cell Sci. 2019, 133, jcs232702. [Google Scholar] [CrossRef]
- Pijuan-Thompson, V.; Gladson, C.L. Ligation of Integrin α5β1Is Required for Internalization of Vitronectin by Integrin αvβ3. J. Biol. Chem. 1997, 272, 2736–2743. [Google Scholar] [CrossRef] [Green Version]
- Ly, D.P.; Zazzali, K.M.; Corbett, S.A. De Novo Expression of the Integrin α5β1 Regulates αvβ3-mediated Adhesion and Migration on Fibrinogen. J. Biol. Chem. 2003, 278, 21878–21885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandström, G.; Sjöstedt, A.; Tärnvik, A. The First International Conference on Tularemia: Umeå, Sweden, 23–25 August 1995. FEMS Immunol. Med. Microbiol. 1996, 13, 179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, D.P.; Caswell, P.T.; Norman, J.C. αvβ3 and α5β1 integrin recycling pathways dictate downstream Rho kinase signaling to regulate persistent cell migration. J. Cell Biol. 2007, 177, 515–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bharadwaj, M.; Strohmeyer, N.; Colo, G.P.; Helenius, J.; Beerenwinkel, N.; Schiller, H.B.; Fässler, R.; Müller, D.J. αV-class integrins exert dual roles on α5β1 integrins to strengthen adhesion to fibronectin. Nat. Commun. 2017, 8, 14348. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, T.; Jones, J.C.R.; Lee, C.K.; Schnaper, H.W. Loss of β1-Integrin Enhances TGF-β1-induced Collagen Expression in Epithelial Cells via Increased αvβ3-Integrin and Rac1 Activity*. J. Biol. Chem. 2010, 285, 30741–30751. [Google Scholar] [CrossRef] [Green Version]
- Huck, L.; Pontier, S.; Zuo, D.M.; Muller, W.J. β1-integrin is dispensable for the induction of ErbB2 mammary tumors but plays a critical role in the metastatic phase of tumor progression. Proc. Natl. Acad. Sci. USA 2010, 107, 15559–15564. [Google Scholar] [CrossRef] [Green Version]
- Parvani, J.G.; Galliher-Beckley, A.J.; Schiemann, B.J.; Schiemann, W.P. Targeted inactivation of β1 integrin induces β3 integrin switching, which drives breast cancer metastasis by TGF-β. Mol. Biol. Cell 2013, 24, 3449–3459. [Google Scholar] [CrossRef]
- Truong, H.H.; Xiong, J.; Ghotra, V.P.S.; Nirmala, E.; Haazen, L.; Le Dévédec, S.E.; Lu, H.E.B.; He, S.; Snaar-Jagalska, B.E.; Vreugdenhil, E.; et al. β1 Integrin Inhibition Elicits a Prometastatic Switch Through the TGFβ-miR-200-ZEB Network in E-Cadherin-Positive Triple-Negative Breast Cancer. Sci. Signal. 2014, 7, ra15. [Google Scholar] [CrossRef]
- Bui, T.; Rennhack, J.; Mok, S.; Ling, C.; Perez, M.; Roccamo, J.; Andrechek, E.R.; Moraes, C.; Muller, W.J. Functional Redundancy between β1 and β3 Integrin in Activating the IR/Akt/mTORC1 Signaling Axis to Promote ErbB2-Driven Breast Cancer. Cell Rep. 2019, 29, 589–602.e6. [Google Scholar] [CrossRef] [Green Version]
- Ambriović-Ristov, A.; Gabrilovac, J.; Čimbora-Zovko, T.; Osmak, M. Increased adenoviral transduction efficacy in human laryngeal carcinoma cells resistant to cisplatin is associated with increased expression of integrin αvβ3 and coxsackie adenovirus receptor. Int. J. Cancer 2004, 110, 660–667. [Google Scholar] [CrossRef]
