Identification of Novel Fusion Genes in Bone and Soft Tissue Sarcoma and Their Implication in the Generation of a Mouse Model
Abstract
1. Introduction
2. Results
2.1. Disease Characteristics
2.2. Fusion Gene Profiles of 29 BSTS Cases
2.3. Deduced Structure of Novel Fusion Proteins
2.3.1. AHRR-NCOA3 (Case #43)
2.3.2. PAK2-RAF1 (Case #2)
2.3.3. EPC1-KDM2B (Case #55)
2.3.4. TAOK1-CRYBA1 (Case #14)
2.3.5. UST-DUSP22 (Case #50)
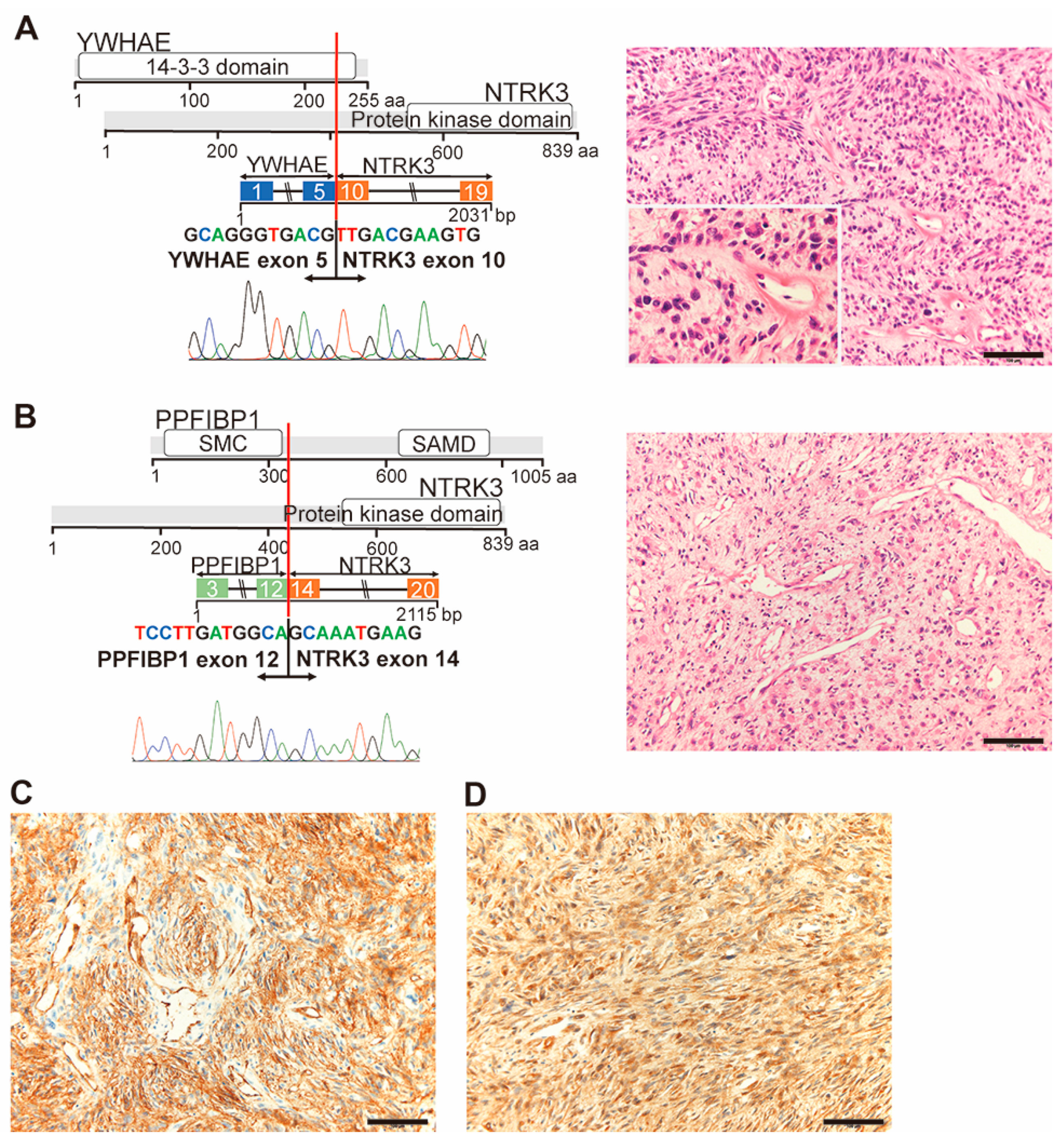
2.3.6. YWHAE-NTRK3 (Case #32) and PPFIBP1-NTRK3 (Case #42)
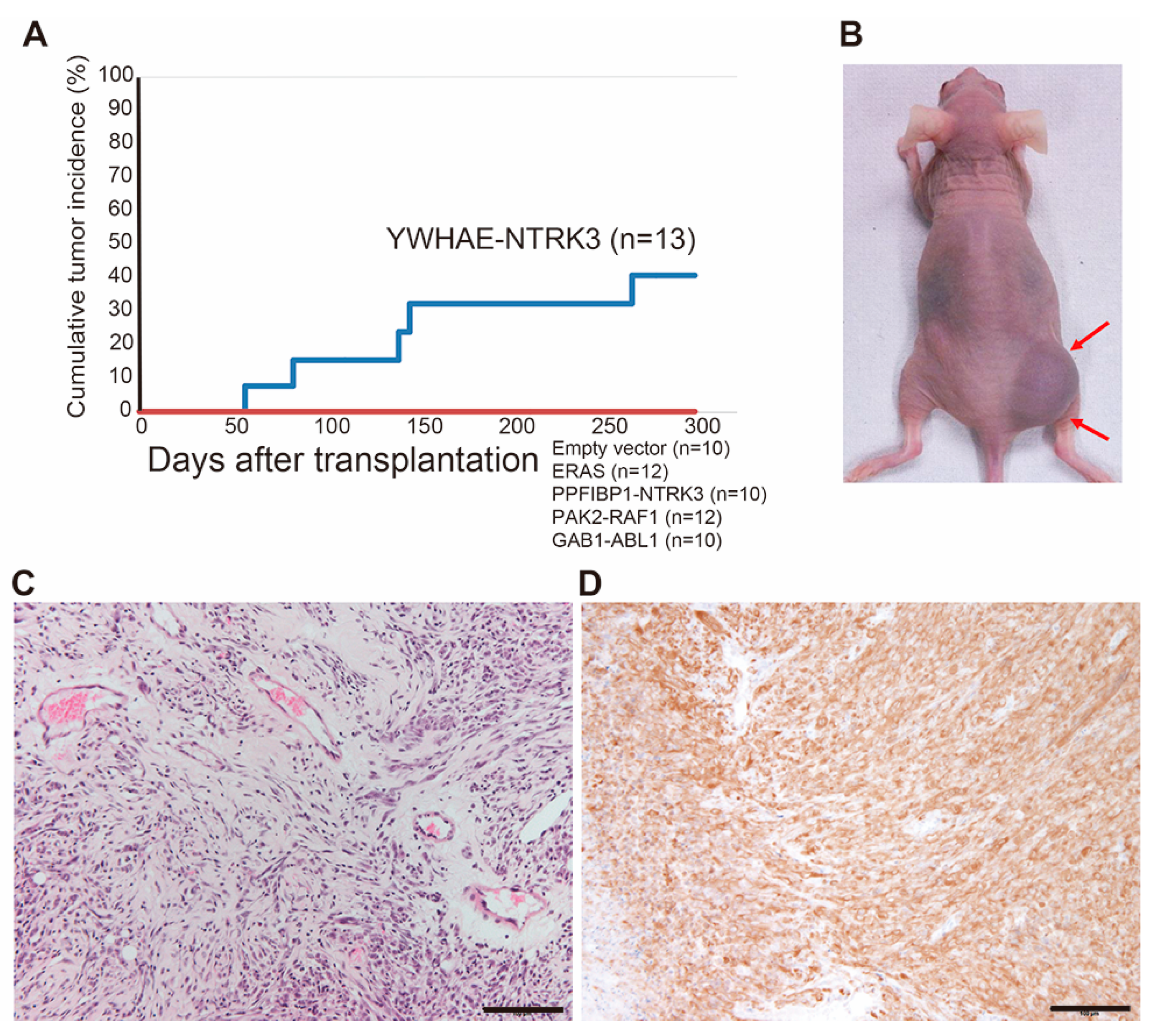
2.4. Generation of an Animal Model for NTRK3 Fusion-Associated Sarcomas
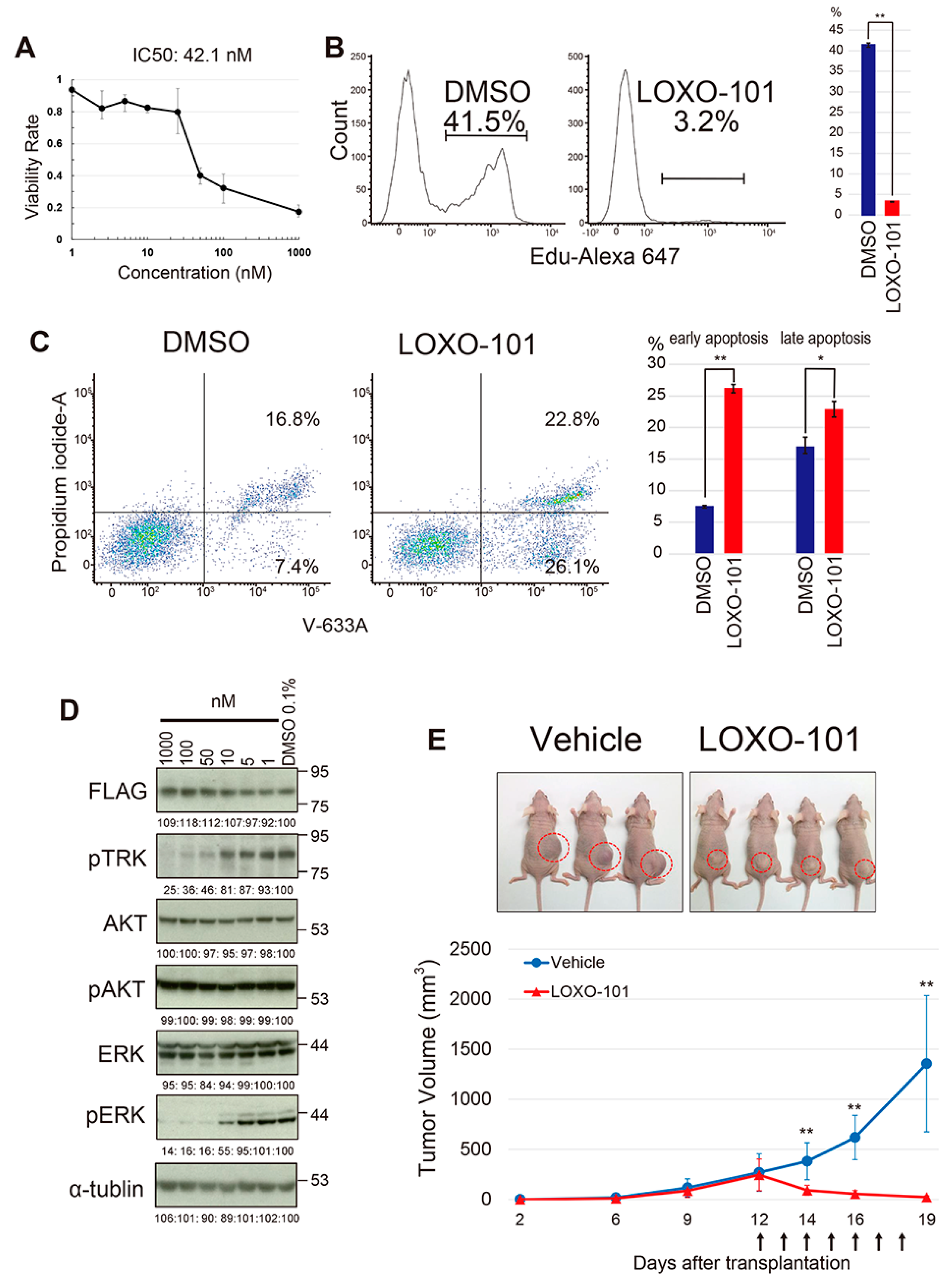
2.5. The Effect of the NTRK Inhibitor on the YWHAE-NTRK3 Sarcoma Model
3. Discussion
4. Materials and Methods
4.1. Patients and Clinical Samples
4.2. Targeted RNA Sequencing and Validation of Fusion Gene Expression by RT-PCR
4.3. Histopathology and Immunohistochemistry
4.4. Generation of the Model Mouse Expressing Novel Fusion Genes
4.5. Pharmacological Experiments
4.6. Western Blotting
4.7. Flow Cytometry
4.8. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- The WHO Classification of Tumours Editorial Board (Ed.) WHO Classification of Tumours: Soft Tissue and Bone Tumours, 5th ed.; International Agency for Research on Cancer: Lyon, France, 2020; pp. 1–607. [Google Scholar]
- Ogura, K.; Higashi, T.; Kawai, A. Statistics of bone sarcoma in Japan: Report from the bone and soft tissue tumor registry in Japan. J. Orthop. Sci. 2017, 22, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Ogura, K.; Higashi, T.; Kawai, A. Statistics of soft-tissue sarcoma in Japan: Report from the bone and soft tissue tumor registry in Japan. J. Orthop. Sci. 2017, 22, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Groisberg, R.; Roszik, J.; Conley, A.; Patel, S.R.; Subbiah, V. The role of next-generation sequencing in sarcomas: Evolution from light microscope to molecular microscope. Curr. Oncol. Rep. 2017, 19, 78. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Garbutt, C.C.; Hornicek, F.; Guo, Z.; Duan, Z. Advances in chromosomal translocations and fusion genes in sarcomas and potential therapeutic applications. Cancer Treat. Rev. 2018, 63, 61–70. [Google Scholar] [CrossRef]
