Prognostic Value of CXCR2 in Breast Cancer
Abstract
1. Introduction
2. Results
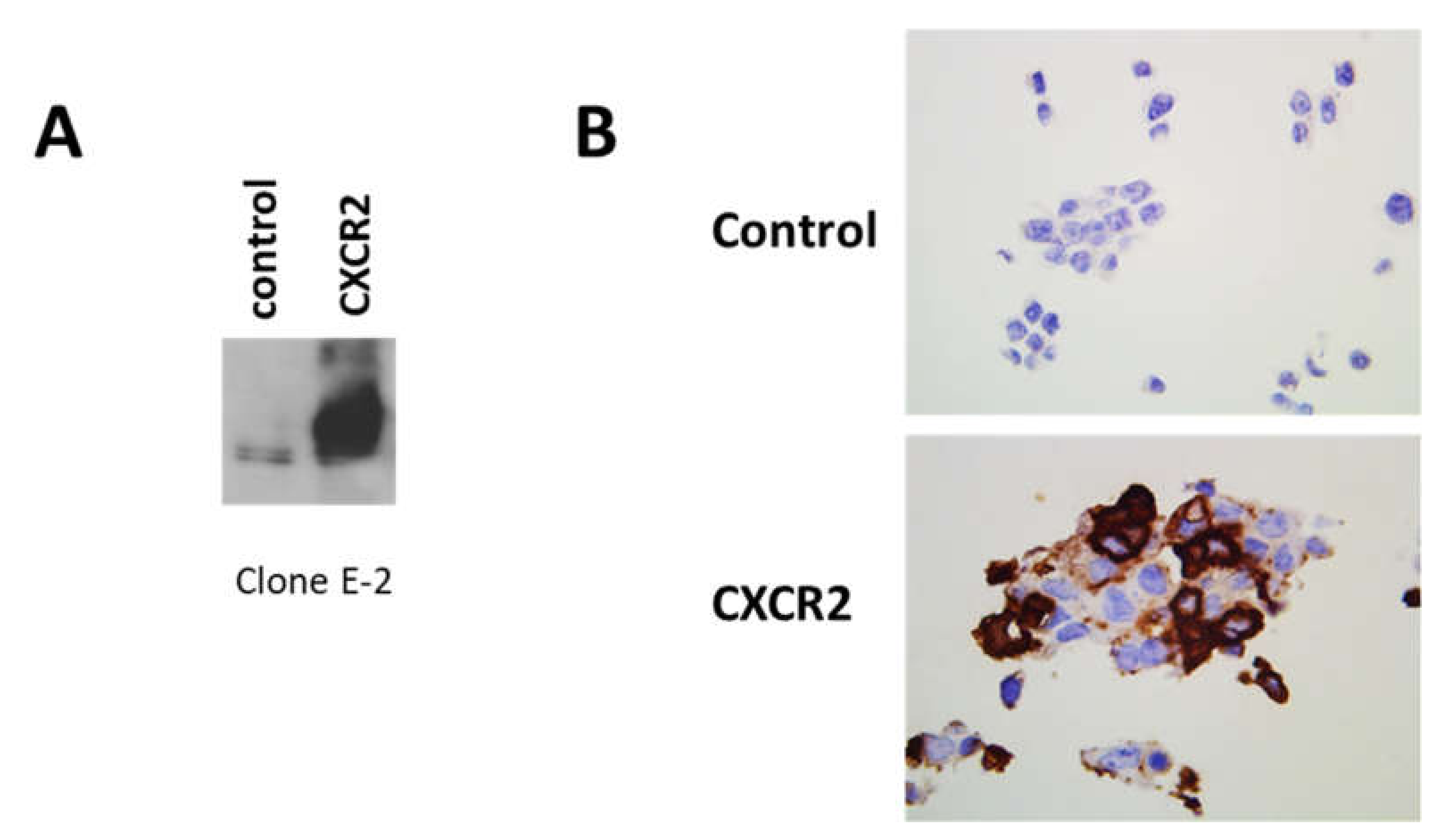
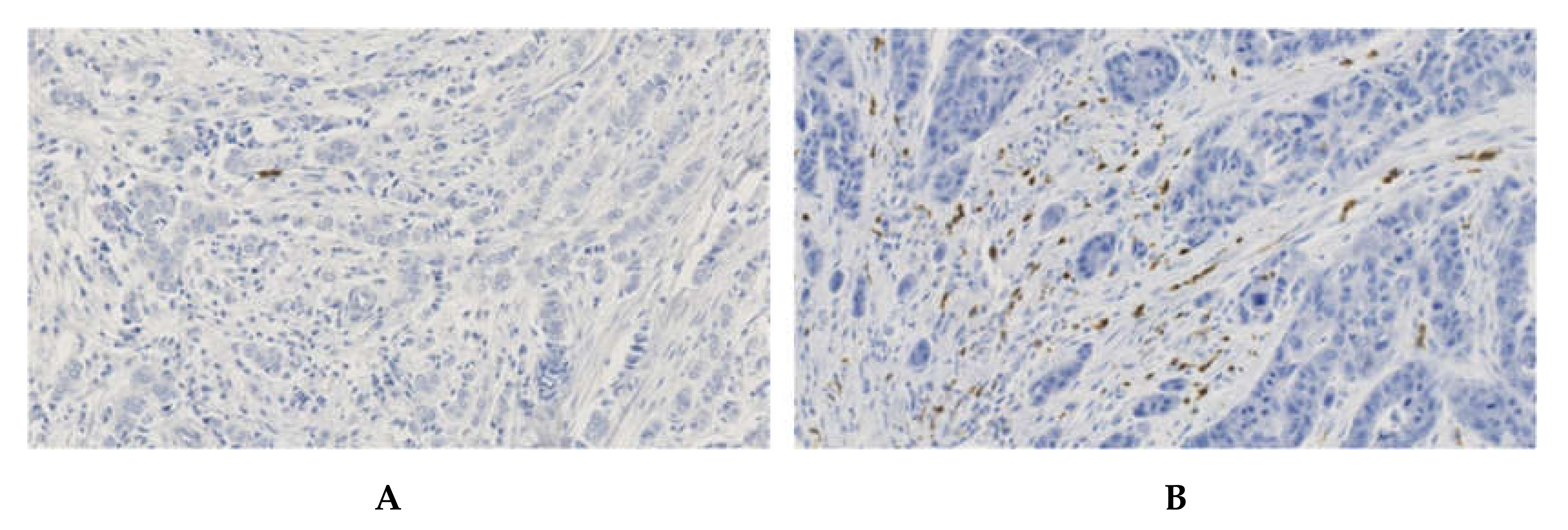
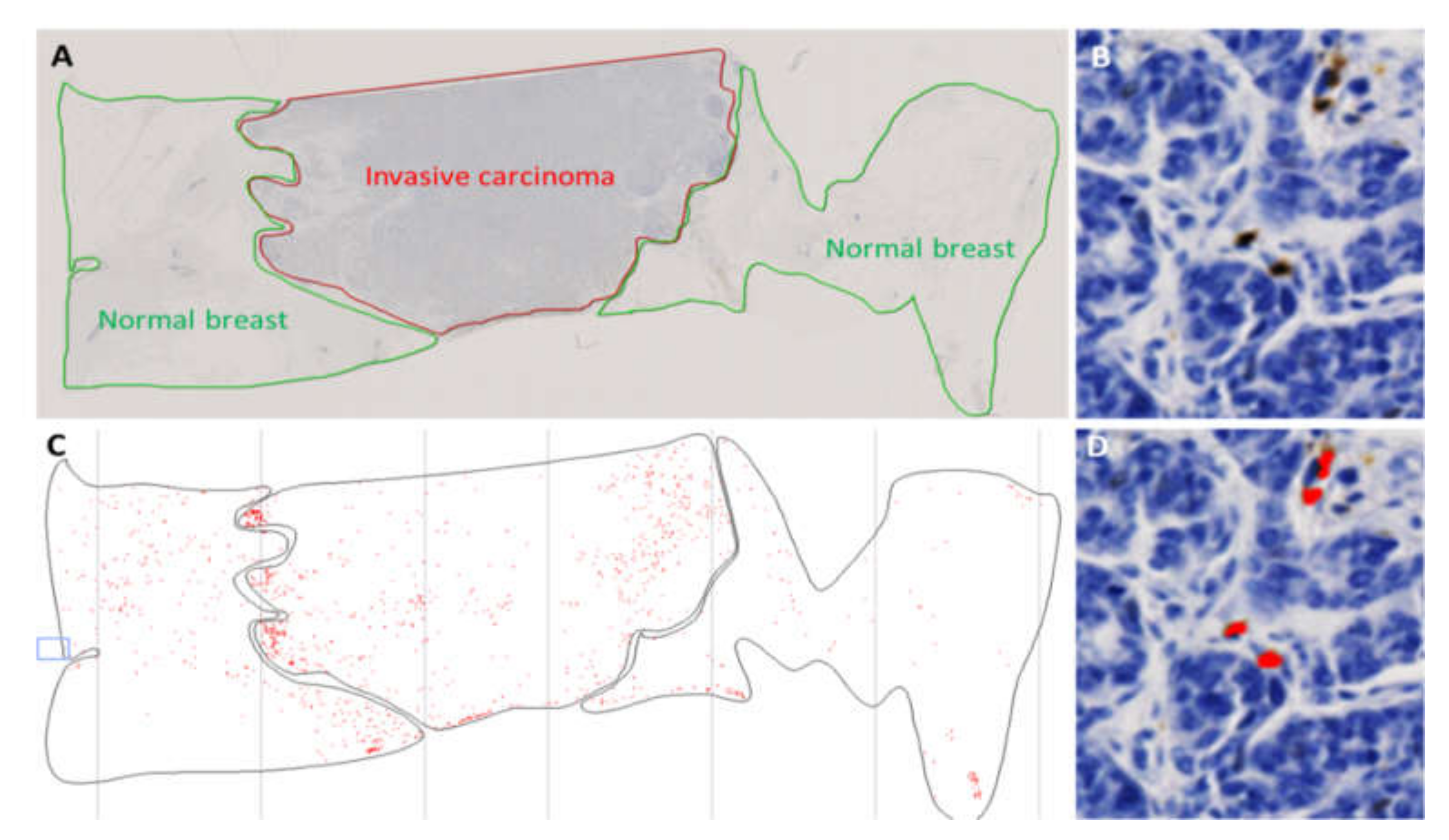
2.1. Validation of the Detection of CXCR2 Expression
2.2. Analysis of CXCR2, CD66b and CD11b with Clinical Parameters of Breast Tumors
2.3. Analysis of the Correlation of CXCR2, CD11b and CD66b with Immune Infiltration of Tumors
2.4. CXCR2 Levels Are Correlated to AP-1 Levels
2.5. High CXCR2 Expression Is an Independent Prognostic Factor of Time to Relapse (TTR)
3. Discussion
4. Materials and Methods
4.1. Patients and Tumor Samples
4.2. Ethics Approval and Consent to Participate
4.3. Immunohistochemistry
4.4. In Situ Hybridization
4.5. Immunofluorescence
4.6. Transfection of Human CXCR2 in HEK-293 Cells
4.7. Protein Extraction and Western Blot
4.8. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Ab | antibody |
| CI | Confidence interval |
| ER | Estrogen Receptor |
| Her2 | Human Epidermal Growth Factor Receptor-2 |
| HR | hormone receptor |
| HdR | hazard ratio |
| IF | immunofluorescence |
| IHC | Immunohistochemistry |
| ISH | in situ hybridization |
| PR | Progesterone Receptor |
| ROI | Region of interest |
| SBR | Scarff–Bloom–Richardson |
| TNBC | Triple-Negative Breast Cancer |
| TTR | Time To Relapse |
References
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Korkaya, H.; Liu, S.; Wicha, M.S. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J. Clin. Investig. 2011, 121, 3804–3809. [Google Scholar] [CrossRef]
- Lazennec, G.; Lam, P.Y. Recent discoveries concerning the tumor—Mesenchymal stem cell interactions. Biochim. Biophys. Acta 2016, 1866, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Gieniec, K.A.; Butler, L.M.; Worthley, D.L.; Woods, S.L. Cancer-associated fibroblasts-heroes or villains? Br. J. Cancer 2019, 121, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Schioppa, T.; Porta, C.; Allavena, P.; Sica, A. Role of tumor-associated macrophages in tumor progression and invasion. Cancer Metastasis Rev. 2006, 25, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Cassatella, M.A.; Costantini, C.; Jaillon, S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 2011, 11, 519–531. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Bronte, V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012, 12, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Lazennec, G. Chemokines: Novel targets for breast cancer metastasis. Cancer Metastasis Rev. 2007, 26, 401–420. [Google Scholar] [CrossRef]