- Majhen, D.; Nemet, J.; Richardson, J.; Gabrilovac, J.; Hajsig, M.; Osmak, M.; Eloit, M.; Ambriović-Ristov, A. Differential role of αvβ3 and αvβ5 integrins in internalization and transduction efficacies of wild type and RGD4C fiber-modified adenoviruses. Virus Res. 2009, 139, 64–73. [Google Scholar] [CrossRef]
- Stojanović, N.; Brozovic, A.; Majhen, A.; Bosnar, M.H.; Fritz, G.; Osmak, M.; Ambriović-Ristov, A. Integrin αvβ3 expression in tongue squamous carcinoma cells Cal27 confers anticancer drug resistance through loss of pSrc(Y418). Biochim. Biophys. Acta (BBA) Bioenerg. 2016, 1863, 1969–1978. [Google Scholar] [CrossRef] [PubMed]
- Lichtner, R. Negative Cooperativity between α3β1and α2β1 Integrins in Human Mammary Carcinoma MDA MB 231 Cells. Exp. Cell Res. 1998, 240, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Laplantine, E.; Vallar, L.; Mann, K.; Kieffer, N.; Aumailley, M. Interaction between the cytodomains of the α3 and β1 integrin subunits regulates remodelling of adhesion complexes on laminin. J. Cell Sci. 2000, 113, 1167–1176. [Google Scholar]
- Dogic, D.; Rousselle, P.; Aumailley, M. Cell adhesion to laminin 1 or 5 induces isoform-specific clustering of integrins and other focal adhesion components. J. Cell Sci. 1998, 111, 793–802. [Google Scholar]
- Jaspers, M.; De Meirsman, C.; Schollen, E.; Vekemans, S.; Cassiman, J.-J. Stable expression of VLA-4 and increased maturation of the β1-integrin precursor after transfection of CHO cells with α4m cDNA. FEBS Lett. 1994, 353, 239–242. [Google Scholar] [CrossRef] [Green Version]
- Porter, J.C.; Hogg, N. Integrin Cross Talk: Activation of Lymphocyte Function-associated Antigen-1 on Human T Cells Alters α4β1- and α5β1-mediated Function. J. Cell Biol. 1997, 138, 1437–1447. [Google Scholar] [CrossRef]
- Kligys, K.R.; Wu, Y.; Hopkinson, S.B.; Kaur, S.; Platanias, L.C.; Jones, J.C. α6β4 Integrin, a Master Regulator of Expression of Integrins in Human Keratinocytes*. J. Biol. Chem. 2012, 287, 17975–17984. [Google Scholar] [CrossRef] [Green Version]
- Longmate, W.M.; Lyons, S.P.; Chittur, S.V.; Pumiglia, K.M.; Van De Water, L.; DiPersio, C.M. Suppression of integrin α3β1 by α9β1 in the epidermis controls the paracrine resolution of wound angiogenesis. J. Cell Biol. 2017, 216, 1473–1488. [Google Scholar] [CrossRef]
- Defilles, C.; Lissitzky, J.-C.; Montero, M.-P.; André, F.; Prévot, C.; Delamarre, E.; Marrakchi, N.; Luis, J.; Rigot, V. αvβ5/β6 integrin suppression leads to a stimulation of α2β1 dependent cell migration resistant to PI3K/Akt inhibition. Exp. Cell Res. 2009, 315, 1840–1849. [Google Scholar] [CrossRef]
- Gonzalez, A.M.; Gonzales, M.; Herron, G.S.; Nagavarapu, U.; Hopkinson, S.B.; Tsuruta, D.; Jones, J.C.R. Complex interactions between the laminin 4 subunit and integrins regulate endothelial cell behavior in vitro and angiogenesis in vivo. Proc. Natl. Acad. Sci. USA 2002, 99, 16075–16080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vial, D.; McKeown-Longo, P.J. PAI1 stimulates assembly of the fibronectin matrix in osteosarcoma cells through crosstalk between the αvβ5 and α5β1 integrins. J. Cell Sci. 2008, 121, 1661–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hang, Q.; Isaji, T.; Hou, S.; Zhou, Y.; Fukuda, T.; Gu, J. N-Glycosylation of integrin α5 acts as a switch for EGFR-mediated complex formation of integrin α5β1 to α6β4. Sci. Rep. 2016, 6, 33507. [Google Scholar] [CrossRef] [Green Version]
