Abstract
High mobility group box B (HMGB) proteins are overexpressed in different types of cancers such as epithelial ovarian cancers (EOC). We have determined the first interactome of HMGB1 and HMGB2 in epithelial ovarian cancer (the EOC-HMGB interactome). Libraries from the SKOV-3 cell line and a primary transitional cell carcinoma (TCC) ovarian tumor were tested by the Yeast Two Hybrid (Y2H) approach. The interactome reveals proteins that are related to cancer hallmarks and their expression is altered in EOC. Moreover, some of these proteins have been associated to survival and prognosis of patients. The interaction of MIEN1 and NOP53 with HMGB2 has been validated by co-immunoprecipitation in SKOV-3 and PEO1 cell lines. SKOV-3 cells were treated with different anti-tumoral drugs to evaluate changes in HMGB1, HMGB2, MIEN1 and NOP53 gene expression. Results show that combined treatment of paclitaxel and carboplatin induces a stronger down-regulation of these genes in comparison to individual treatments. Individual treatment with paclitaxel or olaparib up-regulates NOP53, which is expressed at lower levels in EOC than in non-cancerous cells. On the other hand, bevacizumab diminishes the expression of HMGB2 and NOP53. This study also shows that silencing of these genes affects cell-viability after drug exposure. HMGB1 silencing causes loss of response to paclitaxel, whereas silencing of HMGB2 slightly increases sensitivity to olaparib. Silencing of either HMGB1 or HMGB2 increases sensitivity to carboplatin. Lastly, a moderate loss of response to bevacizumab is observed when NOP53 is silenced.
1. Introduction
Ovarian cancer is nowadays the 7th most common cancer in women, with only a 30–40% average 5-year relative survival rate. Early diagnosis, when the tumor is still localized in the ovaries, is a clear advantage, since this rate then increases up to 92% [1]. More than 90% of malignant ovarian tumors are epithelial ovarian cancers (EOC) and the rest derive from stromal or germ cells. The histology of malignant EOC, or carcinomas, is heterogeneous [1] and they have been classified in five main histotypes: high-grade serous (HGSOC with an incidence of 70% among total EOC), low-grade serous (LGSOC; incidence <5%); endometrioid (ENOC; incidence of 10%), clear cell (CCOC; incidence of 10%), and mucinous (MOC; incidence 3%). This classification takes into account the resemblance to normal gynecological cell line; serous: resembling epithelium lining the Fallopian tubes; mucinous: resembling epithelium lining endocervix, and containing intra-cytoplasmic mucin; endometrioid: resembling epithelium of uterine corpus; clear cell: comprising clear cells and hobnail cells [1]. Each histotype has been associated to a particular set of somatic mutations. HGSOC type is associated to BRCA1/2 and TP53 mutations; LGSOC type, to BRAF and, KRAS mutations; MOC type, to KRAS mutation; ENOC type to PTEN, CTNNB1, ARID1A and PIK3CA mutations; and finally CCOC type, to ARID1A and PIK3CA mutations [1].
High mobility group box-B proteins (HMGB), non-histone components of chromatin, exert global regulatory functions on gene expression [2]. Their release from cancerous cells to the extracellular medium promotes tumor growth and metastasis, and their overexpression is associated to ovarian cancer, among others [3]. Increased HMGB1 expression has been associated to TLR4 expression and activated NF-κB signaling pathway [4,5]. This is accompanied by worse clinical outcomes in EOC patients, which suggests that signaling by endogenous ligands may contribute to an inflammatory microenvironment, which worsens the disease [4]. In the ovarian cancer cell line SKOV-3, HMGB1 silencing diminishes the expression of VEGF and CXCL12, revealing its involvement in angiogenesis [6].
HMGB1 has been repeatedly proposed as a diagnostic and prognostic biomarker for human ovarian cancer [7,8,9,10,11]. HMGB1 immunostaining in serous, mucinous, endometrioid, and clear-cell ovarian carcinomas [12] shows that HMGB1 expression is observed in different EOC histotypes. HMGB1 overexpression in early stages of the disease is also an advantage for its use as diagnostic biomarker [12]. HMGB1 and HMGB2 proteins have been also related to chemotherapy resistance in serous EOC [13]. High expression of HMGB1 has been related to cisplatin resistance [14] and downregulation of HMGB1 re-sensitizes ovarian cancer cells to carboplatin [15]. HMGB2 expression has been associated to resistance against oxaliplatin and cisplatin [16].
Protein interactomes associated to a particular disease are valuable tools to understand molecular mechanisms of pathogenesis and to re-define diagnostic panels of specific biomarkers [17]. The low rates of survival after first diagnosis, clearly show that clinical management of ovarian cancer patients needs of earlier diagnosis and more specific therapies [18,19]. Considering the relevance of HMGB proteins in EOC, deciphering the HMGB1/2 interactome in ovarian cancerous cells is an attractive tool to achieve this goal. We have followed a Yeast-Two-Hybrid (Y2H) approach using libraries derived from SKOV-3 cells, a widely used ovarian cancer cell line; and also libraries from cancer tissue from a patient diagnosed of primary transitional cell carcinoma (TCC) of the ovary, a relatively rare subtype of epithelial ovarian cancer, which represents approximately 2% of all ovarian tumors [20]. Common targets in both libraries could discover important interactions, which are independent of the histological subtypes and their specific mutations. Functional significance of the discovered targets in relation to ovarian cancer is discussed, with special focus on MIEN1 and NOP53. Expression changes of HMGB1, HMGB2, MIEN1 and NOP53 genes have been evaluated in response to drugs usually employed in ovarian cancer treatments: bevacizumab, olaparib, paclitaxel or carboplatin. The effects of HMGB1, HMGB2, MIEN1 and NOP53 silencing on cell sensitivity to these drugs are also reported.
2. Results
2.1. HMGB1 and HMGB2 Y2H-Interactomes in Epithelial Ovarian Cancer
Protein interactions were assayed by the yeast two-hybrid (Y2H) approach. The baits (HMGB1 or HMGB2) were cloned in fusion to the DNA-binding domain of the yeast transcriptional activator GAL4, while libraries of prey proteins were fused in frame to the GAL4 activation domain. Plasmids expressing bait and prey fusion proteins were used to transform two different haploid yeast strains able to form diploids. After mating, the transcriptional activator GAL4 is reconstructed in the diploids only if interaction between proteins exists, and therefore reporter genes under GAL4-activated promoters are expressed. This technique is highly sensitive and, in order to diminish the appearance of false positives, a screening using independent markers was used as explained in Materials and Methods. Two cDNA libraries were constructed using total RNA extracted from SKOV-3 cells or from tissue obtained from a tumor diagnosed as primary transitional cell carcinoma (TCC) of the ovary. TCC is grouped with high-grade serous carcinoma (HGSC) in the current World Health Organization classification and it is also associated with BRCA mutations [21]. Y2H assays and screening were carried out as described in Materials and Methods and clones, identified as positives using at least three different reporter genes, were sequenced. In the libraries derived from SKOV-3 cells and using the HMGB1 bait (Table 1), a positive clone showed homology to lncRNA MALAT1 and 5 to coding sequences (AKIP1, KRT7, ATF71P, UHRF2, WDR60); using the HMGB2 bait, 7 coding sequences were identified (BCCIP, COMMD1, NOP53, MIEN1, ROCK1, U2AF1, ZNF668).

Table 1.
Clones from the SKOV-3 libraries, which interact with HMGB1 or HMGB2 in the Y2H assays.
In the library prepared from ovarian tumor tissue (Table 2), 5 clones with coding sequences were identified using the HMGB1 bait (C1QA, DAG1, RPL29, RSF1, TGM2), and 6 (COMMD1, MIEN1, PCBP1, TBC1D25, ZFR, ZNF428) were identified with the HMGB2 bait. Table 1 and Table 2 summarize the details of each detected interaction with HMGB1 or HMGB2, as well as a brief functional description of the proteins identified in these EOC-HMGB-interactomes.

Table 2.
Clones from the cancerous ovarian tissue libraries, which interact with HMGB1 or HMGB2 in the Y2H assays.
As above reported, MALAT1, a lncRNA, previously related to ovarian cancer [22,23,24,25,26,27], and to epithelial to mesenchymal transition (EMT) [28], was detected in our results of EOC-HMGB1-interactome (Table 1). We verified by sequencing that the micropeptide derived from MALAT1 lncRNA, MTEVEMKLLHGVKNVFKRKLRERTTEPRINTNRRAMLLD, is in frame and fused to GAL4 in the recovered Y2H clone. Therefore, it is possible that the interaction of this micropeptide with HMGB1 might be responsible of the positive result obtained in the Y2H screening. In support of this explanation, translation of this micropeptide from lncRNA MALAT1 has been previously reported in several ribosome-profiling experiments using human colorectal cancer cells HCT116 [29] and human embryonic kidney HEK293 cells [30].
Two proteins that interact with HMGB2, COMMD1 and MIEN1, are identified in SKOV-3 and tumor tissue libraries, which cross-validate these results. The interactions of HMGB1 with RLP29 and ZNF428, and the interaction of HMGB2 with ZNF428 were already described in non-cancerous ovarian HOSEpiC cells from epithelial origin [31]. The interaction between HMGB1 and KRT7 was also detected in cells from healthy ovarian tissue [31].
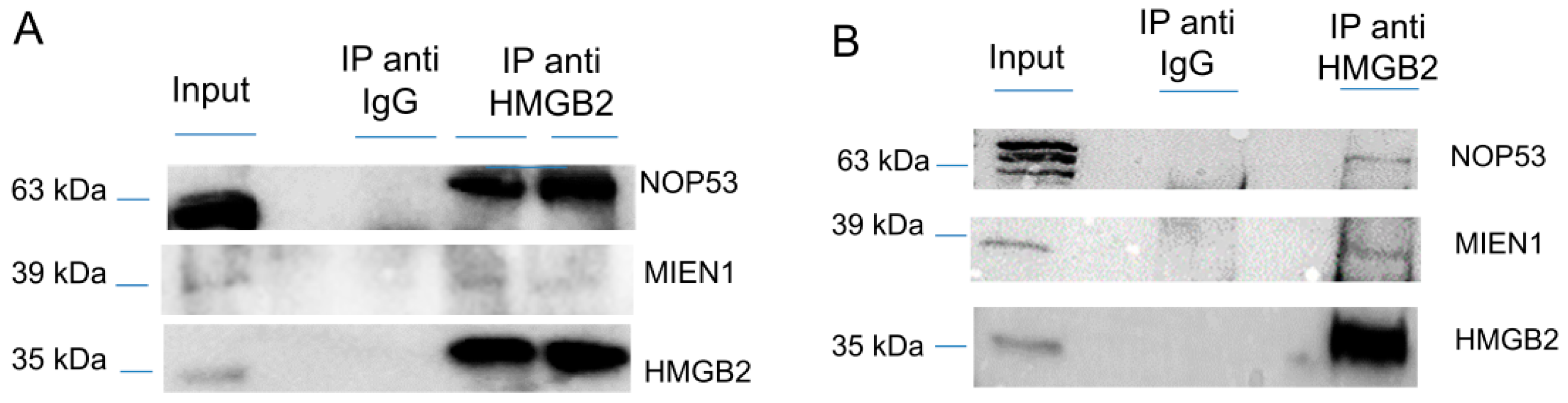
Since the Y2H interactome was obtained from SKOV-3 cells, not considered as a cell line representative of the more frequent HGSOC, although extensively used in EOC studies [32], and from tissue extracted from primary transitional cell carcinoma (TCC) of the ovary, which is a relatively unfrequently diagnosed serous EOC, we decided to validate HMGB2 interactions with MIEN1 and NOP53 by co-immunoprecipitation, an orthogonal method to the Y2H approach, and using two EOC cell lines, SKOV-3 and PEO1 representatives of ENOC/CCOC and HGSOC respectively [33]. Results (Figure 1 and Figure S1) corroborate the Y2H data. Apparently the co-immunoprecipitation of NOP53 with HMGB2 is much more efficient in SKOV-3 than in PEO1, perhaps due to the different characteristics of both cell lines, although we cannot attribute this observation to a specific cause.
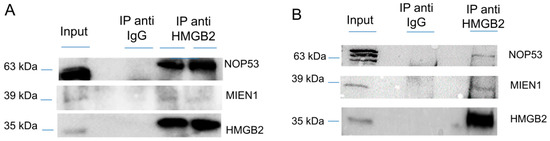
Figure 1.
Co-immunoprecipitation of HMGB2 and MIEN1 or NOP53 (A) in SKOV-3 cells (B) in PEO1 cells. Proteins extracted from cells (Input) were immunoprecipitated with HMGB2 antibodies (IP anti HMMGB2) or IgG (IP anti IgG) and the Western blot were incubated with HMGB2, MIEN1 and NOP53 antibodies.