- Dickson, B.C.; Swanson, D. Targeted RNA sequencing: A routine ancillary technique in the diagnosis of bone and soft tissue neoplasms. Gene Chromosome Cancer 2019, 58, 75–87. [Google Scholar] [CrossRef]
- Kawamura-Saito, M.; Yamazaki, Y.; Kaneko, K.; Kawaguchi, N.; Kanda, H.; Mukai, H.; Gotoh, T.; Motoi, T.; Fukayama, M.; Aburatani, H.; et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum. Mol. Genet. 2006, 15, 2125–2137. [Google Scholar] [CrossRef]
- Pierron, G.; Tirode, F.; Lucchesi, C.; Reynaud, S.; Ballet, S.; Cohen-Gogo, S.; Perrin, V.; Coindre, J.M.; Delattre, O. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat. Genet. 2012, 44, 461–466. [Google Scholar] [CrossRef]
- Mantilla, J.G.; Ricciotti, R.W.; Chen, E.; Hoch, B.L.; Liu, Y.J. Detecting disease-defining gene fusions in unclassified round cell sarcomas using anchored multiplex PCR/targeted RNA next-generation sequencing-Molecular and clinicopathological characterization of 16 cases. Gene Chromosome Cancer 2019, 58, 713–722. [Google Scholar] [CrossRef]
- de Bruijn, D.R.H.; Nap, J.P.; van Kessel, A.G. The (Epi)genetics of human synovial sarcoma. Gene Chromosome Cancer 2007, 46, 107–117. [Google Scholar] [CrossRef]
- Bode-Lesniewska, B.; Frigerio, S.; Exner, U.; Abdou, M.T.; Moch, H.; Zimmermann, D.R. Relevance of translocation type in myxoid liposarcoma and identification of a novel EWSR1-DDIT3 fusion. Gene Chromosome Cancer 2007, 46, 961–971. [Google Scholar] [CrossRef]
- Tanaka, M.; Yamazaki, Y.; Kanno, Y.; Igarashi, K.; Aisaki, K.; Kanno, J.; Nakamura, T. Ewing’s sarcoma precursors are highly enriched in embryonic osteochondrogenic progenitors. J. Clin. Investig. 2014, 121, 3061–3074. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Homme, M.; Yamazaki, Y.; Shimizu, R.; Takazawa, Y.; Nakamura, T. Modeling alveolar soft part sarcoma unveils novel mechanisms of metastasis. Cancer Res. 2017, 77, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, T.; Tanaka, M.; Homme, M.; Yamazaki, Y.; Takazawa, Y.; Antonescu, C.R.; Nakamura, T. CIC-DUX4 induces small round cell sarcoma distinct from Ewing sarcoma. Cancer Res. 2017, 77, 2927–2937. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Homme, M.; Yamazaki, Y.; Ae, K.; Matsumoto, S.; Subramanian, S.; Nakamura, T. Cooperation between SS18-SSX1 and miR-214 in synovial sarcoma development and progression. Cancers 2020, 12, 324. [Google Scholar] [CrossRef] [PubMed]
- Fujino, T.; Suzuki, A.; Ito, Y.; Ohyashiki, K.; Hatano, Y.; Miura, I.; Nakamura, T. Single-translocation and double-chimeric transcripts: Detection of NUP98-HOXA9 in myeloid leukemias with HOXA11 or HOXA13 breraks of the chromosomal translocation t(7;11)(p15;p15). Blood 2002, 99, 1428–1433. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Moller, E.; Nord, K.H.; Mandahl, N.; Von Steyern, F.V.; Domanski, H.A.; Marino-Enriquez, A.; Magnusson, L.; Nilsson, J.; Sciot, R.; et al. Fusion of the AHRR and NCOA2 genes through a recurrent translocation t(5;8)(p15;q13) in soft tissue angiofibroma results in upregulation of aryl hydrocarbon receptor target genes. Gene Chromosome Cancer 2012, 51, 510–520. [Google Scholar] [CrossRef]
- Suurmeijer, A.J.H.; Dickson, B.C.; Swanson, D.; Zhang, L.; Sung, Y.S.; Cotzia, P.; Fletcher, C.D.M.; Antonescu, C.R. A novel group of spindle cell tumors defined by S100 and CD34 co-expression shows recurrent fusions involving RAF1, BRAF, and NTRK1/2 genes. Gene Chromosome Cancer 2018, 57, 611–621. [Google Scholar] [CrossRef]
- Wang, Y.; Alla, V.; Goody, D.; Gupta, S.K.; Spitschak, A.; Wolkenhauer, O.; Putzer, B.M.; Engelmann, D. Epigenetic factor EPC1 is a master regulator of DNA damage response by interacting with E2F1 to silence death and activate metastasis-related gene signatures. Nucl. Acids Res. 2016, 44, 117–133. [Google Scholar] [CrossRef] [PubMed]
- Searle, N.E.; Pillus, L. Critical genomic regulation mediated by enhancer of polycomb. Curr. Genet. 2018, 64, 147–154. [Google Scholar] [CrossRef]
- Micci, F.; Panagopoulos, I.; Bjerkehagen, B.; Heim, S. Consistent rearrangement of chromosomal band 6p21 with generation of fusion genes JAZF1/PHF1 and EPC1/PHF1 in endometrial stromal sarcoma. Cancer Res. 2006, 66, 107–112. [Google Scholar] [CrossRef]