- Lazennec, G.; Richmond, A. Chemokines and chemokine receptors: New insights into cancer-related inflammation. Trends Mol. Med. 2010, 16, 133–144. [Google Scholar] [CrossRef]
- Zlotnik, A.; Yoshie, O. Chemokines: A new classification system and their role in immunity. Immunity 2000, 12, 121–127. [Google Scholar] [CrossRef]
- Wang, J.; Knaut, H. Chemokine signaling in development and disease. Development 2014, 141, 4199–4205. [Google Scholar] [CrossRef] [PubMed]
- Bieche, I.; Chavey, C.; Andrieu, C.; Busson, M.; Vacher, S.; Le Corre, L.; Guinebretiere, J.M.; Burlinchon, S.; Lidereau, R.; Lazennec, G. CXC chemokines located in the 4q21 region are up-regulated in breast cancer. Endocr. Relat. Cancer 2007, 14, 1039–1052. [Google Scholar] [CrossRef] [PubMed]
- Chavey, C.; Bibeau, F.; Gourgou-Bourgade, S.; Burlinchon, S.; Boissiere, F.; Laune, D.; Roques, S.; Lazennec, G. Estrogen-receptor negative breast cancers exhibit a high cytokine content. Breast Cancer Res. 2007, 9, R15. [Google Scholar] [CrossRef]
- Freund, A.; Chauveau, C.; Brouillet, J.P.; Lucas, A.; Lacroix, M.; Licznar, A.; Vignon, F.; Lazennec, G. IL-8 expression and its possible relationship with estrogen-receptor-negative status of breast cancer cells. Oncogene 2003, 22, 256–265. [Google Scholar] [CrossRef]
- Freund, A.; Jolivel, V.; Durand, S.; Kersual, N.; Chalbos, D.; Chavey, C.; Vignon, F.; Lazennec, G. Mechanisms underlying differential expression of interleukin-8 in breast cancer cells. Oncogene 2004, 23, 6105–6114. [Google Scholar] [CrossRef]
- Escobar, P.; Bouclier, C.; Serret, J.; Bieche, I.; Brigitte, M.; Caicedo, A.; Sanchez, E.; Vacher, S.; Vignais, M.L.; Bourin, P.; et al. IL-1beta produced by aggressive breast cancer cells is one of the factors that dictate their interactions with mesenchymal stem cells through chemokine production. Oncotarget 2015, 6, 29034–29047. [Google Scholar] [CrossRef] [PubMed]