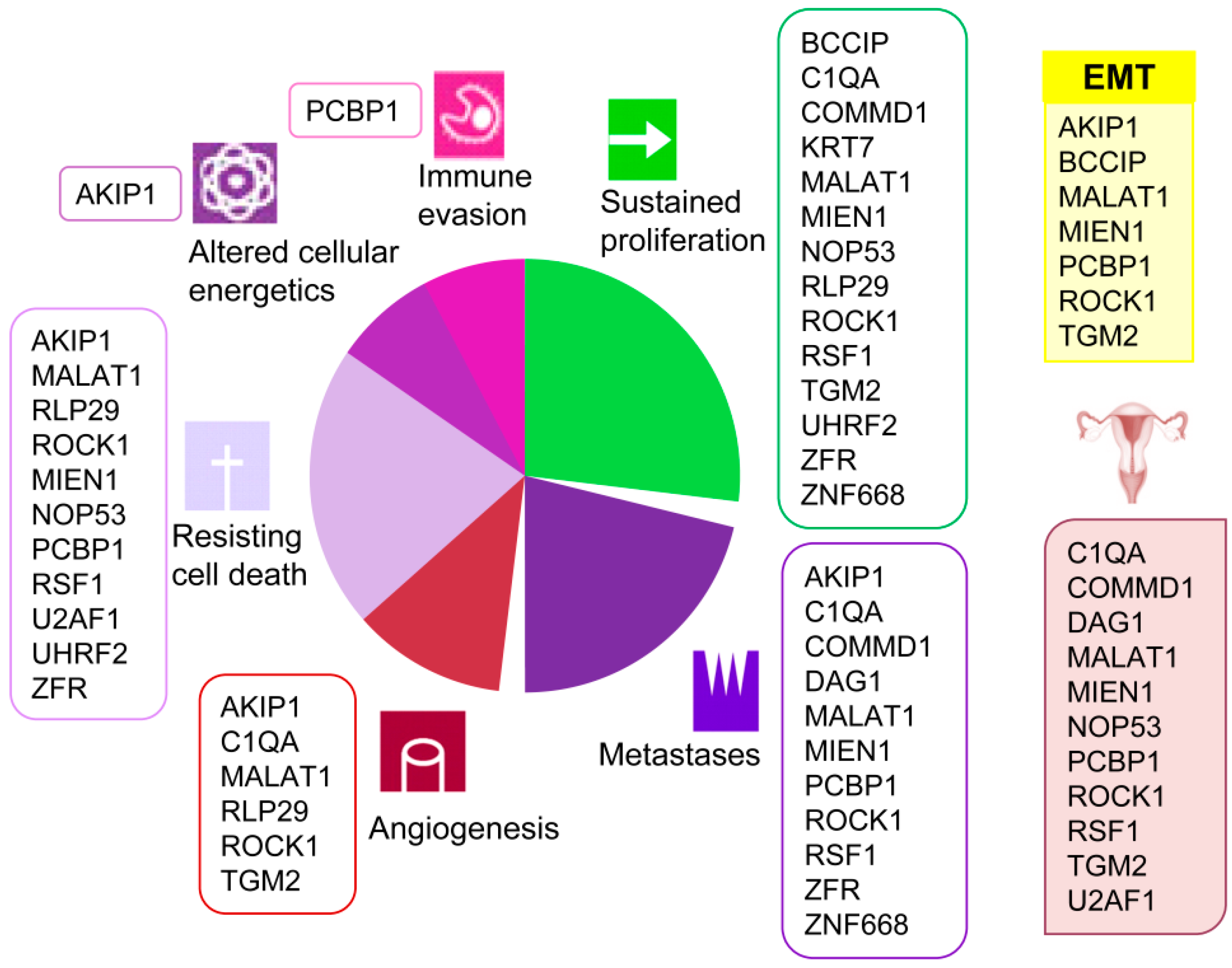
A clear association of these proteins with several current cancer hallmarks such as sustained proliferation, metastasis, angiogenesis, resisting cell death, altered cellular energetics, and immune evasion is evidenced (Figure 2). Besides, and remarkably, several among these proteins have been previously associated to ovarian cancer (COMMD1 [34], NOP53 [35], MIEN1 [36,37], ROCK1 [38,39,40], PCBP1 [41], TGM2 [42], U2AF1 [43], C1QA [44], DAG1 [45] and RSF1 [46,47,48,49]). Furthermore, several detected proteins (BCCIP [50], ROCK1 [51], PCBP1 [52], AKIP1 [53], TGM2 [54], and MIEN1 [55]) are involved in the epithelial to mesenchymal transition (EMT), typical of epithelial cells in malignant differentiation processes. However, none of them had been cited for their interaction with HMGB proteins until now, which add value to the present work and reveals the importance of the HMGB interactome in EOC. We have reviewed in the literature the experimentally confirmed functions of the proteins detected in our EOC-HMGB-interactome study (Figure 2).
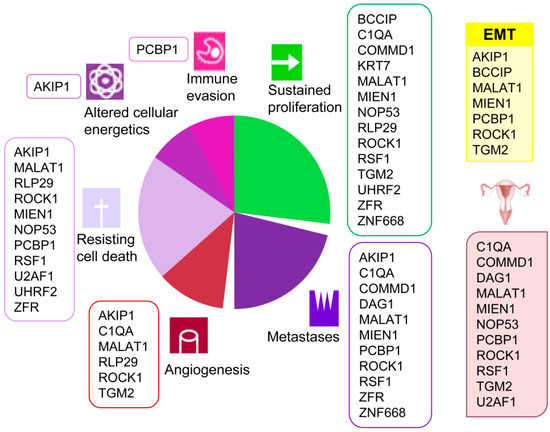
Figure 2.
Associations of proteins detected in the Y2H EOC-HMGB-interactome to cancer hallmarks, EOC and EMT. Proteins previously related to cancer from ovarian origin are shown under the ovary pictogram; those involved in epithelial to mesenchymal transition under the yellow EMT box. References supporting the scheme are indicated for each protein as follows. AKIP1 [56,57,58,59,60], BCCIP [61,62], COMMD1 [63,64,65], C1QA [66], DAG1 [67], KRT7 [68], MALAT1 [24,69,70], MIEN1 [55,71,72], NOP53 [73,74,75], PCBP1 [76,77,78], RLP29 [79,80], ROCK1 [38,81,82,83,84,85,86], RSF1 [47,48,87,88], TGM2 [89,90], UHRF2 [91,92], U2AF1 [93], ZNF668 [94,95], ZRF [96,97,98].
2.2. Analysis of the EOC-HMGB-Interactome According to Differential Expression and Clinical Outcome
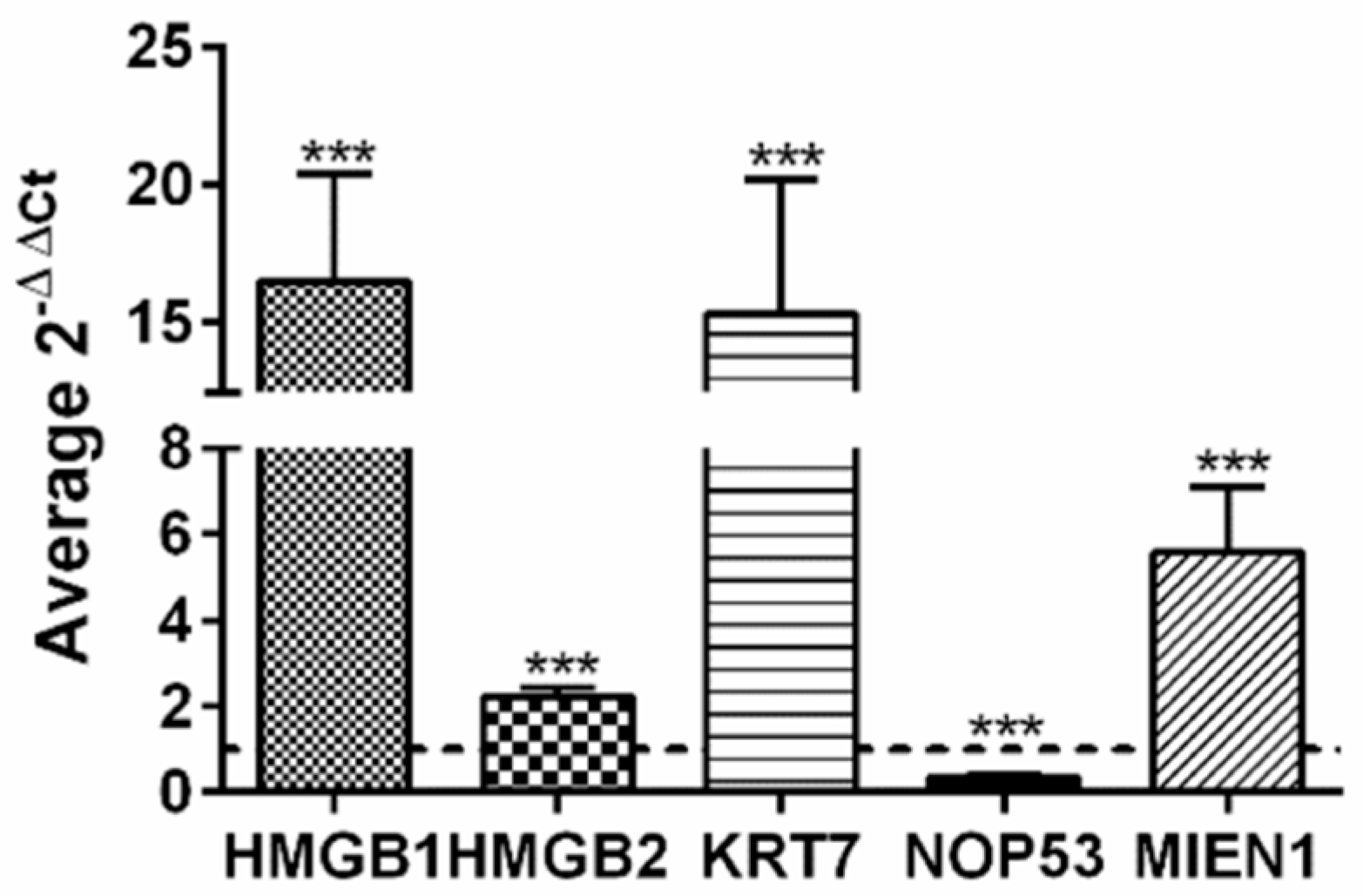
Taking advantage of public available data, accessible though Expression Atlas [99] at European Bioinformatics Institute (https://www.ebi.ac.uk, accessed on 15-5-2020) we have compared gene expression levels of HMGB1, HMGB2 and all the genes found in our EOC-HMGB-interactome in ovary tissue from healthy individuals (39 samples from GTEx Project [100]) and public data extracted from Pan-Cancer Analysis of Whole Genomes (PCAWG) corresponding to 110 tumors of ovarian adenocarcinomas (Table 3). Data indicate that HMGB1, HMGB2, as well as most of their targets identified in our Y2H study are expressed at higher levels in ovarian adenocarcinoma than in normal ovarian tissue, following a pattern of co-regulation that is frequently found among genes encoding proteins that interact with each other [101]. The highest ratios of RNA changes in cancerous versus healthy ovarian samples correspond to KRT7 (ratio >1700), C1QA (ratio 12.5) and MIEN1 (ratio 6.3). Only two genes, NOP53 and MALAT1 are less expressed in cancerous than in healthy ovarian cells in this comparison. Selecting a subgroup of genes we have also directly observed in experiments carried in our laboratory that HMGB1, HMGB2, MIEN1 and KRT7 are expressed at higher levels in SKOV-3 cancerous cells than in HOSEpiC normal ovary cells (Figure 3); while, also in accordance with patient data (Table 3), NOP53 is expressed at lower levels in the cancerous cell line than in the healthy ovary cell line (Figure 3). Although it is generally accepted that the majority of HGSOC arise from an extra-ovarian site we wanted to discriminate non-cancerous ovarian cells from tumor cells. For this reason we preferred to use ovarian non-cancerous cells as a pair-matched control.

Table 3.
Differential expression of HMGB1, HMGB2 and their interacting partners in healthy individuals and ovary cancer patients. mRNA levels obtaining by RNAseq are expressed in TPM (Transcripts per million).
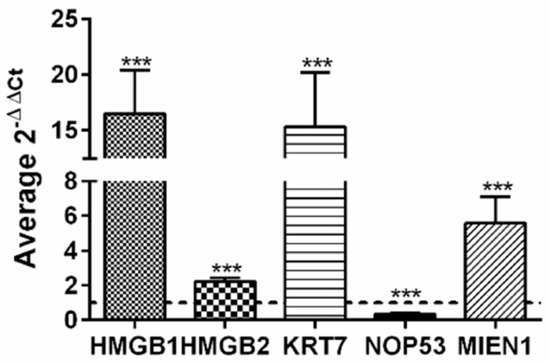
Figure 3.
Relative expression of HMGB1, HMGB2, KRT7, MIEN1 and NOP53 genes in SKOV-3 cells versus non-cancerous human ovarian HOSEpiC cells. The dotted line indicates no variation, boxes upper the line show genes over-expressed in SKOV-3 cells, and those under the line are genes under-expressed in SKOV-3 cells. *** (p < 0.001).