- Antonescu, C.R.; Sung, Y.S.; Chen, C.L.; Zhang, L.; Chen, H.W.; Singer, S.; Agaram, N.P.; Sboner, A.; Fletcher, J.D. Novel ZC3H7B-BCOR, MEAF6-PHF1, and EPC1-PH1 fusions in ossifying fibromyxoid tumors—Molecular characterization shows genetic overlap with endometrial stromal sarcoma. Gene Chromosome Cancer 2014, 53, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Banito, A.; Laporte, A.N.; Roe, J.S.; Sanchez-Vega, F.; Huang, C.H.; Dancsok, A.R.; Hatzi, K.; Chen, C.C.; Tschaharganeh, D.F.; Chandwani, R.; et al. The SS18-SSX1 oncoprotein hijacks KDM2B-PRC1.1 to drive synovial sarcoma. Cancer Cell 2018, 33, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Isshiki, Y.; Iwama, A. Emerging role of noncanonical polycomb repressive complexes in normal and malignant hematopoiesis. Exp. Hematol. 2018, 68, 10–14. [Google Scholar] [CrossRef]
- Schulte, I.; Batty, E.M.; Pole, J.C.M.; Blood, K.A.; Mo, S.; Cooke, S.L.; Ng, C.; Howe, K.L.; Chin, S.F.; Brenton, J.D.; et al. Structural analysis of the genome of breast cancer cell line ZR-75-30 identifies twelve expressed fusion genes. BMC Genom. 2012, 13, 719. [Google Scholar] [CrossRef]
- Koo, C.Y.; Giacomini, C.; Reyes-Corral, M.; Olmos, Y.; Tavares, I.A.; Marson, C.M.; Linardopoulos, S.; Tutt, A.N.; Morris, J.D.H. Targeting TAO kinases using a new inhibitor compound delays mitosis and induces mitotic cell death in centrosome amplified breast cancer cells. Mol. Cancer Ther. 2017, 16, 2410–2421. [Google Scholar] [CrossRef] [PubMed]
- Jobby, M.K.; Sharma, Y. Calcium-binding to lens bB2- and bA3-crystallins suggests that all b-crystallins are calcium-binding proteins. FEBS J. 2007, 274, 4135–4147. [Google Scholar] [CrossRef]
- Parthasarathy, G.; Ma, B.; Zhang, C.; Gongora, C.; Zigler, J.S., Jr.; Duncan, M.K.; Sinha, D. Expression of bA3/A1-crystallin in the developing and adult rat eye. J. Mol. Hist. 2011, 42, 59–69. [Google Scholar] [CrossRef]
- Hegde, S.; Kesterson, R.A.; Srivastava, O.P. CRYbA3/A1-crystallin knockout develops nuclear cataract and causes impaired lysosomal cargo clearance and calpain activation. PLoS ONE 2016, 11, e0149027. [Google Scholar] [CrossRef]
