The Regulatory Role of Mitochondrial MicroRNAs (MitomiRs) in Breast Cancer: Translational Implications Present and Future
Abstract
Simple Summary
Abstract
1. Current Perspectives of Breast Cancer
2. MicroRNAs in Breast Cancer
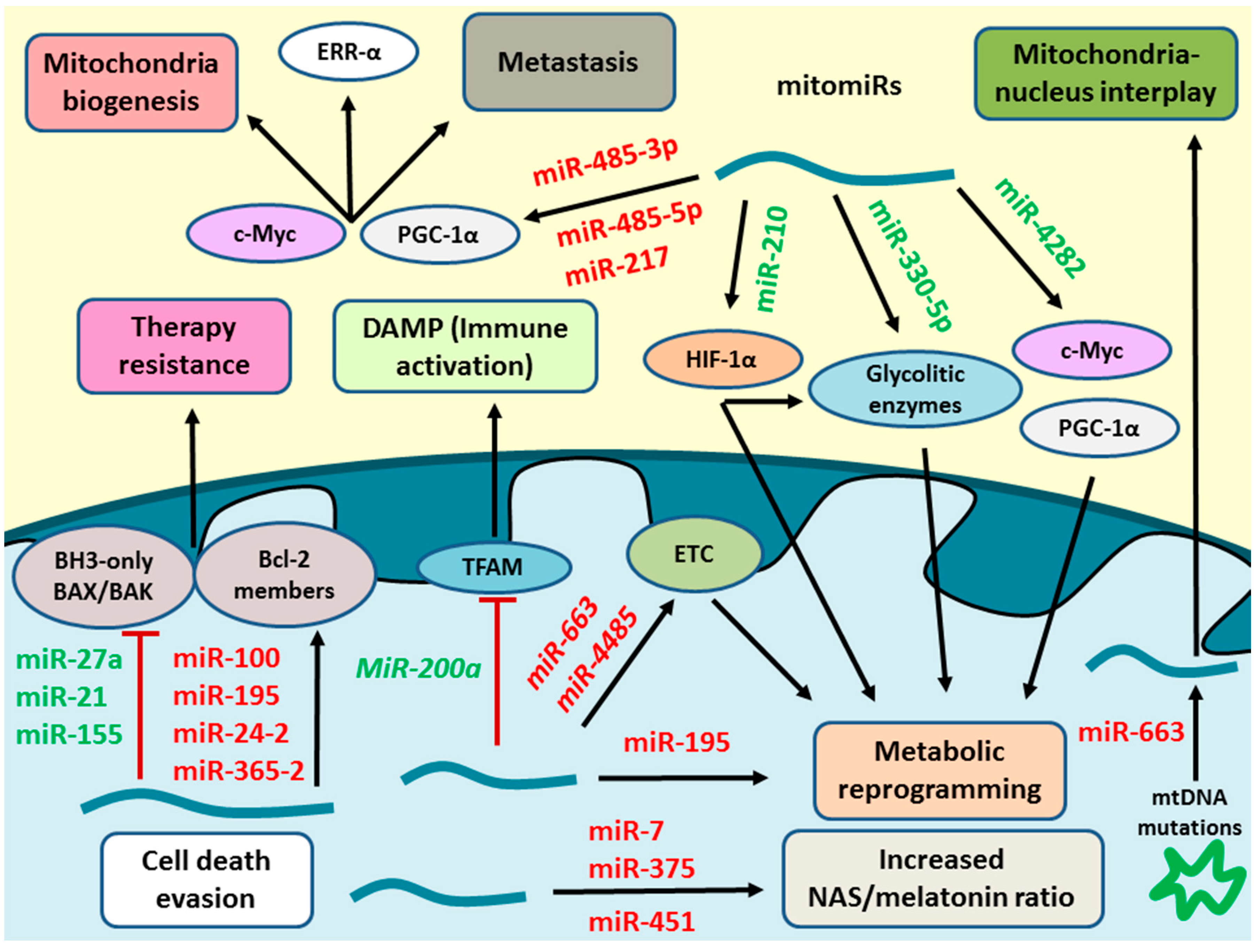
3. Mitochondrial MicroRNAs in Breast Cancer
3.1. Regulatory Mechanisms of MitomiRs
3.2. Purification, Isolation, and Detection of MitomiRs
3.3. Role of Mitochondrial MicroRNAs in Breast Cancer
3.3.1. Mitochondrial MicroRNAs in Tumour Metabolism
3.3.2. Mitochondria, MicroRNAs, and Cell Death Evasion
3.3.3. Impact of the Mitochondria Components in the Carcinogenic Process
3.3.4. MicroRNAs in Mitochondria Biogenesis and Degradation
3.3.5. MicroRNAs in Mitochondria-Nucleus Interactions
3.3.6. MitomiR in Mitochondrial Fission and Fusion Dynamics
3.3.7. Mitochondria and Immune System Interactions
4. From Basic to Translational Research. MicroRNAs and the Management of Patients with Breast Cancer: Present and Future
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ER | Oestrogen Receptor |
| PR | Progesterone receptor |
| miR | MicroRNA |
| mitomiR | Mitochondrial microRNA |
| pri-microRNA | Primary microRNA |
| DGCR8 | DiGeorge Syndrome Critical Region 8 |
| pre-microRNA | Precursor microRNA |
| XPO5 | Exportin 5 |
| ds-microRNA | Double-stranded microRNA |
| RISC | RNA-induced silencing complex |
| TRBP | Trans-activation response RNA-binding protein |
| AGO | Argonaute |
| MRE | MicroRNA response element |
| mRNA | Messenger RNA |
| IPO8 | Importin 8 |
| mtDNA | Mitochondrial DNA |
| PET | Positron emission tomography |
| HIF-1α | Hypoxia-induced factor 1α |
| PDK1 | Pyruvate dehydrogenase kinase 1 |
| CAF | Cancer-associated fibroblasts |
| PKM2 | Pyruvate Kinase M2 |
| LncRNA | Long non-coding RNA |
| NFE2L2 | Nuclear factor (erythroid-derived 2)-like 2 |
| MTC01 | Subunit 1 of cytochrome C oxidase gene |
| TLE3 | Transducin-like enhancer of split3 |
| GPCR | G protein coupled receptor |
| NAS | N-acetyl serotonin |
| CYP27B1 | Cytochrome P450 family 27, subfamily B, polypeptide 1 |
| ACACA | Acetyl-CoA carboxylase 1/alpha |
| FASN | Fatty acid synthase |
| HMGCR | 3-Hydroxy-3-methylglutaryl-CoA reductase |
| mPTP | Mitochondrial permeability transition pore |
| MOMP | Mitochondrial outer membrane permeabilization |
| APAF-1 | Apoptotic protease activation factor 1 |
| TRAIL | Tumour necrosis factor-related apoptosis-inducing ligand |
| ERRα | Oestrogen related receptor alpha |
| TCA | Tricarboxylic acid cycle |
| BCSCs | Breast cancer stem cells |
| ROS | Reactive oxygen species |
| POLG1 | DNA polymerase gamma |
| Drp1 | Dynamin-related protein 1 |
| MFN-1/2 | Mitofusins 1 and 2 |
| DAMPs | Damage-associated molecular patterns |
| IL-6 | Interleukin 6 |
| CRP | C reactive protein |
| TFAM | Mitochondrial transcription factor A |
| (pCR) | Pathological complete response |
References