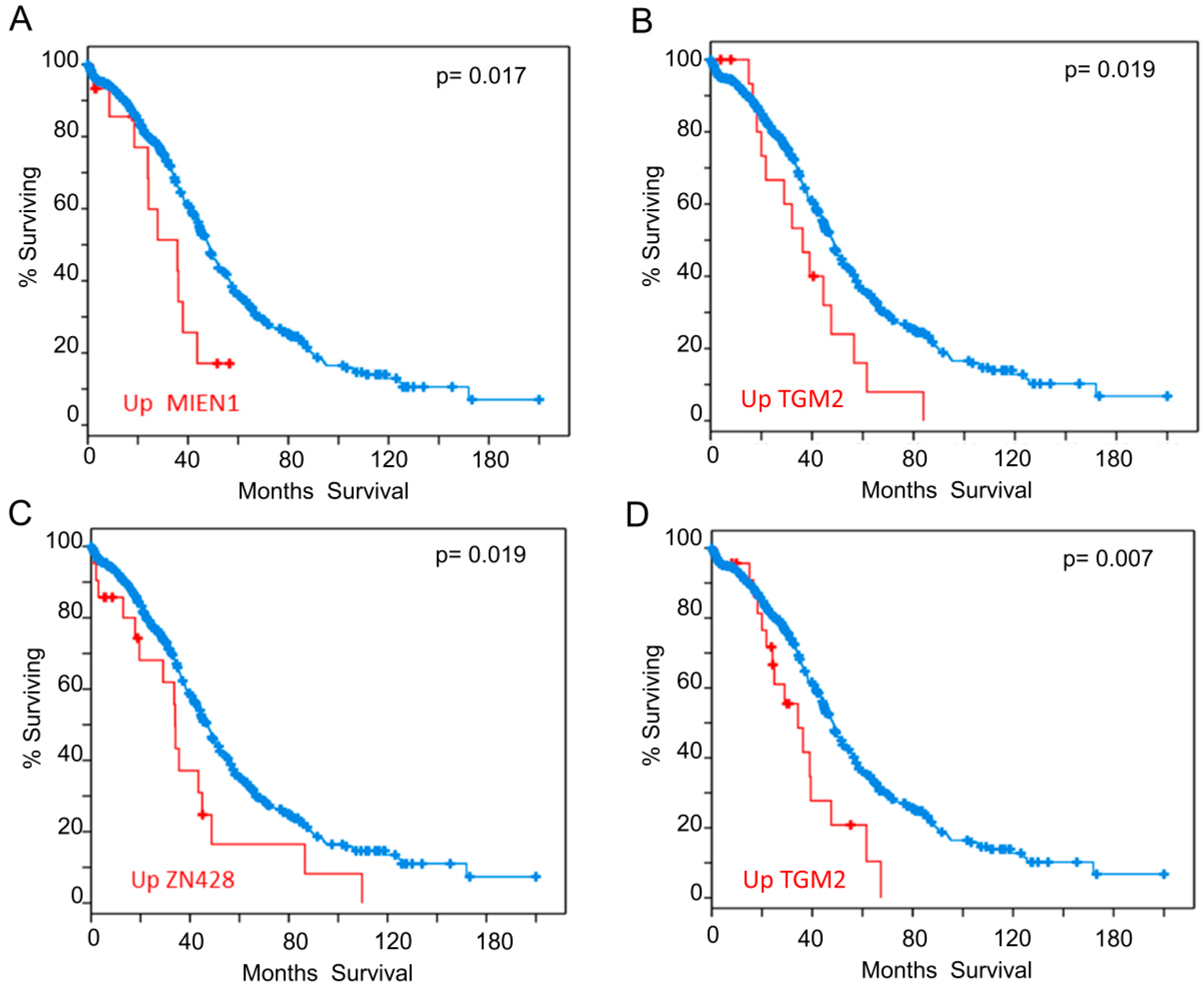
We also analyzed our interactome components (listed in Table 1 and Table 2) with the tools available in cBioportal (http://www.cbioportal.org/ accessed on 03-06-2020) [102,103] looking for correlations between expression levels and clinical outcome. To carry out this analysis, we selected samples from the study “Ovarian Serous Cystadenocarcinoma (TCGA)” with information of 606 samples from 594 patients (https://www.cbioportal.org/study/summary?id=ov_tcga. accessed on 03-06-2020). We found that patients who have higher expression of some of the genes identified in our study have lower survival expectation than the rest. Analysis of expression data based on microarray technology revealed that up-regulation of mRNA levels of MIEN1 (Figure 4A) or TGM2 (Figure 4B), negatively correlated to survival. Analysis of expression data based on RNAseq technology showed that up-regulation of ZN428 (Figure 4C) or TGM2 (Figure 4D) worsens survival outcomes. Additionally, it has also been reported that patients with RSF1 amplification or overexpression had a significantly shorter overall survival than those without [104], although for RSF1 the p-value obtained in the Logrank Test analysis did not reveal statistical significance.
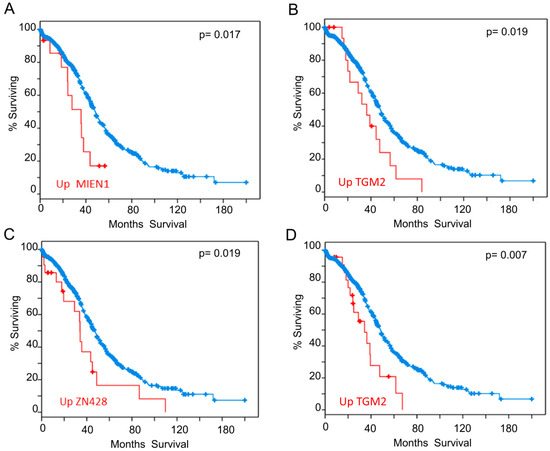
Figure 4.
Overall Survival Kaplan-Meier Estimate. Significant differences found in the analysis carried out with the proteins from Table 1 and Table 2. (A) Influence of MIEN1 overexpression according to array data (B) Influence of TGM2 overexpression according to array data (C) Influence of ZN428 overexpression according to RNAseq data (D) Influence of TGM2 overexpression according to RNAseq data. Red line represents cases with alterations and blue line cases without alterations.
2.3. Effect of HMGB1 and HMGB2 Silencing on the Expression of Genes Encoding Proteins Detected in the EOC-HMGB-Interactome
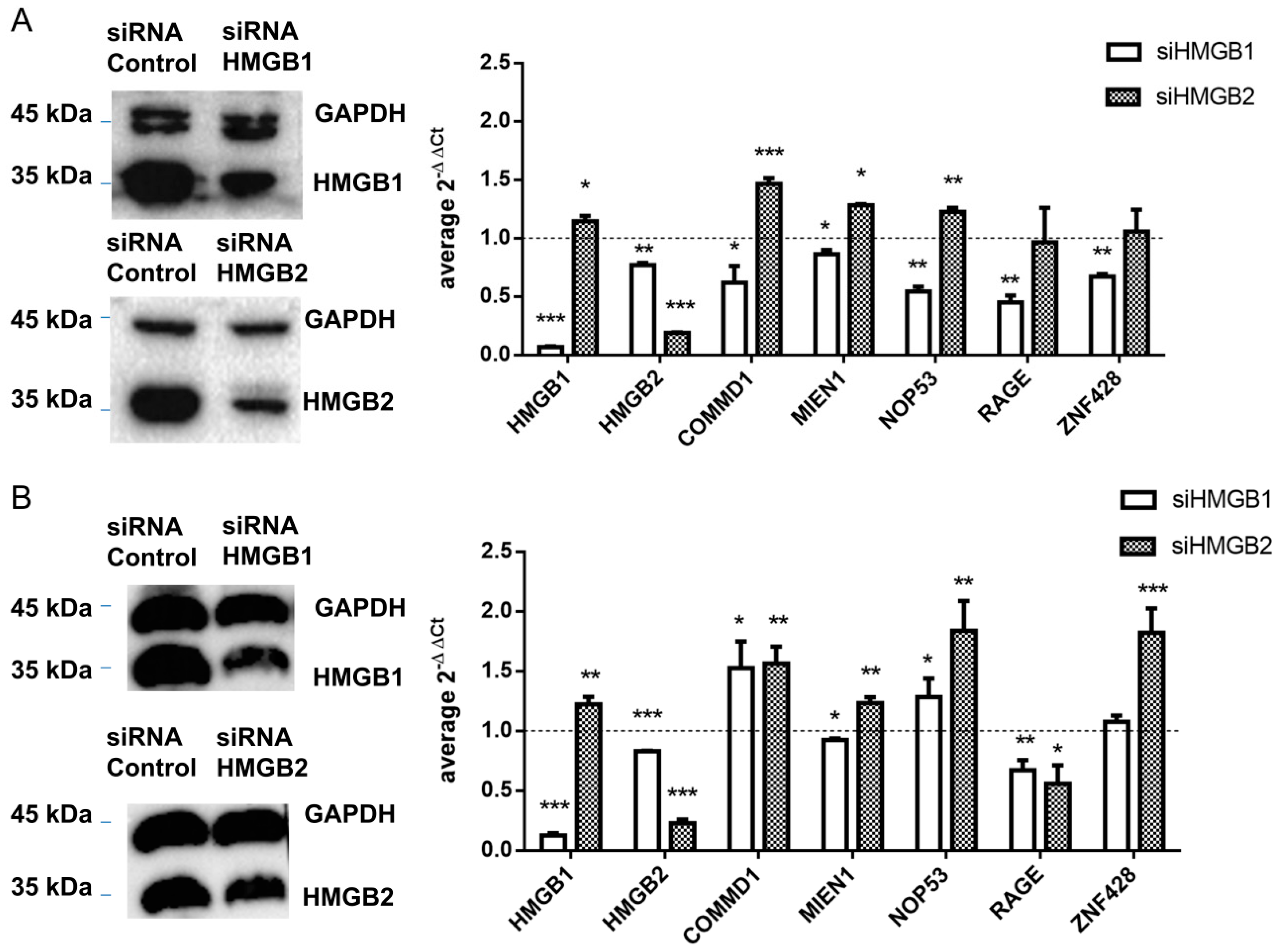
HMGB1 and HMGB2 genes were silenced in SKOV-3 and PEO1 EOC cell lines and levels of mRNA from 4 detected interacting partners of HMGB1 or HMGB2 in ovary cancer, COMMD1, MIEN1, NOP53 and ZNF428, were analyzed by qRT-PCR as explained in Materials and Methods. Changes in gene expression (siHMGB/HMGB) are shown in Figure 5. In this analysis we also included RAGE, one of the membrane receptors in the extracellular signaling function of HMGB1 [97] as a positive control. HMGB1-induced signaling can activate NFκB, which can subsequently induce the expression of HMGB1 receptors [105]. Accordingly, silencing of HMGB1 causes a decrease of RAGE expression, as well as for the other tested genes in SKOV-3 (Figure 5A); in PEO1 cells this decrease was not so evident, which might be explained by a lesser degree of the silencing achieved in this cell line (Figure 5B and Figure S2). Oppositely, we observed that expression of three of the tested genes, COMMD1, MIEN1 and NOP53, was increased after HMGB2 silencing in the two EOC cell lines analyzed, while the increase in expression of ZNF428 was statistically significant only in PEO1 cells.
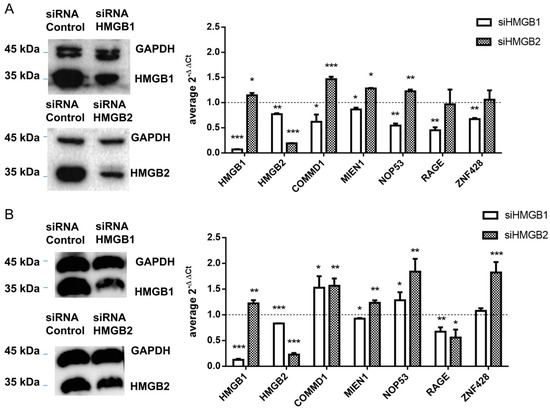
Figure 5.
Control of gene expression by HMGB1 and HMGB2 (A) in SKOV-3 cells (B) in PEO1 cells. Left panel shows the western blot confirming HMGB1 and HMGB2 silencing; complete blots and quantification are provided in Figure S3. The right panel shows relative expression of tested genes after HMGB1 or HMGB2 silencing. The dotted line indicates no variation of relative expression in the silenced line versus the line treated with cRNA, boxes upper the line show genes over-expressed in silenced cells, and those under the line are genes under-expressed in silenced cells. * (p < 0.05); ** (p < 0.01); *** (p < 0.001).
2.4. The Involvement of Proteins Detected in the EOC-HMGB-Interactome in the Response to Drugs Used in Cancer Chemotherapy
Considering that HMGB1 and HMGB2 proteins have been associated to drug resistance during cancer treatment [13,16] we also reviewed available literature to see whether the proteins detected in our interactome study could also be related to this unfavorable event in ovary cancer treatment. We found that at least 13 of these proteins had been previously cited in reference to drug resistance or sensitivity. MIEN1 has been associated to cisplatin resistance [36]. BCCIP (aliases CDKN1A or p21) is involved in resistance to carboplatin [106] and paclitaxel [107]. RSF1 is also involved in resistance to carboplatin and paclitaxel [46,49]. PCBP1 binds and stabilizes p27 mRNA and promotes cell apoptosis under paclitaxel treatment [108]. TGM2 modulates chemosensitivity of breast cancer to docetaxel [109]. DAG1 improves sensitivity to dasatinib, a tyrosine kinase inhibitor of the Src-family kinases, in EOC [45]. Besides, comparing gene expression in SKOV-3 cells and a paclitaxel resistant derived cell line (available in the GEO accession GSE54772) [110], six of the genes encoding proteins detected in our EOC-HMGB-interactomes (ATF7IP, DAG1, PCBP1, TGM2, U2AF1, and ZNF668) are expressed at higher levels in sensitive than in resistant cells, and one (WDR60) is less expressed in sensitive than in resistant cells (Figure S2).
Although the role of HMGB proteins in cisplatin resistance is widely accepted [3], there is scarce information about the role of HMGB proteins in the resistance towards its derivatives like carboplatin, or other drugs used in ovarian cancer treatment. Indeed, no data are available about the role of HMGB1 or HMGB2 in the resistance to olaparib or bevacizumab in the treatment of ovarian cancer. About the involvement of MIEN1 and NOP53 in drug resistance, only MIEN1 has been previously related to cisplatin resistance [36]. With these precedents we decided to investigate the role of HMGB1, HMGB2, MIEN1 and NOP53 in cell viability as well as in response and sensitivity to drugs currently used in ovarian cancer therapy.
We tested the effect of four compounds, used in ovary-cancer therapy in the expression of the genes HMGB1, HMGB2, MIEN1 and NOP53 in cultured cancerous SKOV-3 cells and in non-cancerous IOSE-80 ovarian cells. Each selected compound has a different mechanism of action. Carboplatin (Paraplatin®), a derivative of cisplatin, generates lesions in DNA, thereby inhibiting replication and transcription and leading to cell death [111,112]. Olaparib (AZD-2281, Lynparza®) inhibits poly ADP ribose polymerase (PARP), an enzyme necessary in DNA repair, leading to apoptosis of cancer cells [113]. Olaparib is FDA approved, for women with advanced ovarian cancer and a BRCA1/2 mutation, after they have completed the first line of platinum-based chemotherapy [114]. Bevacizumab (Avastin®), a humanized anti-vascular endothelial growth factor (VEGF) monoclonal antibody for cancer therapy is used as anti-angiogenic [115]; and paclitaxel (Taxol®), a cyclodecane first isolated from Taxus brevifolia, stabilizes microtubules in their polymerized form, leading to cell death [116]. Cells were exposed to drug concentrations selected according to previous studies, as explained in Materials and Methods, for 48 h. A comparative analysis between the effects caused by these drugs on SKOV-3 and non-cancerous ovarian IOS-80 cells is shown in Table 4.