- Strieter, R.M.; Burdick, M.D.; Gomperts, B.N.; Belperio, J.A.; Keane, M.P. CXC chemokines in angiogenesis. Cytokine Growth Factor Rev. 2005, 16, 593–609. [Google Scholar] [CrossRef] [PubMed]
- Petering, H.; Gotze, O.; Kimmig, D.; Smolarski, R.; Kapp, A.; Elsner, J. The biologic role of interleukin-8: Functional analysis and expression of CXCR1 and CXCR2 on human eosinophils. Blood 1999, 93, 694–702. [Google Scholar] [CrossRef]
- Acharyya, S.; Oskarsson, T.; Vanharanta, S.; Malladi, S.; Kim, J.; Morris, P.G.; Manova-Todorova, K.; Leversha, M.; Hogg, N.; Seshan, V.E.; et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell 2012, 150, 165–178. [Google Scholar] [CrossRef]
- Lee, J.; Horuk, R.; Rice, G.C.; Bennett, G.L.; Camerato, T.; Wood, W.I. Characterization of two high affinity human interleukin-8 receptors. J. Biol. Chem. 1992, 267, 16283–16287. [Google Scholar]
- Morohashi, H.; Miyawaki, T.; Nomura, H.; Kuno, K.; Murakami, S.; Matsushima, K.; Mukaida, N. Expression of both types of human interleukin-8 receptors on mature neutrophils, monocytes, and natural killer cells. J. Leukoc. Biol. 1995, 57, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Murdoch, C.; Monk, P.N.; Finn, A. Cxc chemokine receptor expression on human endothelial cells. Cytokine 1999, 11, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Petzelbauer, P.; Watson, C.A.; Pfau, S.E.; Pober, J.S. IL-8 and angiogenesis: Evidence that human endothelial cells lack receptors and do not respond to IL-8 in vitro. Cytokine 1995, 7, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tu, L.; Du, C.; Xie, X.; Liu, Y.; Wang, J.; Li, Z.; Jiang, M.; Cao, D.; Yan, X.; et al. CXCR2 is a novel cancer stem-like cell marker for triple-negative breast cancer. Onco Targets Ther. 2018, 11, 5559–5567. [Google Scholar] [CrossRef]
- Xu, H.; Lin, F.; Wang, Z.; Yang, L.; Meng, J.; Ou, Z.; Shao, Z.; Di, G.; Yang, G. CXCR2 promotes breast cancer metastasis and chemoresistance via suppression of AKT1 and activation of COX2. Cancer Lett. 2018, 412, 69–80. [Google Scholar] [CrossRef]
- Romero-Moreno, R.; Curtis, K.J.; Coughlin, T.R.; Cristina Miranda-Vergara, M.; Dutta, S.; Natarajan, A.; Facchine, B.A.; Jackson, K.M.; Nystrom, L.; Li, J.; et al. The CXCL5/CXCR2 axis is sufficient to promote breast cancer colonization during bone metastasis. Nat. Commun. 2019, 10, 4404. [Google Scholar] [CrossRef]