- Luchtel, R.A.; Dasari, S.; Oishi, N.; Pedersen, M.B.; Hu, G.; Rech, K.L.; Ketterling, R.P.; Sidhu, J.; Wang, X.; Katoh, R.; et al. Molecular profiling reveals immunogenic cues in anaplastic large cell lymphomas with DUSP22 rearrangements. Blood 2018, 132, 1386–1398. [Google Scholar] [CrossRef]
- Kobayashi, M.; Sugumaran, G.; Liu, J.; Shworak, N.W.; Silbert, J.E.; Rosenberg, R.D. Molecular cloning and characterization of a human uronyl 2-sulfotransferase that sulfates iduronyl and glucuronyl residues in dermatan/chondroitin sulfate. J. Biol. Chem. 1999, 274, 10474–10480. [Google Scholar] [CrossRef]
- Melard, P.; Idrissi, Y.; Andrique, L.; Poglio, S.; Prochazkova-Carlotti, M.; Berhouet, S.; Boucher, C.; Laharanne, E.; Chevret, E.; Pham-Ledard, A.; et al. Molecular alterations and tumor suppressive function of the DUSP22 (dual specificity phosphatase 22) gene in peripheral T-cell lymphoma subtypes. Oncotarget 2016, 7, 68734–68748. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Ou, W.B.; Marino-Enriquez, A.; Zhu, M.; Mayeda, M.; Wang, Y.; Guo, X.; Brunner, A.L.; Amant, F.; French, C.A.; et al. 14-3-3- fusion oncogenes in high-grade endometrial stromal sarcoma. Proc. Natl. Acad. Sci. USA 2012, 109, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Hoang, L.N.; Yip, S.; Reyes, C.; Marino-Enriquez, A.; Eilers, G.; Tao, D.; Chiang, S.; Fletcher, J.A.; Soslow, R.A.; et al. Frequent expression of KIT in endometrial stromal sarcoma with YWHAE genetic rearrangement. Mod. Pathol. 2014, 27, 751–757. [Google Scholar] [CrossRef] [PubMed]
- O’Meara, E.; Stack, D.; Lee, C.H.; Garvin, A.J.; Morris, T.; Argani, P.; Han, J.S.; Karlsson, J.; Gisselson, D.; Leuschner, I.; et al. Characterization of the chromosomal translocation t(10;17)(q22;p13) in clear cell sarcoma of kidney. J. Pathol. 2012, 227, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Soda, M.; Togashi, Y.; Sugawara, E.; Hatano, S.; Asaka, R.; Okumura, S.; Nakagawa, K.; Mano, H.; Ishikawa, Y. Pulmonary inflammatory myofibroblastic tumor expressing a novel fusion, PPFIBP1-ALK: Reappraisal of anti-ALK immunohistochemistry as a tool for novel ALK fusion identification. Clin. Cancer Res. 2011, 17, 3341–3348. [Google Scholar] [CrossRef]