Table 4.
Comparative effect of treatments on gene expression in cancerous (SKOV-3) versus non-cancerous (IOSE80) ovarian cells.
Relative RNA expression after these treatments was measured by qRT-PCR in reference to cells cultured in absence of the drugs, but treated with the corresponding buffer used in the preparation of drug-solutions. In general, significant effects were more frequently observed in cancerous than in non-cancerous cells. For carboplatin or paclitaxel treatments, which are generally used in first line therapy of EOC, results indicate that they cause down regulation of the genes that are over-expressed in EOC cells (HMGB1, HMGB2 and MIEN1). Combined treatment with paclitaxel and carboplatin potentiates down-regulation of these genes in comparison to individual treatments, as deduced from the fold-changes observed. NOP53, which is expressed at lower levels in EOC than in non-cancerous cells, was up-regulated after 48 h treatment with paclitaxel. Among the genes assayed, the treatment with olaparib in cancerous cells only affected NOP53, increasing its expression (Table 4). Bevacizumab had also minor effects on HMGB2 and NOP53 expression, in this case diminishing their expression (Table 4).
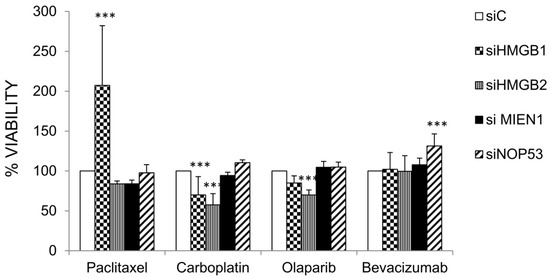
2.5. Effect HMGB1, HMGB2, MIEN1 and NOP53 Silencing on Drug Sensitivity
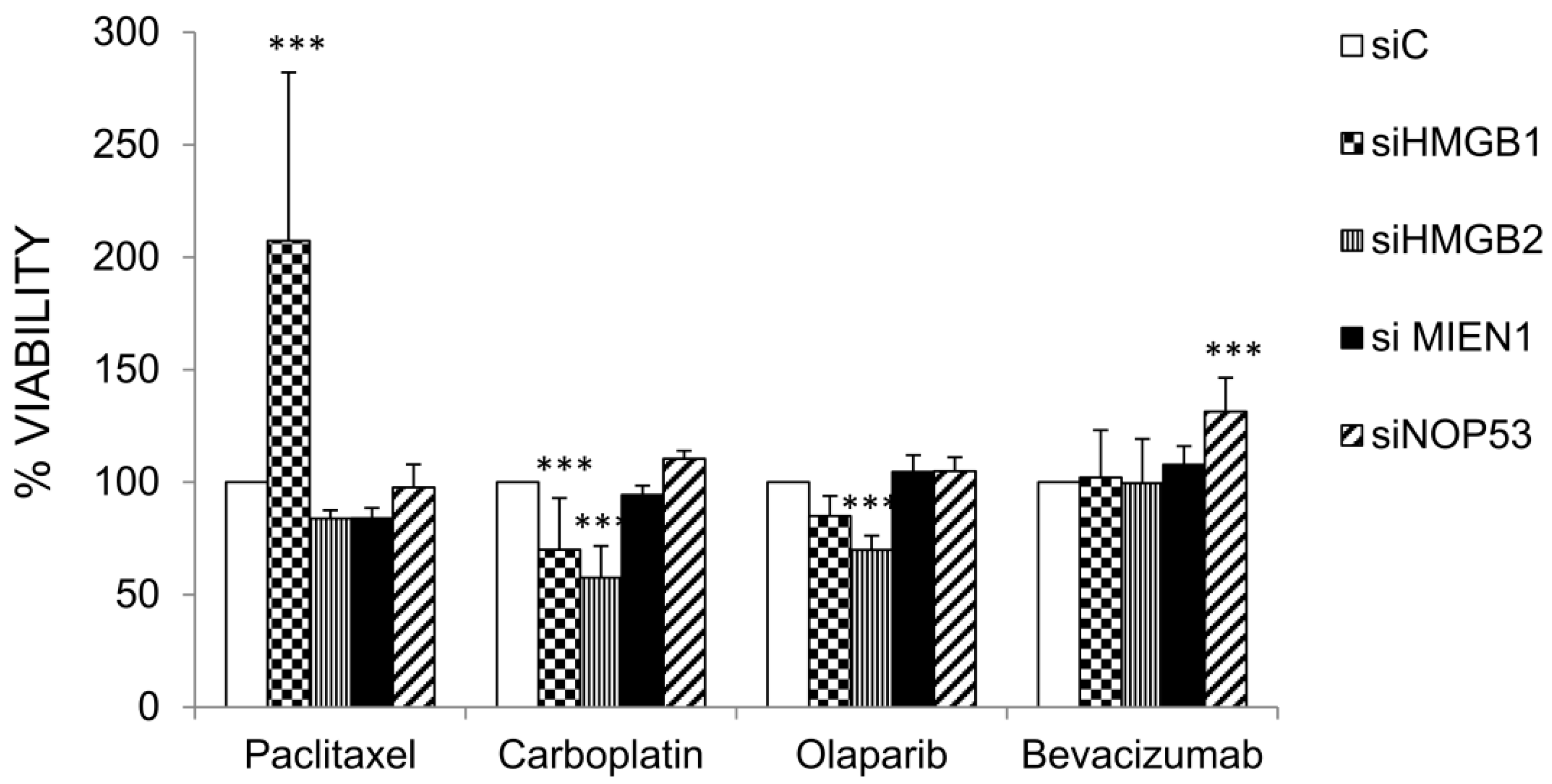
The genes HMGB1, HMGB2, MIEN1 and NOP53 were silenced by siRNA as described in Materials and Methods and the effect on cell viability after treatments with paclitaxel, carboplatin, olaparib and bevacizumab were compared in cells transfected with the corresponding specific siRNAs and siC (unrelated Control). Results are shown in Figure 6. Silencing of HMGB1 or HMGB2 diminished SKOV-3 cell viability after treatment with carboplatin. However, silencing of HMGB1 increased cell viability after treatment with paclitaxel. Cell viability after treatment with olaparib diminished with HMGB2 silencing. Finally, silencing of NOP53 increased cell viability after treatment with bevacizumab.
3. Discussion
Considering the relevance of HMGB proteins in Epithelial Ovary Cancer (EOC), we have determined for the first time the interactome of HMGB1 and HMGB2 related to this gynecological cancer. To this purpose, we have screened Y2H libraries, prepared from SKOV-3 cells and from tumor tissue diagnosed as primary transitional cell carcinoma (TCC) of the ovary, with HMGB1 and HMGB2 baits.
Supporting the functional significance of proteins detected in this EOC-HMGB-interactome study, we have found in the literature that all of them are experimentally associated to cancer hallmarks (Figure 2) and, in a high proportion, they had been directly related to ovary cancer (Figure 2). Furthermore, ROCK1 [51], PCBP1 [52], AKIP1 [53], TGM2 [54], BCCIP [50], and MIEN1 [55] proteins have been cited in relation to the epithelial to mesenchymal transition (EMT). Since EMT is an important step in carcinogenesis, which precedes metastasis, the relevance of these proteins for early EOC diagnosis is open to further analysis. Also reinforcing the significance of the interactions detected in our study in relation to clinics, data of gene expression according to Pan-Cancer Analysis of Whole Genomes (PCAWG), and corresponding to 110 tumors of ovarian adenocarcinomas, show that HMGB1, HMGB2 and >90% of their preys detected in this EOC-HMGB-interactome are up-regulated in the comparison between tumor tissue and adjacent non-tumor tissue (Table 3). Besides, according to the “Ovarian Serous Cystadenocarcinoma (TCGA)” study and using the tools from cBioportal, up-regulation of MIEN1, TGM2 or ZN428 in samples from these patients is correlated to poorer survival outcomes (Figure 4).
Although the proteins identified in the study, as well as HMGB1 and HMGB2, had been previously and independently related to EOC, the implication of a direct interaction with HMGB proteins, as part of their mechanism of action in cancer progression, had not been previously envisaged. Remarkably, this association is functionally reinforced by their confluence in a common signaling pathway; the function of HMGB1 in EOC has been previously associated to NF-kB signaling [4,5] and the functions of several of the EOC-HMGB-interactome components found in our analysis are also related to this signaling pathway. AKIP1 is a binding partner of NF-kappa B p65 subunit, which enhances the NF-kappa B-mediated gene expression [117]. MALAT1 and NF-κB signaling crosstalk during cancer and other diseases [118]. BCCIP binds to the protein LYRIC/AEG-1, which promotes tumor cell migration and invasion through activation of NF-kappaB [119]. COMMD1 inhibits NF-κB by promoting the ubiquitination and subsequent proteasomal degradation of RELA, component of NF-κB dimer, RELA/p50, bound to chromatin [120]. MIEN1 (C35) functionally enhances migration and invasion via NF-κB/Akt activity [121]. Rho-kinase isoform ROCK1 and its downstream target p38 MAPK regulate nuclear translocation of NF-κB RelA/p65 and subsequent DNA binding activity [122]. Over expression of C1qA up-regulates nuclear factor-κB reporters [123]. RSF1-overexpressing paclitaxel-resistant ovarian cancer cell lines were found to express elevated levels of genes regulated by NF-κB [46].
The interaction of HMGB2 with MIEN1 and NOP53 has been validated by co-immunoprecipitation in our study. The structure of MIEN1 and their emerging functions in relation to cancer have been recently reviewed [37], although the role of MIEN1 in pathophysiology of ovarian cancer had not been previously explored in depth. NOP53 regulates the activation of the tumor suppressor p53 when ribosome biogenesis is perturbed or DNA damage is produced [124,125,126]. Previous studies on the role of NOP53 in ovarian cancer were scarce, but down-regulation was observed in invasive serous ovarian tumors compared with benign and normal tissues [35] a feature also confirmed by our experiments in SKOV-3 cells versus noncancerous cells (Figure 3). NOP53 had been identified as a tumor suppressor downregulated in brain tumor cells [127,128]; however, in other cancers (esophagus or colon), NOP53 behaves as an oncogene that increments its expression in malignant cells [129]. Our data show that MIEN1 and NOP53 genes are regulated by HMGB1 and HMGB2 expression in EOC cell lines (Figure 5). HMGB2 negatively regulates and HMGB1 mostly positively regulates these genes, and in general the partners selected, which also correlates with the positive effect caused by HMGB2 silencing on HMGB1 expression. This could indicate that imbalance between HMGB1/HMGB2 is an important issue to consider in EOC, although further evidences are needed to clarify this mechanism.
Interestingly, we have shown in our study that HMGB1, HMGB2 and their EOC-HMGB-interactome partners MIEN1 and NOP53 are involved in the response to carboplatin, or drugs nowadays used in ovarian cancer treatment, such as bevacizumab, olaparib and paclitaxel. Expression levels of the pro-oncogenic genes HMGB1, HMGB2, and MIEN1 are down-regulated after treatment with paclitaxel, carboplatin or a combination of both (Table 4). Accordingly, HMGB1 or HMGB2 silencing decrease cell-viability of SKOV-3 cells exposed to carboplatin (Figure 6). Our data support the role of HMGB1 in resistance to carboplatin that was previously reported [15]. No data were previously available about the role of HMGB1 or HMGB2 in the resistance to paclitaxel, olaparib or bevacizumab in EOC treatment, although HMGB1 had been reported as a possible prognosis biomarker of bevacizumab treatment in non-small-cell lung cancer [130] and bevacizumab and HMGB1 have been related to malignant mesothelioma [131]. We have not detected changes in HMGB1 gene expression or effects of HMGB1 silencing in ovarian cancerous cells viability after bevacizumab treatment in the assayed conditions. However, we have found that HMGB1 is related to sensitivity to paclitaxel and HMGB2 is related to sensitivity to olaparib. NOP53, considered a tumor suppressor in brain tumor cells [127,128], is overexpressed after treatment with paclitaxel and bevacizumab, and NOP53 silencing decrease sensitivity to bevacizumab in SKOV-3 cells. According to our data, downregulation of HMGB1 causes loss of response to paclitaxel; down regulation of HMGB1 or HMGB2 increases sensitivity to carboplatin and, only in the case of HMGB2, slightly increases sensitivity to olaparib; finally, down regulation of NOP53 causes a moderate loss of response to bevacizumab. Levels of these genes might be analyzed in the clinical laboratory to guide treatment strategy in order to improve prognosis and avoid resistances.
4. Materials and Methods
4.1. Yeast Two Hybrid Methodology
HMGB1 and HMGB2 interacting partners were identified using Matchmaker Gold Yeast Two-Hybrid System (Clontech. Fremont, CA, USA). Sacchacomyces cerevisiae strains were Y187 (MATα, ura3-52, his3-200, ade2-101, trp1-901, leu2-3, 112, gal4Δ, gal80Δ, met-, URA3::GALuas-GAL1TATA-LacZ MEL1) and Y2HGold (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UAS-GAl1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-AUR1-C-MEL1). This is a very reliable Y2H system in which the three used promoters, controlling the four reporter genes HIS3, ADE2, AUR1-C and MEL1 in Y2HGold, are unrelated except for the short sequence in the upstream activation site (UAS) that are specifically bound by the Gal4 DNA binding domain. Thus, library proteins that interact with unrelated sequences, flanking or within the UAS, (i.e., false positives) are automatically screened out.