- Koivisto, L.; Heino, J.; Häkkinen, L.; Larjava, H. The size of the intracellular β1-integrin precursor pool regulates maturation of β1-integrin subunit and associated α-subunits. Biochem. J. 1994, 300, 771–779. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, A.M.; Claiborne, J.; Jones, J.C.R. Integrin Cross-talk in Endothelial Cells Is Regulated by Protein Kinase A and Protein Phosphatase 1. J. Biol. Chem. 2008, 283, 31849–31860. [Google Scholar] [CrossRef] [Green Version]
- Retta, S.F.; Balzac, F.; Ferraris, P.; Belkin, A.M.; Fässler, R.; Humphries, M.J.; De Leo, G.; Silengo, L.; Tarone, G. β1-Integrin Cytoplasmic Subdomains Involved in Dominant Negative Function. Mol. Biol. Cell 1998, 9, 715–731. [Google Scholar] [CrossRef]
- Danen, E.H.; Sonneveld, P.; Brakebusch, C.; Fässler, R.; Sonnenberg, A. The fibronectin-binding integrins α5β1 and αvβ3 differentially modulate RhoA–GTP loading, organization of cell matrix adhesions, and fibronectin fibrillogenesis. J. Cell Biol. 2002, 159, 1071–1086. [Google Scholar] [CrossRef] [Green Version]
- Retta, S.F.; Cassarà, G.; D’Amato, M.; Alessandro, R.; Pellegrino, M.; Degani, S.; De Leo, G.; Silengo, L.; Tarone, G. Cross Talk between β1 and αVIntegrins: β1 Affects β3 mRNA Stability. Mol. Biol. Cell 2001, 12, 3126–3138. [Google Scholar] [CrossRef]
- Werr, J.; Eriksson, E.E.; Hedqvist, P.; Lindbom, L. Engagement of β2 integrins induces surface expression of β1 integrin receptors in human neutrophils. J. Leukoc. Biol. 2000. [Google Scholar] [CrossRef]
- Uotila, L.; Jahan, F.; Hinojosa, L.S.; Melandri, E.; Grӧnholm, M.; Gahmberg, C. Specific Phosphorylations Transmit Signals from Leukocyte β2 to β1 Integrins and Regulate Adhesion*. J. Biol. Chem. 2014, 289, 32230–32242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worth, D.C.; Hodivala-Dilke, K.M.; Robinson, S.D.; King, S.J.; Morton, P.E.; Gertler, F.B.; Humphries, M.J.; Parsons, M. αvβ3 integrin spatially regulates VASP and RIAM to control adhesion dynamics and migration. J. Cell Biol. 2010, 189, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, D.A.; Tai, V.; Di Paolo, G.; De Camilli, P.; Ginsberg, M.H. Competition for Talin Results in Trans-dominant Inhibition of Integrin Activation. J. Biol. Chem. 2004, 279, 28889–28895. [Google Scholar] [CrossRef] [Green Version]
- Cantor, D.; Slapetova, I.; Kan, A.; McQuade, L.R.; Baker, M.S. Overexpression of αvβ6 Integrin Alters the Colorectal Cancer Cell Proteome in Favor of Elevated Proliferation and a Switching in Cellular Adhesion That Increases Invasion. J. Proteome Res. 2013, 12, 2477–2490. [Google Scholar] [CrossRef]
- Rossetti, G.; Collinge, M.; Molteni, R.; Bender, J.R.; Pardi, R. Integrin-dependent regulation of gene expression in leukocytes. Immunol. Rev. 2002, 186, 189–207. [Google Scholar] [CrossRef]