- Doebel, R.C.; Davis, L.E.; Vaishnavi, A.; Le, A.T.; Estrada-Bernal, A.; Keysar, S.; Jimeno, A.; Varella-Garcia, M.; Aisner, D.L.; Li, Y.; et al. An oncogenic NTRK fusion in a patient with soft-tissue sarcoma with response to the tropomyosin-related kinase inhibitor LOXO-101. Cancer Discov. 2015, 5, 1049–1057. [Google Scholar] [CrossRef]
- Chmielecki, J.; Crago, A.M.; Rosenberg, M.; O’Conner, R.; Walker, S.R.; Ambrogio, L.; Auclair, D.; McKenna, A.; Heinrich, M.C.; Frank, D.A.; et al. Whol-exome sequencing identifies a recurrent NAB2-STAT6 fusion in solitary fibrous tumors. Nat. Genet. 2013, 45, 131–132. [Google Scholar] [CrossRef]
- Robinson, D.R.; Wu, Y.M.; Kalyana-Sundaram, S.; Cao, X.; Lonigro, R.J.; Sung, Y.S.; Chen, C.L.; ZZhang, L.; Wang, R.; Su, F.; et al. Identification of recurrent NAB2-STAT6 gene fusions in solitary fibrous tumor by integrative sequencing. Nat. Genet. 2013, 45, 180–185. [Google Scholar] [CrossRef]
- Cocco, E.; Scaltriti, M.; Drilon, A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat. Rev. Clin. Oncol. 2018, 15, 731–747. [Google Scholar] [CrossRef]
- Patel, N.; Chrisinger, J.S.A.; Demicco, E.G.; Sarabia, S.F.; Reuther, J.; Kumar, E.; Oliveira, A.M.; Billings, S.D.; Bovee, J.V.M.G.; Roy, A.; et al. USP6 activation in nodular fasciitis by promoter-swapping gene fusions. Mod. Pathol. 2017, 30, 1277–1588. [Google Scholar] [CrossRef]
- Teramura, Y.; Yamazaki, Y.; Tanaka, M.; Sugiura, Y.; Takazawa, Y.; Takeuchi, K.; Nakayama, T.; Kaneko, T.; Musha, Y.; Funauchi, Y.; et al. Case of mesenchymal tumor with the PPP6R3-USP6 fusion, possible nodular fasciitis with malignant transformation. Pathol. Int. 2019, 69, 706–709. [Google Scholar] [CrossRef] [PubMed]
- Aman, P. Fusion genes in solid tumors. Semin. Cancer Biol. 1999, 9, 303–318. [Google Scholar] [CrossRef] [PubMed]
- Hirata, M.; Asano, N.; Katayama, K.; Yoshida, A.; Tsuda, Y.; Sekimizu, M.; Mitani, S.; Kobayashi, E.; Komiyama, M.; Fujimoto, J.; et al. Integrated exome and RNA sequencing of dedifferentiated liposarcoma. Nat. Commun. 2019, 10, 5683. [Google Scholar] [CrossRef] [PubMed]