- Li, A.; Varney, M.L.; Singh, R.K. Expression of interleukin 8 and its receptors in human colon carcinoma cells with different metastatic potentials. Clin. Cancer Res. 2001, 7, 3298–3304. [Google Scholar]
- Kuwada, Y.; Sasaki, T.; Morinaka, K.; Kitadai, Y.; Mukaida, N.; Chayama, K. Potential involvement of IL-8 and its receptors in the invasiveness of pancreatic cancer cells. Int. J. Oncol. 2003, 22, 765–771. [Google Scholar] [CrossRef]
- Liu, Q.; Li, A.; Tian, Y.; Wu, J.D.; Liu, Y.; Li, T.; Chen, Y.; Han, X.; Wu, K. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev. 2016, 31, 61–71. [Google Scholar] [CrossRef]
- Wendt, M.K.; Cooper, A.N.; Dwinell, M.B. Epigenetic silencing of CXCL12 increases the metastatic potential of mammary carcinoma cells. Oncogene 2008, 27, 1461–1471. [Google Scholar] [CrossRef] [PubMed]
- Sauve, K.; Lepage, J.; Sanchez, M.; Heveker, N.; Tremblay, A. Positive feedback activation of estrogen receptors by the CXCL12-CXCR4 pathway. Cancer Res. 2009, 69, 5793–5800. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, L.V.; Short, S.P.; Neel, N.F.; Salvo, V.A.; Zhu, Y.; Elliott, S.; Wei, Y.; Yu, D.; Sun, M.; Muir, S.E.; et al. Cytokine receptor CXCR4 mediates estrogen-independent tumorigenesis, metastasis, and resistance to endocrine therapy in human breast cancer. Cancer Res. 2011, 71, 603–613. [Google Scholar] [CrossRef]
- Bernard, S.; Myers, M.; Fang, W.B.; Zinda, B.; Smart, C.; Lambert, D.; Zou, A.; Fan, F.; Cheng, N. CXCL1 Derived from Mammary Fibroblasts Promotes Progression of Mammary Lesions to Invasive Carcinoma through CXCR2 Dependent Mechanisms. J. Mammary Gland Biol. Neoplasia 2018, 23, 249–267. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.; Pandey, N.B.; Popel, A.S. Crosstalk between stromal components and tumor cells of TNBC via secreted factors enhances tumor growth and metastasis. Oncotarget 2017, 8, 60210–60222. [Google Scholar] [CrossRef]
- Mendonca, M.A.; Souto, F.O.; Micheli, D.C.; Alves-Filho, J.C.; Cunha, F.Q.; Murta, E.F.; Tavares-Murta, B.M. Mechanisms affecting neutrophil migration capacity in breast cancer patients before and after chemotherapy. Cancer Chemother. Pharmacol. 2014, 73, 317–324. [Google Scholar] [CrossRef]