RNAs from human samples used to prepare the Y2H libraries were provided by Biobanco de Andalucía (Granada, Spain). Tumor and paired non-tumor tissue were obtained from a 63-year-old woman diagnosed with grade III ovarian transitional cell carcinoma without previous chemotherapy treatment. RNA was extracted from frozen tissue sections in OCT (Optimal Cutting Temperature) compound, using the Qiacube robot (Qiagen. Hilden, Germany) based on ion exchange columns with silica membrane. RNA was obtained with the miRNeasy mini kit (Qiagen, Hilden, Germany). The samples were finally treated with RNase-Free DNAase (Qiagen, Hilden, Germany). The quantity of RNA obtained was evaluated at 260 nm and 280 nm by spectrophotometry using the Infinite F200 equipment (Tecan Group Ltd. Männedorf. Switzerland) with a Nanoquant plate; finally, the integrity of the samples was evaluated by the 2200 Tape Station apparatus (Agilent Technologies, Inc. Santa Clara, CA, USA), being the RIN (RNA Integrity Number) parameter greater than 8. Total RNA from the ovarian cell line SKOV-3, was also used to prepare cDNA libraries. Library construction, bait construction and Yeast Two-Hybrid library screening were done as recommended by the vendor of the Matchmaker Gold Yeast Two-Hybrid System (Clontech, Mountain View, CA, USA) and details are already published [132]. After plasmid rescue, inserts were sequenced with primer T7 (5′-TAATACGACTCACTATAGGG-3′). Homology searches were done with BlastN and BlastX at NCBI (https://blast.ncbi.nlm.nih.gov/ accessed on 02-02-2020).
4.2. Cell Lines, Treatments and Cell Viability Assays
The SKOV-3 and PEO1 cell lines (originally derived from human EOC) and regularly tested for mycoplasma by Eurofins Scientific (Luxembourg, Luxembourg), were grown in McCoy’s-5A or RPMI-1640 medium, respectively, supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin-streptomycin (Thermo Fisher Scientific Inc. Waltham, MA, USA). The non-cancerous immortalized human ovarian cell line IOS3-80 (RRID:CVCL_5546) was obtained from Canadian Ovarian Tissue Bank (University of British Columbia, Vancouver, BC, Canada) and grown in RMPI-1640 medium supplemented as described above. Cells were cultured in a humidified incubator at 37 °C at 5% CO2.
SKOV-3 and IOSE-80 cells at 80% confluence were exposed during 48 h to different treatments, using drug concentrations and conditions selected according to previous studies. Paclitaxel was used at 25 μM [133]; carboplatin at 25 μg/mL [15]; olaparib to 2 μM [134]; and bevacizumab at 100 µg/mL [135] Paclitaxel was purchased to Sigma Aldrich Inc. (St. Louis, MO, USA) and olaparib, bevacizumab and carboplatin were provided by the Pharmacy Service of the Teresa Herrera hospital (INIBIC). In parallel, cells were grown with the same amount of vehicle-buffer used to prepare drug solutions, or with an unspecific IgG not directed to vascular endothelial growth factor as control of bevacizumab treatment. Cell viability-cytotoxicity assays were done using the Cell Counting Kit-8, CCK-8 (Tebu-Bio. Le-Perray-en-Yvelines, France).
4.3. Cross-Linking and HMGB2 Co-Immunoprecipitation
After reaching 70–80% confluence of SKOV-3 and PEO1 cultures, medium was removed and substituted by medium without fetal bovine serum and supplemented with 1% formaldehyde used as cross-linker. The cells were incubated with formaldehyde during 10 min at room temperature. To stop the cross-linking reaction, the medium with formaldehyde was substituted with a solution containing 0.125 M glycine in PBS (Phosphate Buffered saline) pH 7.4 (NZYTech, Lda. Lisbon, Portugal) and incubated during 5 min at room temperature. Cells were washed three times with PBS and harvested by scraping. After subsequent collection by centrifugation at 1200 rpm for 10 min at 4 °C, cells were re-suspended in lysis buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 0.1% NP-40, 1 mM EDTA, 2 Mm MgCl2) containing a cocktail of EDTA-free protease inhibitors (Roche Diagnostics, Laval, QC, Canada). Cell lysates were clarified for 15 min at 14000 rpm to pellet cell debris. Supernatants were collected and protein quantified using the Bradford reagent. 16 mg of total protein extracts from cells were immunoprecipitated (IPs) using 10 µg HMGB2 rabbit polyclonal ab67282 (Abcam, Cambridge, UK) bound to 50 μL dynabeads-protein A (Thermo Fisher Scientific Inc. Waltham, MA, USA) following manufacturer’s instructions. The presence of MIEN1 and NOP53 in the immunoprecipitations (IPs) was confirmed by western blot using the antibodies against MIEN1 (PA1-31180 from Thermo-Fisher Scientific Inc. Waltham, MA, USA) and NOP53, (sc517088 Santa Cruz, Dallas, TX, USA). After second incubation with 1:5000 G-protein HRP-linked (18-161.Millipore-Merck-KGaA, Darmstadt, Germany), 5% (w/v) non-fat milk diluted in PBST, PSB containing 0.1% Tween 20 (P1379 from Sigma Aldrich Inc.) was used as blocking solution. Western blots were developed using LuminataTMCrescendo Western HRP Substrate (Millipore Corporation. Burlington, MA, USA), and visualized in a ChemiDocTM imager (Bio-Rad Laboratories. Hercules, CA, USA).
4.4. Gene Expression Analysis by Quantitative Retrotranscription and Polymerase Chain Reaction (qRT-PCR)
RNA samples from cell cultures were obtained using GeneJET RNA Purification Kit (Thermo Fisher Scientific Inc., Waltham, MA, USA). The samples were treated with RNase-Free DNAase (Thermo Fisher Scientific Inc., Waltham, MA, USA) and purified using GeneJET RNA Cleanup and Concentration kit (Thermo Fisher Scientific Inc. Waltham, MA, USA). RNA from human ovarian surface epithelial cells (HOSEpiC) was provided by Innoprot (Derio, Vizcaya, Spain). RNA samples were retro-transcribed into cDNA and labeled with the KAPA SYBR FAST universal one-step qRT-PCR kit (Kappa Biosystems Inc., Woburn, MA, USA). The primers for qPCR were designed with the characteristics shown in Table S1 (Supplementary Material). Reaction conditions for thermal cycling were: 42 °C for 5 min, 95 °C for 5 s, 40 cycles of 95 °C for 3 s and finally 60 °C for 20 s. The ECO Real-Time PCR System was used for the experiments (Illumina Inc., San Diego, CA, USA), and relative expression was calculated by the 2−ΔΔCt method [136]. A t-test was used to check the statistically significance of differences between samples (p < 0.05 at least). The relative expression of selected genes were calculated by referring to the mRNA levels of the housekeeping gene, GADPH, which had been verified as being expressed constitutively under the assay conditions. For valid quantification using the 2−ΔΔCt method, the target and reference primer pairs were previously tested for PCR efficiencies and differed by <10%. At least, three independent biological replicas and two technical replicas were made for each.
4.5. siRNA Silencing
siRNAs directed against each mRNA and unspecific controls were purchased. siRNA-HMGB1 (s20254 Silencer Select) and siRNA-HMGB2 (s6650) from Life technologies (Thermo Fisher Scientific Inc., Waltham, MA, USA); siRNA-MIEN1 (S228354), siRNA-NOP53, (S26871) and siRNAControl2 (4390846) from Ambion Inc. (Thermo Fisher Scientific Inc., Waltham, MA, USA). Transfection of cells with siRNAs was done using Lipofectamine® 2000 (Life-Technologies-Invitrogen. Thermo Fisher Scientific Inc., Waltham, MA, USA) and following the protocol recommended by the vendor. Silencing was verified by qRT-PCR, with the methods described in the previous section, and Western blot using the antibodies against HMGB1 and HMGB2, NOP53 and MIEN1 already described and anti-GAPDH (60004-I-Ig from Proteintech. Manchester, UK) used for loading control. After second incubation with 1:5000 G-protein HRP-linked (18–161, Millipore-Merck-KGaA), western blot was developed as above described.
4.6. Survival Analysis
The Overall Survival Kaplan-Meier Estimate analysis was performed through cBioPortal (http://www.cbioportal.org/ accessed on 3-6-2020) using the databases Ovarian Serous Cystadenocarcinoma (TCGA, Provisional), composed of 606 samples. Results obtained for the genes giving Logrank Test p < 0.05 were selected for discussion.
5. Conclusions
In conclusion, results from our EOC-HMGB interactome study provides a set of proteins highly correlated with cancer hallmarks, EMT, ovarian cancer, NF-kB signaling and, the expression of some of them has been previously associated to patient’s survival. We have experimentally probed that HMGB1, HMGB2 and two of their partners, MIEN1 and NOP53 are also involved in the response of ovarian cancer cells to several drugs used in chemotherapy against EOC. Although clinical studies are needed before translation to early diagnosis and prognosis of patients, these proteins found in our EOC-HMGB-Interactome study are in the focus for the search of biomarkers and therapeutic targets in the fight against EOC disease.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/9/2435/s1, Table S1: Oligonucleotides used in this study, Figure S1: Co-immunoprecipitation of HMGB2 and MIEN1 or NOP53, Figure S2: Differential expression of components of the EOC-HMGB-Interactome in normal versus paclitaxel resistant cells, Figure S3: Quantification of HMGB1 and HMGB2 silencing, Figure S4: Quantification of MIEN1 and NOP53 silencing in SKOV-3 cells.
Author Contributions
All the authors contributed to conceptualization and design of experiments. A.B.-A. and M.C.-Q. constructed the libraries and run Y2H experiments. M.C.-Q. and M.L.-M. run the treatment of SKOV-3 cells and qPCR experiments. M.C.-Q. and M.L.-M. performed siRNA experiments and co-immunoprecipitations. M.E.C., Á.V.-V. and E.R.-B. performed data curation and M.Q.-V. provided advice for treatment selection. A.B.-A., M.C.-Q., M.L.-M. and M.E.C. wrote the original draft and figures. All authors have read and agreed to the published version of the manuscript.
Funding
This work has been funded by the Projects Nº PI14/01031 and PI18/01714, integrated in the National Plan for Scientific Research, Development and Technological Innovation 2013–2016 of the ISCIII- General Subdirection of Assesment and Promotion of the Research – European Regional Development Fund (FEDER) “A way of making Europe”. Funding is also acknowledged from Xunta de Galicia (Consolidación Grupos Referencia Competitiva Contract no. ED431C 2016–012). Aida Barreiro-Alonso was funded by a predoctoral fellowship from Xunta de Galicia-2013 (Spain) cofinanced by FEDER.