- Stephens, P.J.; Greenman, C.D.; Fu, B.; Yang, F.; Bignell, G.R.; Mudie, L.J.; Pleasance, E.D.; Lau, K.W.; Beare, D.; Stebbings, L.A.; et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 2011, 144, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Ashar, H.R.; Fejzo, M.S.; Tkachenko, A.; Zhou, X.; Fletcher, J.A.; Weremowicz, S.; Morton, C.C.; Chada, K. Disruption of the architectural factor HMGI-C: DNA-binding AT hook motifs fused in lipomas to distinct transcriptional regulatory domains. Cell 1995, 82, 57–65. [Google Scholar] [CrossRef]
- Kazmierczak, B.; Henning, Y.; Wanschura, S.; Rogalla, P.; Bartnitzke, S.; Van de Ven, W.; Bullerdiek, J. Description of a novel fusion transcript between HMGI-C, a gene encoding for a member of the high mobility group proteins, and the mitochondrial aldehyde dehydrogenase gene. Cancer Res. 1995, 55, 6038–6039. [Google Scholar]



| Features | Round Cell Sarcomas (n = 25) | Spindle Cell Sarcomas (n = 30) |
|---|---|---|
| Male, sex, n (%) | 16 (64.0) | 15 (50.0) |
| Age at diagnosis (in years, average ± SD) | 50.0 ± 17.0 | 50.0 ± 19.0 |
| Primary site, n (%) | ||
| Extremities | 13 (52.0) | 20 (66.6) |
| Trunk | 8 (32.0) | 10 (33.3) |
| Head and neck | 3 (12.0) | 0 (0) |
| Others | 1 (4.0) | 0 (0) |
| Original diagnosis (n) | Round cell sarcoma (14) | Solitary fibrous tumor (12) |
| Ewing sarcoma (3) | Synovial sarcoma (6) | |
| Extraskeletal myxoid chondrosarcoma (3) | Low-grade myxofibrosarcoma (3) | |
| Clear cell sarcoma (1) | Low-grade fibromyxoid tumor (2) | |
| Epithelioid hemangioendothelioma (1) | Spindle cell sarcoma (2) | |
| Malignant epithelioid tumor (1) | Angiofibroma (1) | |
| Mesenchymal chondrosarcoma (1) | Fibrosarcoma (1) | |
| Myoepithelial carcinoma (1) | Hemangiopericytoma (1) | |
| Malignant nodular fasciitis (1) | ||
| Proliferative fasciitis (1) |
| Case | Fusion Gene | Chromosome * | Functional Protein |
|---|---|---|---|
| Round Cell Sarcomas | |||
| 2 | PAK2-RAF1 | inv (3) (p25q29) | yes |
| 6 | YTHDF2-GMEB1 | del (1) (p35p35.3) | no |
| 14 | TAOK1-CRYBA1 | inv (17) (q11.2q11.2) | yes |
| 52 | BTBD9-HMGA2 | t (6;12) (p21;q15) | no |
| BTBD9-PVT1 | t (6;8) (p21;q24) | no | |