- Cheng, Y.; Ma, X.L.; Wei, Y.Q.; Wei, X.W. Potential roles and targeted therapy of the CXCLs/CXCR2 axis in cancer and inflammatory diseases. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 289–312. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.K.; Farnie, G.; Bundred, N.J.; Simoes, B.M.; Shergill, A.; Landberg, G.; Howell, S.J.; Clarke, R.B. Targeting CXCR1/2 significantly reduces breast cancer stem cell activity and increases the efficacy of inhibiting HER2 via HER2-dependent and -independent mechanisms. Clin. Cancer Res. 2013, 19, 643–656. [Google Scholar] [CrossRef] [PubMed]
- Ginestier, C.; Liu, S.; Diebel, M.E.; Korkaya, H.; Luo, M.; Brown, M.; Wicinski, J.; Cabaud, O.; Charafe-Jauffret, E.; Birnbaum, D.; et al. CXCR1 blockade selectively targets human breast cancer stem cells in vitro and in xenografts. J. Clin. Investig. 2010, 120, 485–497. [Google Scholar] [CrossRef]
- Boissiere-Michot, F.; Lazennec, G.; Frugier, H.; Jarlier, M.; Roca, L.; Duffour, J.; Du Paty, E.; Laune, D.; Blanchard, F.; Le Pessot, F.; et al. Characterization of an adaptive immune response in microsatellite-instable colorectal cancer. Oncoimmunology 2014, 3, e29256. [Google Scholar] [CrossRef]



| Variables | N | % |
|---|---|---|
| Age | ||
| ≤50 years | 36 | 34.3 |
| 50–65 years | 31 | 29.5 |
| >65 years | 38 | 36.2 |
| Type | ||
| Ductal carcinoma | 77 | 73.3 |
| Others | 28 | 26.7 |
| SBR Grade | ||
| I/II | 73 | 69.5 |
| III | 32 | 30.5 |
| pT | ||
| pT1 | 42 | 40.4 |
| pT2/pT3/pT4 | 62 | 59.6 |
| Missing | 1 | / |
| pN | ||
| pN0 | 50 | 50 |
| pN1/pN2/pN3 | 50 | 50 |
| Missing | 5 | / |
| pM | ||
| pM0 | 80 | 76.2 |
| pM1 | 1 | 1 |
| pMX | 24 | 22.9 |
| ER | ||
| Negative | 36 | 34.3 |
| Positive | 69 | 65.7 |
| PR | ||
| Negative | 52 | 49.5 |
| Positive | 53 | 50.5 |
| HR (if ER+ and/or PR+) | ||
| Negative | 29 | 27.6 |
| Positive | 76 | 72.4 |
| Her2 | ||
| Negative | 94 | 89.5 |
| Positive | 11 | 10.5 |
| Immunophenotype | ||
| ER/PR− Her2− | 23 | 21.9 |
| ER/PR± Her2+ | 11 | 10.5 |
| ER/PR+ Her2− | 71 | 67.6 |
| CD3 | ||
| Negative | 54 | 51.4 |
| Positive | 51 | 48.6 |
| CD20 | ||
| Negative | 74 | 70.5 |
| Positive | 31 | 29.5 |
| CD68 | ||
| Negative | 5 | 4.8 |
| Positive | 100 | 95.2 |
| AP-1 | ||
| Negative | 34 | 33 |
| Positive | 69 | 67 |
| Missing | 2 | / |
| NF-KB | ||
| Negative | 29 | 28.2 |