- Kramer, G.; Shiber, A.; Bukau, B. Mechanisms of Cotranslational Maturation of Newly Synthesized Proteins. Annu. Rev. Biochem. 2019, 88, 337–364. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Hu, W.; Xu, R.; Jin, J.; Szulc, Z.M.; Zhang, G.; Galadari, S.H.; Obeid, L.M.; Mao, C. Alkaline ceramidase 2 regulates β1 integrin maturation and cell adhesion. FASEB J. 2008, 23, 656–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, K.; Hosono, T.; Nakamura, T.; Shiraishi, H.; Maeda, T.; Komano, H.; Yanagisawa, K.; Michikawa, M. Novel Role of Presenilins in Maturation and Transport of Integrin β1. Biochemistry 2008, 47, 3370–3378. [Google Scholar] [CrossRef]
- Salicioni, A.M.; Gaultier, A.; Brownlee, C.; Cheezum, M.K.; Gonias, S.L. Low Density Lipoprotein Receptor-related Protein-1 Promotes 1 Integrin Maturation and Transport to the Cell Surface. J. Biol. Chem. 2003, 279, 10005–10012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Franceschi, N.; Hamidi, H.; Alanko, J.; Sahgal, P.; Ivaska, J. Integrin traffic-the update. J. Cell Sci. 2015, 128, 839–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mana, G.; Valdembri, D.; Serini, G.G. Conformationally active integrin endocytosis and traffic: Why, where, when and how? Biochem. Soc. Trans. 2020, 48, 83–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gough, R.E.; Goult, B.T. The tale of two talins—Two isoforms to fine-tune integrin signalling. FEBS Lett. 2018, 592, 2108–2125. [Google Scholar] [CrossRef] [PubMed]
- Blystone, S.D.; Slater, S.E.; Williams, M.P.; Crow, M.T.; Brown, E.J. A Molecular Mechanism of Integrin Crosstalk: αvβ3 Suppression of Calcium/Calmodulin-dependent Protein Kinase II Regulates α5β1 Function. J. Cell Biol. 1999, 145, 889–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malric, L.; Monferran, S.; Gilhodes, J.; Boyrie, S.; Dahan, P.; Skuli, N.; Sesen, J.; Filleron, T.; Kowalski-Chauvel, A.; Moyal, E.C.-J.; et al. Interest of integrins targeting in glioblastoma according to tumor heterogeneity and cancer stem cell paradigm: An update. Oncotarget 2017, 8, 86947–86968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickreuter, E.; Cordes, N. The cancer cell adhesion resistome: Mechanisms, targeting and translational approaches. Biol. Chem. 2017, 398, 721–735. [Google Scholar] [CrossRef]
- Carlson, P.; Dasgupta, A.; Grzelak, C.A.; Kim, J.; Barrett, A.; Coleman, I.M.; Shor, R.E.; Goddard, E.T.; Dai, J.; Schweitzer, E.M.; et al. Targeting the perivascular niche sensitizes disseminated tumour cells to chemotherapy. Nature 2019, 21, 238–250. [Google Scholar] [CrossRef]
- Lautenschlaeger, T.; Perry, J.; Peereboom, D.M.; Li, B.; Ibrahim, A.; Huebner, A.; Meng, W.; White, J.R.; Chakravarti, A. In vitro study of combined cilengitide and radiation treatment in breast cancer cell lines. Radiat. Oncol. 2013, 8, 246. [Google Scholar] [CrossRef] [Green Version]
- Dickreuter, E.; Eke, I.; Krause, M.; Borgmann, K.; Van Vugt, M.A.; Cordes, N. Targeting of β1 integrins impairs DNA repair for radiosensitization of head and neck cancer cells. Oncogene 2015, 35, 1353–1362. [Google Scholar] [CrossRef]