Acknowledgments
We thank the Biobanco of Andalucía (Spain) for the samples provided to prepare the libraries from ovarian cancer tissue and to the Biobanco of Complejo Hospitalario de Santiago de Compostela for samples from ovarian cancer tissue provided and used in preliminary studies to set up Y2H experimental conditions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Reid, B.M.; Permuth, J.B.; Sellers, T.A. Epidemiology of Ovarian Cancer: A Review. Cancer Biol. Med. 2017, 14, 9–32. [Google Scholar]
- Ueda, T.; Yoshida, M. HMGB Proteins and Transcriptional Regulation. Biochim. Biophys. Acta 2010, 1799, 114–118. [Google Scholar] [CrossRef]
- Barreiro-Alonso, A.; Lamas-Maceiras, M.; Rodriguez-Belmonte, E.; Vizoso-Vazquez, A.; Quindos, M.; Cerdan, M.E. High Mobility Group B Proteins, their Partners, and Other Redox Sensors in Ovarian and Prostate Cancer. Oxidative Med. Cell. Longev. 2016, 2016, 5845061. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Block, M.S.; Vierkant, R.A.; Fogarty, Z.C.; Winham, S.J.; Visscher, D.W.; Kalli, K.R.; Wang, C.; Goode, E.L. The Inflammatory Microenvironment in Epithelial Ovarian Cancer: A Role for TLR4 and MyD88 and Related Proteins. Tumor Biol. 2016, 37, 13279–13286. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Qu, X.; Ke, H.; Gong, W.; Chen, R.; Yang, W.; Cheng, Z. Association between the HMGB1/TLR4 Signaling Pathway and the Clinicopathological Features of Ovarian Cancer. Mol. Med. Rep. 2018, 18, 3093–3098. [Google Scholar] [CrossRef]
- Zhou, L.Y.; Shi, L.Y.; Xiao, Y. Changes of HMGB1 Expression on Angiogenesis of Ovarian Cancer and its Mechanism. J. Biol. Regul. Homeost. Agents 2016, 30, 233–238. [Google Scholar]
- Chen, J.; Xi, B.; Zhao, Y.; Yu, Y.; Zhang, J.; Wang, C. High-Mobility Group Protein B1 (HMGB1) is a Novel Biomarker for Human Ovarian Cancer. Gynecol. Oncol. 2012, 126, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tian, J.; Fu, X.; Chen, Y.; Zhang, W.; Yao, H.; Hao, Q. Serum High Mobility Group Box Protein 1 as Aclinical Marker for Ovarian Cancer. Neoplasma 2014, 61, 579–584. [Google Scholar] [CrossRef][Green Version]
- Wang, H.; Li, Z.; Sun, Y.; Xu, Z.; Han, J.; Song, B.; Song, W.; Qin, C.; Yin, L. Relationship between High-Mobility Group Box 1 Overexpression in Ovarian Cancer Tissue and Serum: A Meta-Analysis. Onco Targets Ther. 2015, 8, 3523–3531. [Google Scholar]
- Paek, J.; Lee, M.; Nam, E.J.; Kim, S.W.; Kim, Y.T. Clinical Impact of High Mobility Group Box 1 Protein in Epithelial Ovarian Cancer. Arch. Gynecol. Obstet. 2016, 293, 645–650. [Google Scholar] [CrossRef]
- Machado, L.R.; Moseley, P.M.; Moss, R.; Deen, S.; Nolan, C.; Spendlove, I.; Ramage, J.M.; Chan, S.Y.; Durrant, L.G. High Mobility Group Protein B1 is a Predictor of Poor Survival in Ovarian Cancer. Oncotarget 2017, 8, 101215–101223. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ju, L.L.; Zhao, C.Y.; Ye, K.F.; Yang, H.; Zhang, J. Expression and Clinical Implication of Beclin1, HMGB1, p62, Survivin, BRCA1 and ERCC1 in Epithelial Ovarian Tumor Tissues. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 1993–2003. [Google Scholar] [PubMed]
- Bernardini, M.; Lee, C.H.; Beheshti, B.; Prasad, M.; Albert, M.; Marrano, P.; Begley, H.; Shaw, P.; Covens, A.; Murphy, J.; et al. High-Resolution Mapping of Genomic Imbalance and Identification of Gene Expression Profiles Associated with Differential Chemotherapy Response in Serous Epithelial Ovarian Cancer. Neoplasia 2005, 7, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wei, Y. Association of HMGB1, BRCA1 and P62 Expression in Ovarian Cancer and Chemotherapy Sensitivity. Oncol. Lett. 2018, 15, 9572–9576. [Google Scholar] [CrossRef]
- Shu, W. Downregulation of High Mobility Group Protein Box-1 Resensitizes Ovarian Cancer Cells to Carboplatin. Oncol. Lett. 2018, 16, 4586–4592. [Google Scholar] [CrossRef]
- Varma, R.R.; Hector, S.M.; Clark, K.; Greco, W.R.; Hawthorn, L.; Pendyala, L. Gene Expression Profiling of a Clonal Isolate of Oxaliplatin-Resistant Ovarian Carcinoma Cell Line A2780/C10. Oncol. Rep. 2005, 14, 925–932. [Google Scholar] [CrossRef]
- Poornima, P.; Kumar, J.D.; Zhao, Q.; Blunder, M.; Efferth, T. Network Pharmacology of Cancer: From Understanding of Complex Interactomes to the Design of Multi-Target Specific Therapeutics from Nature. Pharmacol. Res. 2016, 111, 290–302. [Google Scholar] [CrossRef]
- Eisenhauer, E.A. Real-World Evidence in the Treatment of Ovarian Cancer. Ann. Oncol. 2017, 28, viii61–viii65. [Google Scholar] [CrossRef]
- Henderson, J.T.; Webber, E.M.; Sawaya, G.F. Screening for Ovarian Cancer: Updated Evidence Report and Systematic Review for the US Preventive Services Task Force. JAMA 2018, 319, 595–606. [Google Scholar] [CrossRef]
- Ichigo, S.; Takagi, H.; Matsunami, K.; Murase, T.; Ikeda, T.; Imai, A. Transitional Cell Carcinoma of the Ovary (Review). Oncol. Lett. 2012, 3, 3–6. [Google Scholar] [CrossRef]
- Ali, R.H.; Seidman, J.D.; Luk, M.; Kalloger, S.; Gilks, C.B. Transitional Cell Carcinoma of the Ovary is Related to High-Grade Serous Carcinoma and is Distinct from Malignant Brenner Tumor. Int. J. Gynecol. Pathol. 2012, 31, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Guan, W.; Ren, W.; Zhang, L.; Zhang, J.; Xu, G. MALAT1 Affects Ovarian Cancer Cell Behavior and Patient Survival. Oncol. Rep. 2018, 39, 2644–2652. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, X.; Guo, Y. Long Non-Coding RNA MALAT1 is Upregulated and Involved in Cell Proliferation, Migration and Apoptosis in Ovarian Cancer. Exp. Ther. Med. 2017, 13, 3055–3060. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Feng, S.J.; Qiu, S.; Shao, N.; Zheng, J.H. LncRNA MALAT1 Promotes Proliferation and Metastasis in Epithelial Ovarian Cancer via the PI3K-AKT Pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3176–3184. [Google Scholar]
- Zou, A.; Liu, R.; Wu, X. Long Non-Coding RNA MALAT1 is Up-Regulated in Ovarian Cancer Tissue and Promotes SK-OV-3 Cell Proliferation and Invasion. Neoplasma 2016, 63, 865–872. [Google Scholar] [CrossRef]
- Chen, Q.; Su, Y.; He, X.; Zhao, W.; Wu, C.; Zhang, W.; Si, X.; Dong, B.; Zhao, L.; Gao, Y.; et al. Plasma Long Non-Coding RNA MALAT1 is Associated with Distant Metastasis in Patients with Epithelial Ovarian Cancer. Oncol. Lett. 2016, 12, 1361–1366. [Google Scholar] [CrossRef]
- Liu, S.; Jiang, X.; Li, W.; Cao, D.; Shen, K.; Yang, J. Inhibition of the Long Non-Coding RNA MALAT1 Suppresses Tumorigenicity and Induces Apoptosis in the Human Ovarian Cancer SKOV3 Cell Line. Oncol. Lett. 2016, 11, 3686–3692. [Google Scholar] [CrossRef]
- Shi, B.; Wang, Y.; Yin, F. MALAT1/miR-124/Capn4 Axis Regulates Proliferation, Invasion and EMT in Nasopharyngeal Carcinoma Cells. Cancer Biol. Ther. 2017, 18, 792–800. [Google Scholar] [CrossRef]
- Crappe, J.; Ndah, E.; Koch, A.; Steyaert, S.; Gawron, D.; De Keulenaer, S.; De Meester, E.; De Meyer, T.; Van Criekinge, W.; Van Damme, P.; et al. PROTEOFORMER: Deep Proteome Coverage through Ribosome Profiling and MS Integration. Nucleic Acids Res. 2015, 43, e29. [Google Scholar] [CrossRef]
- Lee, S.; Liu, B.; Lee, S.; Huang, S.X.; Shen, B.; Qian, S.B. Global Mapping of Translation Initiation Sites in Mammalian Cells at Single-Nucleotide Resolution. Proc. Natl. Acad. Sci. USA 2012, 109, E2424–E2432. [Google Scholar] [CrossRef]
- Barreiro-Alonso, A.; Lamas-Maceiras, M.; Garcia-Diaz, R.; Rodriguez-Belmonte, E.; Yu, L.; Pardo, M.; Choudhary, J.S.; Cerdan, M.E. Delineating the HMGB1 and HMGB2 Interactome in Prostate and Ovary Epithelial Cells and its Relationship with Cancer. Oncotarget 2018, 9, 19050–19064. [Google Scholar] [CrossRef] [PubMed]
- Haley, J.; Tomar, S.; Pulliam, N.; Xiong, S.; Perkins, S.M.; Karpf, A.R.; Mitra, S.; Nephew, K.P.; Mitra, A.K. Functional Characterization of a Panel of High-Grade Serous Ovarian Cancer Cell Lines as Representative Experimental Models of the Disease. Oncotarget 2016, 7, 32810–32820. [Google Scholar] [CrossRef] [PubMed]
- Beaufort, C.M.; Helmijr, J.C.A.; Piskorz, A.M.; Hoogstraat, M.; Ruigrok-Ritstier, K.; Bessenlink, N.; Murtaza, M.; van IJcken, W.F.; Heine, A.A.; Smid, M.; et al. Ovarian Cancer Cell Line Panel (OCCP): Clinical Importance of In Vitro Morphological Subtypes. PLoS ONE 2014, 9, e103988. [Google Scholar] [CrossRef] [PubMed]
- Fedoseienko, A.; Wieringa, H.W.; Wisman, G.B.; Duiker, E.; Reyners, A.K.; Hofker, M.H.; van der Zee, A.G.; van de Sluis, B.; van Vugt, M.A. Nuclear COMMD1 is Associated with Cisplatin Sensitivity in Ovarian Cancer. PLoS ONE 2016, 11, e0165385. [Google Scholar] [CrossRef]
- Merritt, M.A.; Parsons, P.G.; Newton, T.R.; Martyn, A.C.; Webb, P.M.; Green, A.C.; Papadimos, D.J.; Boyle, G.M. Expression Profiling Identifies Genes Involved in Neoplastic Transformation of Serous Ovarian Cancer. BMC Cancer 2009, 9, 378. [Google Scholar] [CrossRef]
- Leung, T.H.; Wong, S.C.; Chan, K.K.; Chan, D.W.; Cheung, A.N.; Ngan, H.Y. The Interaction between C35 and DeltaNp73 Promotes Chemo-Resistance in Ovarian Cancer Cells. Br. J. Cancer 2013, 109, 965–975. [Google Scholar] [CrossRef]
- Kushwaha, P.P.; Gupta, S.; Singh, A.K.; Kumar, S. Emerging Role of Migration and Invasion Enhancer 1 (MIEN1) in Cancer Progression and Metastasis. Front. Oncol. 2019, 9, 868. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Fu, X.; Lu, Z. Long Non-Coding RNA NEAT1 Promoted Ovarian Cancer Cells’ Metastasis via Regulating of miR-382-3p/ROCK1 Axial. Cancer Sci. 2018, 109, 2188–2198. [Google Scholar] [CrossRef]
- Park, G.B.; Kim, D. PI3K Catalytic Isoform Alteration Promotes the LIMK1-Related Metastasis through the PAK1 or ROCK1/2 Activation in Cigarette Smoke-Exposed Ovarian Cancer Cells. Anticancer Res. 2017, 37, 1805–1818. [Google Scholar]
- Ohta, T.; Takahashi, T.; Shibuya, T.; Amita, M.; Henmi, N.; Takahashi, K.; Kurachi, H. Inhibition of the Rho/ROCK Pathway Enhances the Efficacy of Cisplatin through the Blockage of Hypoxia-Inducible Factor-1alpha in Human Ovarian Cancer Cells. Cancer. Biol. Ther. 2012, 13, 25–33. [Google Scholar] [CrossRef]
- Wegdam, W.; Argmann, C.A.; Kramer, G.; Vissers, J.P.; Buist, M.R.; Kenter, G.G.; Aerts, J.M.; Meijer, D.; Moerland, P.D. Label-Free LC-MSe in Tissue and Serum Reveals Protein Networks Underlying Differences between Benign and Malignant Serous Ovarian Tumors. PLoS ONE 2014, 9, e108046. [Google Scholar] [CrossRef] [PubMed]
- Sodek, K.L.; Ringuette, M.J.; Brown, T.J. Compact Spheroid Formation by Ovarian Cancer Cells is Associated with Contractile Behavior and an Invasive Phenotype. Int. J. Cancer 2009, 124, 2060–2070. [Google Scholar] [CrossRef] [PubMed]
- Je, E.M.; Yoo, N.J.; Kim, Y.J.; Kim, M.S.; Lee, S.H. Mutational Analysis of Splicing Machinery Genes SF3B1, U2AF1 and SRSF2 in Myelodysplasia and Other Common Tumors. Int. J. Cancer 2013, 133, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Hagemann, A.; DeMichele, A. Immuno-Modulatory Gene Polymorphisms and Outcome in Breast and Ovarian Cancer. Immunol. Investig. 2009, 38, 324–340. [Google Scholar] [CrossRef] [PubMed]
- Pathak, H.B.; Zhou, Y.; Sethi, G.; Hirst, J.; Schilder, R.J.; Golemis, E.A.; Godwin, A.K. A Synthetic Lethality Screen using a Focused siRNA Library to Identify Sensitizers to Dasatinib Therapy for the Treatment of Epithelial Ovarian Cancer. PLoS ONE 2015, 10, e0144126. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.I.; Ahn, J.H.; Lee, K.T.; Shih, I.; Choi, J.H. RSF1 is a Positive Regulator of NF-kappaB-Induced Gene Expression Required for Ovarian Cancer Chemoresistance. Cancer Res. 2014, 74, 2258–2269. [Google Scholar] [CrossRef]