| Positive | 74 | 71.8 |
| Missing | 2 | / |
| Variables | CXCR2 | CD11b | CD66b | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N | Median (Range) | p-Value | N | Median (Range) | p-Value | N | Median (Range) | p-Value | |
| Breast | |||||||||
| Normal | 21 | 145.83 (24.20:714.60) | 0.026 | 21 | 242.90 (48.96:1014.74) | 0.001 | 21 | 85.46 (25.43:375.67) | 0.587 |
| Cancer | 95 | 269.07 (19.38:4539.93) | 100 | 97.60 (0.71:85,867.80) | 98 | 68.96 (0.00:1920.86) | |||
| Variables | CXCR2 | CD11b | CD66b | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N | Median (Range) | p-Value | N | Median (Range) | p-Value | N | Median (Range) | p-Value | |
| Age | |||||||||
| ≤50 years | 35 | 344.81 (31.4:7789.8) | 0.546 | 35 | 125.65 (4.7:13376.5) | 0.142 | 35 | 48.37 (4.4:10032.0) | 0.345 |
| 50–65 years | 31 | 268.28 (40.6:5085.6) | 31 | 79.02 (2.8:2963.4) | 30 | 52.17 (0.0:4842.6) | |||
| >65 years | 34 | 406.60 (38.6:2201.2) | 36 | 146.6 (12.2:1118.5) | 37 | 90.0 (4.3:1501.9) | |||
| Type | |||||||||
| Ductal carcinoma | 73 | 351.60 (31.45:7789.79) | 0.468 | 75 | 146.54 (2.81:13376.50) | 0.357 | 76 | 70.36 (0.00:4842.61) | 0.355 |
| Others | 27 | 295.24 (38.59:4998.79) | 27 | 79.02 (11.99:2579.57) | 26 | 35.29 (0.00:10031.97) | |||
| SBR Grade | |||||||||
| I/II | 69 | 298.45 (31.45:5085.62) | 0.002 | 71 | 111.87 (2.81:4568.04) | 0.032 | 71 | 36.08 (0.00:4842.61) | 0.038 |
| III | 31 | 698.38 (55.23:7789.79) | 31 | 239.05 (9.36:13376.50) | 31 | 89.98 (5.45:10031.97) | |||
| pT | |||||||||
| pT1 | 41 | 263.28 (31.45:4998.79) | 0.102 | 42 | 90.11 (4.69:5192.38) | 0.051 | 42 | 48.56 (0.00:10031.97) | 0.119 |
| pT2/pT3/pT4 | 58 | 379.24 (40.61:7789.79) | 59 | 200.09 (2.81:13376.50) | 59 | 85.58 (0.00:4842.61) | |||
| pN | |||||||||
| pN0 | 49 | 384.97 (31.45:7789.79) | 0.561 | 49 | 151.44 (4.69:13376.50) | 0.391 | 48 | 50.24 (4.41:10031.97) | 0.971 |
| pN1/pN2/pN3 | 46 | 320.31 (38.59:3706.72) | 48 | 125.30 (2.81:9547.40) | 49 | 48.37 (0.00:1501.86) | |||
| ER | |||||||||
| Negative | 34 | 540.59 (83.49:7789.79) | 0.005 | 35 | 373.09 (11.99:13376.50) | <0.001 | 35 | 79.85 (0.00:10031.97) | 0.065 |
| Positive | 66 | 296.85 (31.45:5085.62) | 67 | 85.91 (2.81:4568.04) | 67 | 34.50 (0.00:4842.61) | |||
| PR | |||||||||