- Christmann, M.; Diesler, K.; Majhen, D.; Steigerwald, C.; Berte, N.; Freund, H.; Stojanović, N.; Kaina, B.; Osmak, M.; Ambriović-Ristov, A.; et al. Integrin αVβ3 silencing sensitizes malignant glioma cells to temozolomide by suppression of homologous recombination repair. Oncotarget 2017, 8, 27754–27771. [Google Scholar] [CrossRef] [Green Version]
- Renner, G.; Janouskova, H.; Noulet, F.; Koenig, V.; Guérin, E.; Bär, S.; Nüesch, J.; Rechenmacher, F.; Neubauer, S.; Kessler, H.; et al. Integrin α5β1 and p53 convergent pathways in the control of anti-apoptotic proteins PEA-15 and survivin in high-grade glioma. Cell Death Differ. 2015, 23, 640–653. [Google Scholar] [CrossRef] [Green Version]
- Ruffini, F.; Graziani, G.; Levati, L.; Tentori, L.; D’Atri, S.; Lacal, P.M. Cilengitide downmodulates invasiveness and vasculogenic mimicry of neuropilin 1 expressing melanoma cells through the inhibition of αvβ5 integrin. Int. J. Cancer 2014, 136, E545–E558. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Huang, J.; Vue, M.; Alavian, M.R.; Goel, H.L.; Altieri, D.C.; Languino, L.R.; Fitzgerald, T.J. αvβ3 Integrin Mediates Radioresistance of Prostate Cancer Cells through Regulation of Survivin. Mol. Cancer Res. 2018, 17, 398–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Rekowski, K.W.; König, P.; Henze, S.; Schlesinger, M.; Zawierucha, P.; Januchowski, R.; Bendas, G. The Impact of Integrin-Mediated Matrix Adhesion on Cisplatin Resistance of W1 Ovarian Cancer Cells. Biomolecules 2019, 9, 788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ning, S.; Tian, J.; Marshall, D.J.; Knox, S.J. Anti- v Integrin Monoclonal Antibody Intetumumab Enhances the Efficacy of Radiation Therapy and Reduces Metastasis of Human Cancer Xenografts in Nude Rats. Cancer Res. 2010, 70, 7591–7599. [Google Scholar] [CrossRef] [Green Version]
- Hirata, E.; Girotti, M.R.; Viros, A.; Hooper, S.; Spencer-Dene, B.; Matsuda, M.; Larkin, J.; Marais, R.; Sahai, E. Intravital imaging reveals how BRAF inhibition generates drug-tolerant microenvironments with high integrin β1/FAK signaling. Cancer Cell 2015, 27, 574–588. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.H.; Flores, L.M.; Li, Q.; Zhou, P.; Xu, F.; Krop, I.E.; Hemler, M.E. Disruption of laminin-integrin-CD151-focal adhesion kinase axis sensitizes breast cancer cells to ErbB2 antagonists. Cancer Res. 2010, 70, 2256–2263. [Google Scholar] [CrossRef] [Green Version]
- Navab, R.; Strumpf, D.; To, C.; Pasko, E.; Kim, K.S.; Park, C.J.; Hai, J.; Liu, J.; Jonkman, J.; Barczyk, M.; et al. Integrin α11β1 regulates cancer stromal stiffness and promotes tumorigenicity and metastasis in non-small cell lung cancer. Oncogene 2015, 35, 1899–1908. [Google Scholar] [CrossRef] [Green Version]
- Dallari, S.; Franciotta, D.; Carluccio, S.; Signorini, L.; Gastaldi, M.; Colombo, E.; Bergamaschi, R.; Elia, F.; Villani, S.; Ferrante, P.; et al. Upregulation of integrin expression on monocytes in multiple sclerosis patients treated with natalizumab. J. Neuroimmunol. 2015, 287, 76–79. [Google Scholar] [CrossRef]





| Cells | Intervention | Integrin Crosstalk | Outcome | Reference |
|---|---|---|---|---|