- Sheu, J.J.; Choi, J.H.; Guan, B.; Tsai, F.J.; Hua, C.H.; Lai, M.T.; Wang, T.L.; Shih, I. Rsf-1, a Chromatin Remodelling Protein, Interacts with Cyclin E1 and Promotes Tumour Development. J. Pathol. 2013, 229, 559–568. [Google Scholar] [CrossRef]
- Maeda, D.; Chen, X.; Guan, B.; Nakagawa, S.; Yano, T.; Taketani, Y.; Fukayama, M.; Wang, T.L.; Shih, I. Rsf-1 (HBXAP) Expression is Associated with Advanced Stage and Lymph Node Metastasis in Ovarian Clear Cell Carcinoma. Int. J. Gynecol. Pathol. 2011, 30, 30–35. [Google Scholar] [CrossRef]
- Choi, J.H.; Sheu, J.J.; Guan, B.; Jinawath, N.; Markowski, P.; Wang, T.L.; Shih, I. Functional Analysis of 11q13.5 Amplicon Identifies Rsf-1 (HBXAP) as a Gene Involved in Paclitaxel Resistance in Ovarian Cancer. Cancer Res. 2009, 69, 1407–1415. [Google Scholar] [CrossRef]
- Liu, X.; Cao, L.; Ni, J.; Liu, N.; Zhao, X.; Wang, Y.; Zhu, L.; Wang, L.; Wang, J.; Yue, Y.; et al. Differential BCCIP Gene Expression in Primary Human Ovarian Cancer, Renal Cell Carcinoma and Colorectal Cancer Tissues. Int. J. Oncol. 2013, 43, 1925–1934. [Google Scholar] [CrossRef]
- Luo, H.; Liang, C. MicroRNA-148b Inhibits Proliferation and the Epithelial-Mesenchymal Transition and Increases Radiosensitivity in Non-Small Cell Lung Carcinomas by Regulating ROCK1. Exp. Ther. Med. 2018, 15, 3609–3616. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; Dou, K.F. PCBP1 is an Important Mediator of TGF-Beta-Induced Epithelial to Mesenchymal Transition in Gall Bladder Cancer Cell Line GBC-SD. Mol. Biol. Rep. 2014, 41, 5519–5524. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhao, L.; Cheng, D.; Mu, Q.; Kuang, H.; Feng, K. AKIP1 Promoted Epithelial-Mesenchymal Transition of Non-Small-Cell Lung Cancer via Transactivating ZEB1. Am. J. Cancer. Res. 2017, 7, 2234–2244. [Google Scholar] [PubMed]
- He, W.; Sun, Z.; Liu, Z. Silencing of TGM2 Reverses Epithelial to Mesenchymal Transition and Modulates the Chemosensitivity of Breast Cancer to Docetaxel. Exp. Ther. Med. 2015, 10, 1413–1418. [Google Scholar] [CrossRef]
- Ren, H.; Qi, Y.; Yin, X.; Gao, J. MiR-136 Targets MIEN1 and Involves the Metastasis of Colon Cancer by Suppressing Epithelial-to-Mesenchymal Transition. Onco Targets Ther. 2017, 11, 67–74. [Google Scholar] [CrossRef]
- Mo, D.; Li, X.; Li, C.; Liang, J.; Zeng, T.; Su, N.; Jiang, Q.; Huang, J. Overexpression of AKIP1 Predicts Poor Prognosis of Patients with Breast Carcinoma and Promotes Cancer Metastasis through Akt/GSK-3beta/Snail Pathway. Am. J. Transl. Res. 2016, 8, 4951–4959. [Google Scholar]
- Zhang, W.; Wu, Q.; Wang, C.; Yang, L.; Liu, P.; Ma, C. AKIP1 Promotes Angiogenesis and Tumor Growth by Upregulating CXC-Chemokines in Cervical Cancer Cells. Mol. Cell. Biochem. 2018, 448, 311–320. [Google Scholar] [CrossRef]
- Zimmerman, R.; Peng, D.J.; Lanz, H.; Zhang, Y.H.; Danen-Van Oorschot, A.; Qu, S.; Backendorf, C.; Noteborn, M. PP2A Inactivation is a Crucial Step in Triggering Apoptin-Induced Tumor-Selective Cell Killing. Cell Death Dis. 2012, 3, e291. [Google Scholar] [CrossRef]
- Sastri, M.; Haushalter, K.J.; Panneerselvam, M.; Chang, P.; Fridolfsson, H.; Finley, J.C.; Ng, D.; Schilling, J.M.; Miyanohara, A.; Day, M.E.; et al. A Kinase Interacting Protein (AKIP1) is a Key Regulator of Cardiac Stress. Proc. Natl. Acad. Sci. USA 2013, 110, E387–E396. [Google Scholar] [CrossRef]
- Yu, H.; Tigchelaar, W.; Koonen, D.P.; Patel, H.H.; de Boer, R.A.; van Gilst, W.H.; Westenbrink, B.D.; Sillje, H.H. AKIP1 Expression Modulates Mitochondrial Function in Rat Neonatal Cardiomyocytes. PLoS ONE 2013, 8, e80815. [Google Scholar] [CrossRef]
- Huang, Y.Y.; Dai, L.; Gaines, D.; Droz-Rosario, R.; Lu, H.; Liu, J.; Shen, Z. BCCIP Suppresses Tumor Initiation but is Required for Tumor Progression. Cancer Res. 2013, 73, 7122–7133. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Hu, B.; Ni, W.; Mao, X.; Zhou, H.; Lv, J.; Yin, B.; Shen, Z.; Wu, M.; Ding, W.; et al. Expression Pattern of BCCIP in Hepatocellular Carcinoma is Correlated with Poor Prognosis and Enhanced Cell Proliferation. Tumour Biol. 2016, 37, 16305–16315. [Google Scholar] [CrossRef] [PubMed]
- Van de Sluis, B.; Mao, X.; Zhai, Y.; Groot, A.J.; Vermeulen, J.F.; van der Wall, E.; van Diest, P.J.; Hofker, M.H.; Wijmenga, C.; Klomp, L.W.; et al. COMMD1 Disrupts HIF-1alpha/beta Dimerization and Inhibits Human Tumor Cell Invasion. J. Clin. Investig. 2010, 120, 2119–2130. [Google Scholar] [CrossRef] [PubMed]
- Mu, P.; Akashi, T.; Lu, F.; Kishida, S.; Kadomatsu, K. A Novel Nuclear Complex of DRR1, F-Actin and COMMD1 Involved in NF-kappaB Degradation and Cell Growth Suppression in Neuroblastoma. Oncogene 2017, 36, 5745–5756. [Google Scholar] [CrossRef] [PubMed]
- Yeh, D.W.; Chen, Y.S.; Lai, C.Y.; Liu, Y.L.; Lu, C.H.; Lo, J.F.; Chen, L.; Hsu, L.C.; Luo, Y.; Xiang, R.; et al. Downregulation of COMMD1 by miR-205 Promotes a Positive Feedback Loop for Amplifying Inflammatory- and Stemness-Associated Properties of Cancer Cells. Cell Death Differ. 2016, 23, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Bulla, R.; Tripodo, C.; Rami, D.; Ling, G.S.; Agostinis, C.; Guarnotta, C.; Zorzet, S.; Durigutto, P.; Botto, M.; Tedesco, F. C1q Acts in the Tumour Microenvironment as a Cancer-Promoting Factor Independently of Complement Activation. Nat. Commun. 2016, 7, 10346. [Google Scholar] [CrossRef]
- Brennan, P.A.; Jing, J.; Ethunandan, M.; Gorecki, D. Dystroglycan Complex in Cancer. Eur. J. Surg. Oncol. 2004, 30, 589–592. [Google Scholar] [CrossRef]
- Pohl, M.; Olsen, K.E.; Holst, R.; Donnem, T.; Busund, L.T.; Bremnes, R.M.; Al-Saad, S.; Andersen, S.; Richardsen, E.; Ditzel, H.J.; et al. Keratin 34betaE12/keratin7 Expression is a Prognostic Factor of Cancer-Specific and overall Survival in Patients with Early Stage Non-Small Cell Lung Cancer. Acta Oncol. 2016, 55, 167–177. [Google Scholar] [CrossRef]
- Tee, A.E.; Liu, B.; Song, R.; Li, J.; Pasquier, E.; Cheung, B.B.; Jiang, C.; Marshall, G.M.; Haber, M.; Norris, M.D.; et al. The Long Noncoding RNA MALAT1 Promotes Tumor-Driven Angiogenesis by Up-Regulating Pro-Angiogenic Gene Expression. Oncotarget 2016, 7, 8663–8675. [Google Scholar] [CrossRef]
- Ji, D.G.; Guan, L.Y.; Luo, X.; Ma, F.; Yang, B.; Liu, H.Y. Inhibition of MALAT1 Sensitizes Liver Cancer Cells to 5-Flurouracil by Regulating Apoptosis through IKKalpha/NF-kappaB Pathway. Biochem. Biophys. Res. Commun. 2018, 501, 33–40. [Google Scholar] [CrossRef]
- Kwon, M.J.; Kim, R.N.; Song, K.; Jeon, S.; Jeong, H.M.; Kim, J.S.; Han, J.; Hong, S.; Oh, E.; Choi, J.S.; et al. Genes Co-Amplified with ERBB2 Or MET as Novel Potential Cancer-Promoting Genes in Gastric Cancer. Oncotarget 2017, 8, 92209–92226. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.Q.; Yin, K.; Zhu, S.; Zhang, L.; Wen, P.E.; Li, C.L.; Zhang, D.B.; Liu, M.; Yan, G. Inhibition of C35 Gene Expression by Small Interfering RNA Induces Apoptosis of Breast Cancer Cells. Biosci. Trends 2010, 4, 254–259. [Google Scholar] [PubMed]
- Chen, H.; Duo, Y.; Hu, B.; Wang, Z.; Zhang, F.; Tsai, H.; Zhang, J.; Zhou, L.; Wang, L.; Wang, X.; et al. PICT-1 Triggers a Pro-Death Autophagy through Inhibiting rRNA Transcription and AKT/mTOR/p70S6K Signaling Pathway. Oncotarget 2016, 7, 78747–78763. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Cho, Y.E.; Park, J.H. The Nucleolar Protein GLTSCR2 is an Upstream Negative Regulator of the Oncogenic Nucleophosmin-MYC Axis. Am. J. Pathol. 2015, 185, 2061–2068. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Park, J.H.; Lee, S. GLTSCR2 Contributes to the Death Resistance and Invasiveness of Hypoxia-Selected Cancer Cells. FEBS Lett. 2012, 586, 3435–3440. [Google Scholar] [CrossRef]
- Li, J.; Feng, Q.; Wei, X.; Yu, Y. MicroRNA-490 Regulates Lung Cancer Metastasis by Targeting Poly r(C)-Binding Protein 1. Tumour Biol. 2016, 37, 15221–15228. [Google Scholar] [CrossRef]
- Schiarea, S.; Solinas, G.; Allavena, P.; Scigliuolo, G.M.; Bagnati, R.; Fanelli, R.; Chiabrando, C. Secretome Analysis of Multiple Pancreatic Cancer Cell Lines Reveals Perturbations of Key Functional Networks. J. Proteome Res. 2010, 9, 4376–4392. [Google Scholar] [CrossRef]
- Zhang, W.; Shi, H.; Zhang, M.; Liu, B.; Mao, S.; Li, L.; Tong, F.; Liu, G.; Yang, S.; Wang, H. Poly C Binding Protein 1 Represses Autophagy through Downregulation of LC3B to Promote Tumor Cell Apoptosis in Starvation. Int. J. Biochem. Cell Biol. 2016, 73, 127–136. [Google Scholar] [CrossRef]
- Jones, D.T.; Lechertier, T.; Reynolds, L.E.; Mitter, R.; Robinson, S.D.; Kirn-Safran, C.B.; Hodivala-Dilke, K.M. Endogenous Ribosomal Protein L29 (RPL29): A Newly Identified Regulator of Angiogenesis in Mice. Dis. Model. Mech. 2013, 6, 115–124. [Google Scholar] [CrossRef]
- Li, C.; Ge, M.; Yin, Y.; Luo, M.; Chen, D. Silencing Expression of Ribosomal Protein L26 and L29 by RNA Interfering Inhibits Proliferation of Human Pancreatic Cancer PANC-1 Cells. Mol. Cell. Biochem. 2012, 370, 127–139. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Zhang, Z. Adipokine RBP4 Drives Ovarian Cancer Cell Migration. J. Ovarian Res. 2018, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Montalvo, J.; Spencer, C.; Hackathorn, A.; Masterjohn, K.; Perkins, A.; Doty, C.; Arumugam, A.; Ongusaha, P.P.; Lakshmanaswamy, R.; Liao, J.K.; et al. ROCK1 & 2 Perform Overlapping and Unique Roles in Angiogenesis and Angiosarcoma Tumor Progression. Curr. Mol. Med. 2013, 13, 205–219. [Google Scholar]
- Bryan, B.A.; Dennstedt, E.; Mitchell, D.C.; Walshe, T.E.; Noma, K.; Loureiro, R.; Saint-Geniez, M.; Campaigniac, J.P.; Liao, J.K.; D’Amore, P.A. RhoA/ROCK Signaling is Essential for Multiple Aspects of VEGF-Mediated Angiogenesis. FASEB J. 2010, 24, 3186–3195. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, G.; Zhang, Q.; Tang, Q.; Huang, J.; Hu, C.; Liu, Y.; Wang, Q.; Liu, W.; Gao, N.; et al. Hirsutine Induces mPTP-Dependent Apoptosis through ROCK1/PTEN/PI3K/GSK3beta Pathway in Human Lung Cancer Cells. Cell Death Dis. 2018, 9, 598. [Google Scholar] [CrossRef] [PubMed]
- Guerra, B.; Fischer, M.; Schaefer, S.; Issinger, O.G. The Kinase Inhibitor D11 Induces Caspase-Mediated Cell Death in Cancer Cells Resistant to Chemotherapeutic Treatment. J. Exp. Clin. Cancer Res. 2015, 34, 125. [Google Scholar] [CrossRef]
- Li, G.; Liu, L.; Shan, C.; Cheng, Q.; Budhraja, A.; Zhou, T.; Cui, H.; Gao, N. RhoA/ROCK/PTEN Signaling is Involved in AT-101-Mediated Apoptosis in Human Leukemia Cells in Vitro and in Vivo. Cell Death Dis. 2014, 5, e998. [Google Scholar] [CrossRef]
- Liu, Y.; Li, G.; Liu, C.; Tang, Y.; Zhang, S. RSF1 Regulates the Proliferation and Paclitaxel Resistance via Modulating NF-kappaB Signaling Pathway in Nasopharyngeal Carcinoma. J. Cancer 2017, 8, 354–362. [Google Scholar] [CrossRef][Green Version]
- Li, Q.; Dong, Q.; Wang, E. Rsf-1 is Overexpressed in Non-Small Cell Lung Cancers and Regulates cyclinD1 Expression and ERK Activity. Biochem. Biophys. Res. Commun. 2012, 420, 6–10. [Google Scholar] [CrossRef]
- Lei, Z.; Chai, N.; Tian, M.; Zhang, Y.; Wang, G.; Liu, J.; Tian, Z.; Yi, X.; Chen, D.; Li, X.; et al. Novel Peptide GX1 Inhibits Angiogenesis by Specifically Binding to Transglutaminase-2 in the Tumorous Endothelial Cells of Gastric Cancer. Cell Death Dis. 2018, 9, 579. [Google Scholar] [CrossRef]