- Rojas:, K.; Stuckey, A. Breast Cancer Epidemiology and Risk Factors. Clin. Obstet. Gynecol. 2016, 59, 651–672. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, C.E.; Bray, F.; Ferlay, J.; Lortet-Tieulent, J.; Anderson, B.O.; Jemal, A. International Variation in Female Breast Cancer Incidence and Mortality Rates. Cancer Epidemiol. Biomark. Prev. 2015, 24, 1495–1506. [Google Scholar] [CrossRef] [PubMed]
- Carioli, G.; Malvezzi, M.; Rodriguez, T.; Bertuccio, P.; Negri, E.; La Vecchia, C. Trends and predictions to 2020 in breast cancer mortality in Europe. Breast 2017, 36, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Ruddy, K.J.; Winer, E.P. Male breast cancer: Risk factors, biology, diagnosis, treatment, and survivorship. Ann. Oncol. 2013, 24, 1434–1443. [Google Scholar] [CrossRef] [PubMed]
- Howell, A.; Anderson, A.S.; Clarke, R.; Duffy, S.W.; Evans, D.G.; Garcia-Closas, M.; Gescher, A.J.; Key, T.J.; Saxton, J.M.; Harvie, M. Risk determination and prevention of breast cancer. Breast Cancer Res. 2014, 16, 446. [Google Scholar] [CrossRef] [PubMed]
- Shield, K.D.; Soerjomataram, I.; Rehm, J. Alcohol Use and Breast Cancer: A Critical Review. Alcohol. Clin. Exp. Res. 2016, 40, 1166–1181. [Google Scholar] [CrossRef] [PubMed]
- Brewer, H.R.; Jones, M.E.; Schoemaker, M.J.; Ashworth, A.; Swerdlow, A.J. Family history and risk of breast cancer: An analysis accounting for family structure. Breast Cancer Res. Treat. 2017, 165, 193–200. [Google Scholar] [CrossRef]
- Gray, J.M.; Rasanayagam, S.; Engel, C.; Rizzo, J. State of the evidence 2017: An update on the connection between breast cancer and the environment. Environ. Health 2017, 16, 94. [Google Scholar] [CrossRef]
- Schnitt, S.J. Will Molecular Classification Replace Traditional Breast Pathology? Int. J. Surg. Pathol. 2010, 18, 162–166. [Google Scholar] [CrossRef]
- Abubakar, M.; Figueroa, J.; Ali, H.R.; Blows, F.; Lissowska, J.; Caldas, C.; Easton, D.F.; Sherman, M.E.; Garcia-Closas, M.; Dowsett, M.; et al. Combined quantitative measures of ER, PR, HER2, and KI67 provide more prognostic information than categorical combinations in luminal breast cancer. Mod. Pathol. 2019, 32, 1244–1256. [Google Scholar] [CrossRef]
- Fragomeni, S.M.; Sciallis, A.; Jeruss, J.S. Molecular Subtypes and Local-Regional Control of Breast Cancer. Surg. Oncol. Clin. N. Am. 2018, 27, 95–120. [Google Scholar] [CrossRef]
- Prat, A.; Pineda, E.; Adamo, B.; Galván, P.; Fernández-Martínez, A.; Gaba, L.; Díez, M.; Viladot, M.; Arance, A.; Muñoz, M. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 2015, 24, S26–S35. [Google Scholar] [CrossRef] [PubMed]
- Massarweh, S.A.; Sledge, G.W.; Miller, D.P.; McCullough, D.; Petkov, V.I.; Shak, S. Molecular Characterization and Mortality from Breast Cancer in Men. J. Clin. Oncol. 2018, 36, 1396–1404. [Google Scholar] [CrossRef] [PubMed]
- McDonald, E.S.; Clark, A.S.; Tchou, J.; Zhang, P.; Freedman, G.M. Clinical Diagnosis and Management of Breast Cancer. J. Nucl. Med. 2016, 57, 9–16. [Google Scholar] [CrossRef]