| PTPRR-KSR2 | inv (12) (q15q24.23) | no | |
| TBX5-PTPRR | del (12) (q24.1q15) | no | |
| Spindle cell sarcomas | |||
| 21 | DDX26B-IRF4 | t (X;6) (q26.3;p25.3) | no |
| 22 | NCOA2-SERPINA7 | t (X;8) (q22.2;q13.3) | no |
| 32 | YWHAE-NTRK3 | t (15;17) (q25;p13.3) | yes |
| CNNM2-EGR2 | inv (10) (q24.32q21.3) | no | |
| 36 | RPSAP52-TGFBR3 | t (1;12) (q14.3;p22.1) | no |
| 38 | MSN-ERAS | inv (X) (p11.23q12) | yes ** |
| FRYL-FIP1L1 | del (4) (p11q12) | no | |
| 42 | PPFIBP1-NTRK3 | t (12;15) (p12.1;q25) | yes |
| CRTC1-DOHH | inv (19) (p13.11p13.3) | no | |
| 43 | AHRR-NCOA3 | t (5;20) (q15.3;q12) | yes |
| 50 | UST-DUSP22 | inv (6) (p25.3q25.1) | yes |
| 55 | EPC1-KDM2B | t (10;12) (p11;q24.31) | yes |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teramura, Y.; Tanaka, M.; Yamazaki, Y.; Yamashita, K.; Takazawa, Y.; Ae, K.; Matsumoto, S.; Nakayama, T.; Kaneko, T.; Musha, Y.; et al. Identification of Novel Fusion Genes in Bone and Soft Tissue Sarcoma and Their Implication in the Generation of a Mouse Model. Cancers 2020, 12, 2345. https://doi.org/10.3390/cancers12092345
Teramura Y, Tanaka M, Yamazaki Y, Yamashita K, Takazawa Y, Ae K, Matsumoto S, Nakayama T, Kaneko T, Musha Y, et al. Identification of Novel Fusion Genes in Bone and Soft Tissue Sarcoma and Their Implication in the Generation of a Mouse Model. Cancers. 2020; 12(9):2345. https://doi.org/10.3390/cancers12092345
Chicago/Turabian StyleTeramura, Yasuyo, Miwa Tanaka, Yukari Yamazaki, Kyoko Yamashita, Yutaka Takazawa, Keisuke Ae, Seiichi Matsumoto, Takayuki Nakayama, Takao Kaneko, Yoshiro Musha, and et al. 2020. "Identification of Novel Fusion Genes in Bone and Soft Tissue Sarcoma and Their Implication in the Generation of a Mouse Model" Cancers 12, no. 9: 2345. https://doi.org/10.3390/cancers12092345
APA StyleTeramura, Y., Tanaka, M., Yamazaki, Y., Yamashita, K., Takazawa, Y., Ae, K., Matsumoto, S., Nakayama, T., Kaneko, T., Musha, Y., & Nakamura, T. (2020). Identification of Novel Fusion Genes in Bone and Soft Tissue Sarcoma and Their Implication in the Generation of a Mouse Model. Cancers, 12(9), 2345. https://doi.org/10.3390/cancers12092345