- Hidaka, H.; Seki, N.; Yoshino, H.; Yamasaki, T.; Yamada, Y.; Nohata, N.; Fuse, M.; Nakagawa, M.; Enokida, H. Tumor Suppressive microRNA-1285 Regulates Novel Molecular Targets: Aberrant Expression and Functional Significance in Renal Cell Carcinoma. Oncotarget 2012, 3, 44–57. [Google Scholar] [CrossRef]
- Lu, H.; Hallstrom, T.C. The Nuclear Protein UHRF2 is a Direct Target of the Transcription Factor E2F1 in the Induction of Apoptosis. J. Biol. Chem. 2013, 288, 23833–23843. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Wang, J.; Shan, X.; Li, Y. Selecting Key Genes Associated with Ovarian Cancer Based on Differential Expression Network. J. BUON 2017, 22, 48–57. [Google Scholar] [PubMed]
- Fei, D.L.; Motowski, H.; Chatrikhi, R.; Prasad, S.; Yu, J.; Gao, S.; Kielkopf, C.L.; Bradley, R.K.; Varmus, H. Wild-Type U2AF1 Antagonizes the Splicing Program Characteristic of U2AF1-Mutant Tumors and is Required for Cell Survival. PLoS Genet. 2016, 12, e1006384. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Peng, G.; Dai, H.; Breuer, E.K.; Stemke-Hale, K.; Li, K.; Gonzalez-Angulo, A.M.; Mills, G.B.; Lin, S.Y. ZNF668 Functions as a Tumor Suppressor by Regulating p53 Stability and Function in Breast Cancer. Cancer Res. 2011, 71, 6524–6534. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cheng, Y.; Ren, X.; Zhang, L.; Yap, K.L.; Wu, H.; Patel, R.; Liu, D.; Qin, Z.H.; Shih, I.M.; et al. NAC1 Modulates Sensitivity of Ovarian Cancer Cells to Cisplatin by Altering the HMGB1-Mediated Autophagic Response. Oncogene 2012, 31, 1055–1064. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, C.F.; Chen, R. Zinc Finger RNA-Binding Protein Promotes Non-Small-Cell Carcinoma Growth and Tumor Metastasis by Targeting the Notch Signaling Pathway. Am. J. Cancer Res. 2017, 7, 1804–1819. [Google Scholar]
- Zhao, X.; Chen, M.; Tan, J. Knockdown of ZFR Suppresses Cell Proliferation and Invasion of Human Pancreatic Cancer. Biol. Res. 2016, 49, 26. [Google Scholar] [CrossRef]
- Zwang, Y.; Jonas, O.; Chen, C.; Rinne, M.L.; Doench, J.G.; Piccioni, F.; Tan, L.; Huang, H.T.; Wang, J.; Ham, Y.J.; et al. Synergistic Interactions with PI3K Inhibition that Induce Apoptosis. Elife 2017, 6, e24523. [Google Scholar] [CrossRef]
- Papatheodorou, I.; Fonseca, N.A.; Keays, M.; Tang, Y.A.; Barrera, E.; Bazant, W.; Burke, M.; Fullgrabe, A.; Fuentes, A.M.; George, N.; et al. Expression Atlas: Gene and Protein Expression across Multiple Studies and Organisms. Nucleic Acids Res. 2018, 46, D246–D251. [Google Scholar] [CrossRef]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Wang, J.; Peng, X.; Peng, W.; Wu, F.X. Dynamic Protein Interaction Network Construction and Applications. Proteomics 2014, 14, 338–352. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Shih, I.; Sheu, J.J.; Santillan, A.; Nakayama, K.; Yen, M.J.; Bristow, R.E.; Vang, R.; Parmigiani, G.; Kurman, R.J.; Trope, C.G.; et al. Amplification of a Chromatin Remodeling Gene, Rsf-1/HBXAP, in Ovarian Carcinoma. Proc. Natl. Acad. Sci. USA 2005, 102, 14004–14009. [Google Scholar] [CrossRef] [PubMed]
- Van Beijnum, J.R.; Buurman, W.A.; Griffioen, A.W. Convergence and Amplification of Toll-Like Receptor (TLR) and Receptor for Advanced Glycation End Products (RAGE) Signaling Pathways via High Mobility Group B1 (HMGB1). Angiogenesis 2008, 11, 91–99. [Google Scholar] [CrossRef]
- Rohnalter, V.; Roth, K.; Finkernagel, F.; Adhikary, T.; Obert, J.; Dorzweiler, K.; Bensberg, M.; Muller-Brusselbach, S.; Muller, R. A Multi-Stage Process Including Transient Polyploidization and EMT Precedes the Emergence of Chemoresistent Ovarian Carcinoma Cells with a Dedifferentiated and Pro-Inflammatory Secretory Phenotype. Oncotarget 2015, 6, 40005–40025. [Google Scholar] [CrossRef]
- Xia, X.; Ji, T.; Liu, R.; Weng, Y.; Fang, Y.; Wang, Z.; Xu, H. Cytoplasmic p21 is Responsible for Paclitaxel Resistance in Ovarian Cancer A2780 Cells. Eur. J. Gynaecol. Oncol. 2015, 36, 662–666. [Google Scholar]
- Shi, H.; Li, H.; Yuan, R.; Guan, W.; Zhang, X.; Zhang, S.; Zhang, W.; Tong, F.; Li, L.; Song, Z.; et al. PCBP1 Depletion Promotes Tumorigenesis through Attenuation of p27 (Kip1) mRNA Stability and Translation. J. Exp. Clin. Cancer Res. 2018, 37, 187. [Google Scholar] [CrossRef]
- He, Y.; Ding, Y.; Wang, D.; Zhang, W.; Chen, W.; Liu, X.; Qin, W.; Qian, X.; Chen, H.; Guo, Z. HMGB1 Bound to Cisplatin-DNA Adducts Undergoes Extensive Acetylation and Phosphorylation in Vivo. Chem. Sci. 2015, 6, 2074–2078. [Google Scholar] [CrossRef]
- Ingemarsdotter, C.K.; Tookman, L.A.; Browne, A.; Pirlo, K.; Cutts, R.; Chelela, C.; Khurrum, K.F.; Leung, E.Y.; Dowson, S.; Webber, L.; et al. Paclitaxel Resistance Increases Oncolytic Adenovirus Efficacy via Upregulated CAR Expression and Dysfunctional Cell Cycle Control. Mol. Oncol. 2015, 9, 791–805. [Google Scholar] [CrossRef]
- Knox, R.J.; Friedlos, F.; Lydall, D.A.; Roberts, J.J. Mechanism of Cytotoxicity of Anticancer Platinum Drugs: Evidence that Cis-Diamminedichloroplatinum (II) and Cis-Diammine-(1, 1-Cyclobutanedicarboxylato) Platinum (II) Differ Only in the Kinetics of their Interaction with DNA. Cancer Res. 1986, 46, 1972–1979. [Google Scholar] [PubMed]
- Brabec, V.; Kasparkova, J. Modifications of DNA by Platinum Complexes. Relation to Resistance of Tumors to Platinum Antitumor Drugs. Drug Resist Updat. 2005, 8, 131–146. [Google Scholar] [CrossRef] [PubMed]
- Goulooze, S.C.; Cohen, A.F.; Rissmann, R. Olaparib. Br. J. Clin. Pharmacol. 2016, 81, 171–173. [Google Scholar] [CrossRef] [PubMed]
- Monk, B.J.; Randall, L.M.; Grisham, R.N. The Evolving Landscape of Chemotherapy in Newly Diagnosed Advanced Epithelial Ovarian Cancer. Am. Soc. Clin. Oncol. Educ. Book 2019, 39, e141–e151. [Google Scholar] [CrossRef]
- Ferrara, N.; Hillan, K.J.; Novotny, W. Bevacizumab (Avastin), a Humanized Anti-VEGF Monoclonal Antibody for Cancer Therapy. Biochem. Biophys. Res. Commun. 2005, 333, 328–335. [Google Scholar] [CrossRef]
- Lopes, N.M.; Adams, E.G.; Pitts, T.W.; Bhuyan, B.K. Cell Kill Kinetics and Cell Cycle Effects of Taxol on Human and Hamster Ovarian Cell Lines. Cancer Chemother. Pharmacol. 1993, 32, 235–242. [Google Scholar] [CrossRef]
- Gao, N.; Asamitsu, K.; Hibi, Y.; Ueno, T.; Okamoto, T. AKIP1 Enhances NF-kappaB-Dependent Gene Expression by Promoting the Nuclear Retention and Phosphorylation of p65. J. Biol. Chem. 2008, 283, 7834–7843. [Google Scholar] [CrossRef]
- Gupta, S.C.; Awasthee, N.; Rai, V.; Chava, S.; Gunda, V.; Challagundla, K.B. Long Non-Coding RNAs and Nuclear Factor-kappaB Crosstalk in Cancer and Other Human Diseases. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188316. [Google Scholar] [CrossRef]
- Ash, S.C.; Yang, D.Q.; Britt, D.E. LYRIC/AEG-1 Overexpression Modulates BCCIPalpha Protein Levels in Prostate Tumor Cells. Biochem. Biophys. Res. Commun. 2008, 371, 333–338. [Google Scholar] [CrossRef]
- Maine, G.N.; Mao, X.; Komarck, C.M.; Burstein, E. COMMD1 Promotes the Ubiquitination of NF-kappaB Subunits through a Cullin-Containing Ubiquitin Ligase. EMBO J. 2007, 26, 436–447. [Google Scholar] [CrossRef]
- Chang, K.S.; Tsui, K.H.; Lin, Y.H.; Hou, C.P.; Feng, T.H.; Juang, H.H. Migration and Invasion Enhancer 1 is an NF-kB-Inducing Gene Enhancing the Cell Proliferation and Invasion Ability of Human Prostate Carcinoma Cells in Vitro and in Vivo. Cancers 2019, 11, 1486. [Google Scholar] [CrossRef] [PubMed]
- Matoba, K.; Kawanami, D.; Tsukamoto, M.; Kinoshita, J.; Ito, T.; Ishizawa, S.; Kanazawa, Y.; Yokota, T.; Murai, N.; Matsufuji, S.; et al. Rho-Kinase Regulation of TNF-Alpha-Induced Nuclear Translocation of NF-kappaB RelA/p65 and M-CSF Expression via p38 MAPK in Mesangial Cells. Am. J. Physiol. Renal Physiol. 2014, 307, F571–F580. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tong, X.; Zhang, J.; Ye, X. The Complement C1qA Enhances Retinoic Acid-Inducible Gene-I-Mediated Immune Signalling. Immunology 2012, 136, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Seok, K.O.; Kim, Y.J.; Bae, W.K.; Lee, S.; Park, J.H. Involvement of GLTSCR2 in the DNA Damage Response. Am. J. Pathol. 2011, 179, 1257–1264. [Google Scholar] [CrossRef]
- Sloan, K.E.; Bohnsack, M.T.; Watkins, N.J. The 5S RNP Couples p53 Homeostasis to Ribosome Biogenesis and Nucleolar Stress. Cell. Rep. 2013, 5, 237–247. [Google Scholar] [CrossRef]
- Chen, H.; Han, L.; Tsai, H.; Wang, Z.; Wu, Y.; Duo, Y.; Cao, W.; Chen, L.; Tan, Z.; Xu, N.; et al. PICT-1 is a Key Nucleolar Sensor in DNA Damage Response Signaling that Regulates Apoptosis through the RPL11-MDM2-p53 Pathway. Oncotarget 2016, 7, 83241–83257. [Google Scholar] [CrossRef][Green Version]
- Kim, Y.J.; Cho, Y.E.; Kim, Y.W.; Kim, J.Y.; Lee, S.; Park, J.H. Suppression of Putative Tumour Suppressor Gene GLTSCR2 Expression in Human Glioblastomas. J. Pathol. 2008, 216, 218–224. [Google Scholar] [CrossRef]
- Okahara, F.; Itoh, K.; Nakagawara, A.; Murakami, M.; Kanaho, Y.; Maehama, T. Critical Role of PICT-1, a Tumor Suppressor Candidate, in Phosphatidylinositol 3, 4, 5-Trisphosphate Signals and Tumorigenic Transformation. Mol. Biol. Cell 2006, 17, 4888–4895. [Google Scholar] [CrossRef][Green Version]
- Sasaki, M.; Kawahara, K.; Nishio, M.; Mimori, K.; Kogo, R.; Hamada, K.; Itoh, B.; Wang, J.; Komatsu, Y.; Yang, Y.R.; et al. Regulation of the MDM2-P53 Pathway and Tumor Growth by PICT1 via Nucleolar RPL11. Nat. Med. 2011, 17, 944–951. [Google Scholar] [CrossRef]
- Niki, M.; Yokoi, T.; Kurata, T.; Nomura, S. New Prognostic Biomarkers and Therapeutic Effect of Bevacizumab for Patients with Non-Small-Cell Lung Cancer. Lung Cancer 2017, 8, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Bononi, A.; Napolitano, A.; Pass, H.I.; Yang, H.; Carbone, M. Latest developments in our understanding of the pathogenesis of mesothelioma and the design of targeted therapies. Expert Rev Respir Med. 2015, 9, 633–654. [Google Scholar] [CrossRef] [PubMed]
- Barreiro-Alonso, A.; Camara-Quilez, M.; Salamini-Montemurri, M.; Lamas-Maceiras, M.; Vizoso-Vazquez, A.; Rodriguez-Belmonte, E.; Quindos-Varela, M.; Martinez-Iglesias, O.; Figueroa, A.; Cerdan, M.E. Characterization of HMGB1/2 Interactome in Prostate Cancer by Yeast Two Hybrid Approach: Potential Pathobiological Implications. Cancers 2019, 11, 1729. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wu, Y.; Yoshizawa, T.; Yan, X.; Morohashi, S.; Seino, H.; Kato, Y.; Kijima, H. Basic Helix-Loop-Helix Transcription Factor DEC2 Functions as an Anti-Apoptotic Factor during Paclitaxel-Induced Apoptosis in Human Prostate Cancer Cells. Int. J. Mol. Med. 2016, 38, 1727–1733. [Google Scholar] [CrossRef] [PubMed]
- Kukolj, E.; Kaufmann, T.; Dick, A.E.; Zeillinger, R.; Gerlich, D.W.; Slade, D. PARP Inhibition Causes Premature Loss of Cohesion in Cancer Cells. Oncotarget 2017, 8, 103931–103951. [Google Scholar] [CrossRef]
- Zhao, Z.; Xia, G.; Li, N.; Su, R.; Chen, X.; Zhong, L. Autophagy Inhibition Promotes Bevacizumab-Induced Apoptosis and Proliferation Inhibition in Colorectal Cancer Cells. J. Cancer 2018, 9, 3407–3416. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data using Real-Time Quantitative PCR and the 2 (-Delta Delta C (T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).