| Negative | 51 | 472.48 (38.59:7789.79) | 0.002 | 51 | 262.77 (11.99:13376.50) | <0.001 | 51 | 75.46 (0.00:10031.97) | 0.08 |
| Positive | 49 | 250.94 (31.45:2025.37) | 51 | 78.25 (2.81:4568.04) | 51 | 32.98 (0.00:1501.86) | |||
| Her2 | |||||||||
| Negative | 90 | 343.64 (31.45:7789.79) | 0.37 | 92 | 146.08 (2.81:13376.50) | 0.84 | 92 | 53.00 (0.00:10031.97) | 0.653 |
| Positive | 10 | 285.67 (141.87:1067.14) | 10 | 103.49 (16.43:515.78) | 10 | 52.12 (5.45:570.98) | |||
| Immunophenotype | |||||||||
| ER/PR− Her2− | 23 | 1082.20 (154.6:7789.8) | <0.001 | 23 | 555.08 (12.0:13376.5) | <0.001 | 23 | 108.13 (6.4:10032.0) | 0.043 |
| ER/PR± Her2+ | 10 | 285.67 (141.9:1067.1) | 10 | 103.49 (16.4:515.8) | 10 | 52.12 (5.5:571.0) | |||
| ER/PR+ Her2− | 67 | 295.2 (31.4:5085.6) | 69 | 85.9 (2.8:4568.0) | 69 | 34.5 (0.0:4842.6) | |||
| Variables | CXCR2 | CD11b | CD66b | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N | Median (Range) | p-Value | N | Median (Range) | p-Value | N | Median (Range) | p-Value | |
| CD3 | |||||||||
| Negative | 52 | 253.21 (31.45:5085.62) | 53 | 85.91 (2.81:5192.38) | 0.013 | 52 | 48.44 (0.00:4842.61) | 0.828 | |
| Positive | 48 | 490.63 (55.23:7789.79) | <0.001 | 49 | 205.85 (14.95:13376.50) | 50 | 60.56 (0.00:10031.97) | ||
| CD20 | |||||||||
| Negative | 70 | 292.12 (31.45:7789.79) | 0.007 | 71 | 98.49 (2.81:13376.50) | 0.003 | 71 | 48.37 (0.00:4842.61) | 0.608 |
| Positive | 30 | 481.76 (156.36:4998.79) | 31 | 291.71 (19.02:9547.40) | 31 | 75.46 (6.41:10031.97) | |||
| CD68 | |||||||||
| Negative | 5 | 151.78 (31.45:725.11) | 0.053 | 5 | 41.97 (4.69:151.44) | 0.033 | 4 | 19.96 (4.41:159.43) | 0.196 |
| Positive | 95 | 344.81 (38.59:7789.79) | 97 | 146.54 (2.81:13376.50) | 98 | 55.73 (0.00:10031.97) | |||
| AP-1 | |||||||||
| Negative | 31 | 268.59 (38.59:2201.23) | 0.05 | 32 | 88.07 (9.36:4568.04) | 0.079 | 32 | 32.40 (4.34:1416.34) | 0.311 |
| Positive | 67 | 384.97 (31.45:7789.79) | 68 | 178.18 (2.81:13376.50) | 68 | 77.65 (0.00:10031.97) | |||
| NF-kB | |||||||||
| Negative | 29 | 288.99 (55.23:5085.62) | 0.516 | 29 | 124.96 (9.36:9547.40) | 0.64 | 28 | 42.22 (0.00:10031.97) | 0.857 |
| Positive | 69 | 362.21 (31.45:7789.79) | 71 | 146.54 (2.81:13376.50) | 72 | 53.00 (0.00:2748.86) | |||