| MSC | lentivirus expressing α1 shRNA | ↑ α2, α11 mRNA and protein expression | changes in cell adhesion and migration were not ascribed to changes in individual integrins α2 or α11 knockdown resulted in cell death | [72] |
| lentivirus expressing α2 shRNA | ↑ α1, α11 mRNA and protein expression | |||
| lentivirus expressing α11 shRNA | ↑ α1 mRNA and protein expression | |||
| MDA-MB-231 | α3-blocking Ab | ↑ α2β1 activation | ↑ adhesion to COL | [93] |
| B10-renal cells from E18 α3-defficient mouse | de novo expression of human α3 subunit | ↓ activation of αv | ↓ cell adhesion to the α3 noncollagenous (NC1) domain of COL IV | [73] |
| A549, HT144, HUVEC | α3-blocking Ab | ↑ αvβ3 activation | ↑ adhesion to the α3 noncollagenous (NC1) domain of COL IV | |
| Wi26 | microinjection of the α3 peptide representing the cytoplasmic domain of the α3 integrin | disengagement of the α6β1 | reduced size of α6β1-focal adhesions | [94] |
| primary human skin fibroblasts | α3β1-function blocking Ab | ↓ α6β1 integrin clustering | reduced size of α6β1-focal adhesions | [95] |
| CHO | stable transfection of murine α4 cDNA | ↑ maturation of β1 precursor | [96] | |
| Jurkat α4-defficient cells | de novo expression of α4 | ↑ αLβ2 activation | ↑ migration that was αLβ2-dependent and VCAM-dependent | [75] |
| lymphocytes isolated from α4(S988A) bearing mice | ↑ α4 activation (S988A) which precludes PKA-mediated α4 phosphorylation | ↑ αLβ2 activation | ↑ migration on ICAM-1 ↑ homing to B16 melanoma in vivo | [76] |
| Jurkat | crosslinking of α4β1 | clustering αLβ2 | ↑ adhesion to ICAM-1 | [74] |
| human T-cells | inhibition of α4β1 with blocking MoAb | α5β1 activation or expression was not analysed | ↑ migration mediated by α5β1 | [97] |
| interaction of the αLβ2 with its ligand ICAM-1 or αLβ2-activation Ab | ↓ binding of α4β1 and to a lesser extent α5β1 | ↓ adhesion mediated by α4β1 to FN and VCAM-1 and, to a lesser extent, α5β1 to FN | ||
| HSC-3 | α4-blocking Ab | ↑ α5 and αv | ↑ in vitro migration | [77] |
| overexpression of α4 using transient transfection with plasmid containing α4 gene | ↓ α5 and αv | ↓ in vitro migration | ||
| α5-blocking Ab | ↑ αv and α4 | = in vitro migration | ||
| αv-blocking Ab | ↑ α5 and α4 | = in vitro migration | ||
| CHO-B3 (negative for α5β1 and positive for αvβ3) | de novo expression of α5 using stable transfection with plasmid coding for α5 gene | ↓ αvβ3 activation | ↓ adhesion to FBG and αvβ3-mediated migration on FBG | [81] |
| U-251MG | ligation of α5β1 by plating cells on immobilised α5 MoAb | ↑ αvβ3 activation | ↑ αvβ3-mediated internalisation of VN | [80] |
| HUVEC | α5β1-blocking Ab and small molecule antagonists of α5β1 | ↓ αvβ3 activation | ↓ formation of αvβ3 focal adhesions ↓ αvβ3-mediated in vitro migration on VN | [78] |
| chick CAM | α5β1-blocking Ab | ↓ αvβ3 activation | ↓ angiogenesis in vivo | |
| primary HUVEC | seeding to α5β1 selective substrate | ↑ αvβ3 recruitment | ↑ cell spreading | [79] |
| immortalized epidermal keratinocytes | lentivirus containing α6-specific shRNA | ↓ α3 and α2 mRNA transcription and translation ↓ surface expression of α3β1 and α2β1 | ↓adhesion to LN332 and COL ↓migration | [98] |
| α9β1 null mice keratinocyte cell line | de novo expression of α9β1 using a retrovirus containing human α9 gene | ↓ α3β1 activation | ↓ α3β1-mediated migration in vitro on LN-332 | [99] |
| CHO | αIIbβ3-specific inhibitor (ligand Ro43-5054) | ↓ adhesive function of α5β1 | ↓ adhesion to FN | [29] |
| ↓ adhesive function of α2β1 | ↓ adhesion to COL | |||
| HT29-D4 | reduction of αv expression that is targeted to and degraded in lysosomes (reduces αvβ5/β6 expression) | ↑ α2β1 activation | ↑ cell migration | [100] |
| pKO, pan-ITG deficient murine fibroblasts reconstituted with αv and β1 | binding to FN fragment FNIII7-10 (contains the RGD- and PHSRN-motifs) | αv integrins engagement activates α5β1 to establish additional adhesion sites to FN | ↑ formation of α5β1 mediated adhesion clusters, adhesion strenghthening | [84] |
| HEK293 (β3wt) HUVEC MG-63 | αvβ3-blocking Ab and αvβ3 ligand cyclic G-Pen-GRGDSPC-A (small peptide antagonist) | ↓ α5β1 signallig | ↓ α5β1-mediated migration toward FN, but not attachment to FN | [82] |
| K562 | de novo expression of αvβ3 using stable transfection with plasmid coding for β3 gene | ↓ α5β1 activation | ↓ α5β1-mediated phagocytosis | [71] |
| TrHBMEC and HUVEC | αvβ3-blocking Ab | ↓ α3β1 and α6β1 adhesion | ↓ adhesion to LM5 and α4 LM G domain | [101] |
| β1-blocking Ab | ↓ αvβ3 adhesion | ↓ adhesion to α4 LM G domain | ||
| α3 and α6-blocking Ab | ↓ αvβ3 adhesion | ↓ adhesion to α4 LM G domain | ||
| MG-63 | αvβ5-blocking Ab or cyclic peptide RGDfV | ↑ α5β1 activation | unknown | [102] |
| CHO-B2 HeLa | deletion of α5β1 N-glycosylation site-11 that inhibits EGFR binding | ↑ α6β4-EGFR complex | ↑ cell proliferation | [103] |
| MG-63 | knockdown of β1 using stable transfection with plasmid coding for shβ1 RNA | ↓ protein maturation of α3 and α5 ↑ protein maturation of β1 | =expression of surface β1 | [104] |
| 4T1 | knockdown of β1 using stable transfection with plasmid coding for shβ1 RNA | ↑ β3 mRNA | ↓ in vivo tumor growth and ↑ in vivo metastasis which is not due to compensatory β3 expression | [88] |
| 4T1 and MDA-MB-231 | knockdown of β1 using stable transfection with plasmid coding for shβ1 RNA or exposure to β1-blocking Ab | ↑ β3 mRNA and protein expression | ↑ acinar cell growth, = growth of 3D organotypic culture and = in vivo metastasis due to compensatory β3 expression | [87] |
| HKC | knockdown of β1 using stable transfection with plasmid coding for shβ1 RNA | ↑ αvβ3 activation | ↑ αvβ3 localization to FA ↑ TGF-β1 induced COL mRNA expression | [85] |
| MMTV-NIC mice | mammary-specific deletion of β1 in the NIC model | ↑ β3 mRNA and αvβ3 expression on the cell surface | modest delay of tumor onset and a significant inhibition of lung metastasis | [89] |
| MMTV-NIC mice | mammary-specific deletion of β1 in the NIC model | ↓ β4; ↑ α5, αv, β3, β5 total protein levels measured by western blot | modest delay of tumor onset and a significant inhibition of lung metastasis | [86] |
| TrHBMEC | β1-blocking Ab | ↓ αvβ3 activation | ↓adhesion to a recombinant LM fragment and VN | [105] |