| Parameter | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| HdR (95% CI) | p-Value | HdR (95% CI) | p-Value | |
| SBR Grade | ||||
| I/II | 1.000 | 1.000 | ||
| III | 1.117 (0.465–2.685) | 0.805 | 1.143 (0.382–3.420) | 0.811 |
| Immune Type | ||||
| ER/PR− Her2− | 1.000 | 1.000 | ||
| ER/PR± Her2+ | 2.629 (0.749–9.233) | 0.131 | 1.718 (0.311–9.498) | 0.535 |
| ER/PR+ Her2− | 1.038 (0.358–3.006) | 0.945 | 0.553 (0.137–2.237) | 0.406 |
| Age at Diagnosis | ||||
| <50 | 1.000 | 1.000 | ||
| 50–65 | 0.835 (0.317–2.200) | 0.714 | 0.641 (0.215–1.910) | 0.425 |
| >65 | 0.682 (0.230–2.020) | 0.490 | 0.537 (0.161–1.789) | 0.311 |
| CD20 | ||||
| Negative | 1.000 | 1.000 | ||
| Positive | 0.356 (0.119–1.065) | 0.065 | 0.252 (0.068–0.932) | 0.039 |
| AP-1 | ||||
| Negative | 1.000 | 1.000 | ||
| Positive | 2.270 (0.761–6.773) | 0.142 | 3.404 (0.988–11.723) | 0.052 |
| pT | ||||
| T1 | 1.000 | 1.000 | ||
| T2/T3/T4 | 1.727 (0.704–4.238) | 0.233 | 2.486 (0.909–6.797) | 0.076 |
| CXCR2 invasive | ||||
| low | 1.000 | 1.000 | ||
| medium | 0.231 (0.073–0.731) | 0.013 | 0.168 (0.043–0.650) | 0.010 |
| high | 0.277 (0.100–0.771) | 0.014 | 0.215 (0.054–0.840) | 0.028 |
| CD11b invasive | ||||
| low | 1.000 | |||
| medium | 1.318 (0.457–3.803) | 0.610 | ||
| high | 0.997 (0.342–2.906) | 0.995 | ||
| CD66b invasive | ||||
| low | 1.000 | |||
| medium | 1.626 (0.584–4.529) | 0.352 | ||
| high | 0.874 (0.278–2.749) | 0.818 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boissière-Michot, F.; Jacot, W.; Fraisse, J.; Gourgou, S.; Timaxian, C.; Lazennec, G. Prognostic Value of CXCR2 in Breast Cancer. Cancers 2020, 12, 2076. https://doi.org/10.3390/cancers12082076
Boissière-Michot F, Jacot W, Fraisse J, Gourgou S, Timaxian C, Lazennec G. Prognostic Value of CXCR2 in Breast Cancer. Cancers. 2020; 12(8):2076. https://doi.org/10.3390/cancers12082076
Chicago/Turabian StyleBoissière-Michot, Florence, William Jacot, Julien Fraisse, Sophie Gourgou, Colin Timaxian, and Gwendal Lazennec. 2020. "Prognostic Value of CXCR2 in Breast Cancer" Cancers 12, no. 8: 2076. https://doi.org/10.3390/cancers12082076
APA StyleBoissière-Michot, F., Jacot, W., Fraisse, J., Gourgou, S., Timaxian, C., & Lazennec, G. (2020). Prognostic Value of CXCR2 in Breast Cancer. Cancers, 12(8), 2076. https://doi.org/10.3390/cancers12082076