| GD25 (β1-null) | stable de novo expression of β1B isoform using stable transfection with plasmid coding for β1B gene | ↓ αv adhesion | ↓ αv containing focal adhesions and actin stress fibers ↓ spreading to FN (mediated by αvβ3) | [106] |
| GE11 β1-null | de novo expression of β1 | ↑ surface αvβ5 | unknown | [107] |
| GD25 β1-null | de novo expression of the β1A or B | ↓ β3 mRNA stability; ↓ surface αvβ3 ↑ surface αvβ5 (translational and post-translational level) = surface αv | unknown | [108] |
| PMN | β2-cross-linking Ab | ↑ expression of β1 integrins on the cell surface | ↑ adhesion to FN (through α5β1 and lesser extent α4β1) and COL (through α2β1) | [109] |
| adhesion to COL gel (involvement of β2) | ↑ expression of β1 integrins on the cell surface | ↑ α2β1-dependent migration | ||
| Jurkat | activation of β2 through the T-cell receptor or chemokines | ↓ activation of α4β1 | ↑ in vitro migration ↓ binding to VCAM-1 | [110] |
| MDA-MB-435S | transfection with β3 or β5-specific siRNA | ↑ surface expression of αvβ5 or αvβ3, respectively, = surface expression of αv | = in vitro migration (unlike knockdown of αv which ↓ migration) | [64] |
| Cal27 | de novo expression of β3 using stable transfection with plasmid coding for β3 gene | ↑ β5 mRNA ↑ surface expression of αvβ5 | unknown | [92] |
| HEp2 | de novo expression of β3 using stable transfection with plasmid coding for β3 gene | ↓ αvβ5 on the cell surface | ↓ Adenovirus type 5 transduction efficacy | [90,91] |
| NIH3T3 | deficient mutants β3Y759A and β31-760 that cannot bind protein kinase D1 (PKD1) | ↑ α5β1 recycling and signalling | ↓ peristent and directional migration ↑ random migration | [83] |
| immortalised fibroblasts from β3-null mice vs. WT fibroblasts | depletion of β3 | ↑ β1 activation | ↓ peristent and directional migration ↑adhesion dynamics ↑migration speed | [111] |
| CHO | de novo expression of Tac-β3 constructs with impaired talin binding activity in αIIbβ3 | ↓ α5β1 activation | ↓spreading on FBG | [112] |
| SW480 | de novo expression of β6 using stable transfection with plasmid coding for β6 gene | ↓ α2, α6, β1, β4 and β5 protein expression detected by mass spectrometry | ↓ adhesion through β2, β3 and β4 ↓ adhesion to COL I and II, FN and VN ↑ in vitro invasion | [113] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samaržija, I.; Dekanić, A.; Humphries, J.D.; Paradžik, M.; Stojanović, N.; Humphries, M.J.; Ambriović-Ristov, A. Integrin Crosstalk Contributes to the Complexity of Signalling and Unpredictable Cancer Cell Fates. Cancers 2020, 12, 1910. https://doi.org/10.3390/cancers12071910
Samaržija I, Dekanić A, Humphries JD, Paradžik M, Stojanović N, Humphries MJ, Ambriović-Ristov A. Integrin Crosstalk Contributes to the Complexity of Signalling and Unpredictable Cancer Cell Fates. Cancers. 2020; 12(7):1910. https://doi.org/10.3390/cancers12071910
Chicago/Turabian StyleSamaržija, Ivana, Ana Dekanić, Jonathan D. Humphries, Mladen Paradžik, Nikolina Stojanović, Martin J. Humphries, and Andreja Ambriović-Ristov. 2020. "Integrin Crosstalk Contributes to the Complexity of Signalling and Unpredictable Cancer Cell Fates" Cancers 12, no. 7: 1910. https://doi.org/10.3390/cancers12071910