- Ortega, M.A.; Fraile-Martínez, O.; Asúnsolo, Á.; Buján, J.; Honduvilla, N.; Coca, S. Signal Transduction Pathways in Breast Cancer: The Important Role of PI3K/Akt/mTOR. J. Oncol. 2020, 2020, 9258396. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P. Update on the Treatment of Early-Stage Triple-Negative Breast Cancer. Curr. Treat. Options Oncol. 2018, 19, 22. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.-K.; Liu, R.-T.; Kang, H.-Y. MicroRNA-146b: A Novel Biomarker and Therapeutic Target for Human Papillary Thyroid Cancer. Int. J. Mol. Sci. 2017, 18, 636. [Google Scholar] [CrossRef] [PubMed]
- Qadir, M.I.; Faheem, A. miRNA: A Diagnostic and Therapeutic Tool for Pancreatic Cancer. Crit. Rev. Eukaryot. Gene Expr. 2017, 27, 197–204. [Google Scholar] [CrossRef]
- Matin, F.; Jeet, V.; Moya, L.; Selth, L.; Chambers, S.; BioResource, A.P.C.; Clements, J.A.; Batra, J.; Yeadon, T. A Plasma Biomarker Panel of Four MicroRNAs for the Diagnosis of Prostate Cancer. Sci. Rep. 2018, 8, 6653. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.-H.; Kim, Y.-K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.-H.; Nam, J.-W.; Heo, I.; Rhee, J.-K.; Sohn, S.Y.; Cho, Y.; Zhang, B.-T.; Kim, V.N. Molecular Basis for the Recognition of Primary microRNAs by the Drosha-DGCR8 Complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef] [PubMed]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef]
- Weiss, C.N.; Ito, K. A Macro View of MicroRNAs: The Discovery of MicroRNAs and Their Role in Hematopoiesis and Hematologic Disease. Int. Rev. Cell Mol. Biol. 2017, 334, 99–175. [Google Scholar] [CrossRef] [PubMed]
- Bartel, B. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Forman, J.J.; Coller, H.A. The code within the code: MicroRNAs target coding regions. Cell Cycle 2010, 9, 1533–1541. [Google Scholar] [CrossRef]
- Meijer, H.A.; Kong, Y.W.; Lu, W.-T.; Wilczynska, A.; Spriggs, R.V.; Robinson, S.W.; Godfrey, J.D.; Willis, A.E.; Bushell, M. Translational Repression and eIF4A2 Activity Are Critical for MicroRNA-Mediated Gene Regulation. Science 2013, 340, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liu, C.; Zhang, X. Mitochondrial Damage Mediated by miR-1 Overexpression in Cancer Stem Cells. Mol. Ther. Nucleic Acids 2019, 18, 938–953. [Google Scholar] [CrossRef]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; Da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef]
- Altuvia, Y.; Landgraf, P.; Lithwick, G.; Elefant, N.; Pfeffer, S.; Aravin, A.; Brownstein, M.J.; Tuschl, T.; Margalit, H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005, 33, 2697–2706. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, B. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2008, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Leaderer, D.; Hoffman, A.E.; Zheng, T.; Fu, A.; Weidhaas, J.; Paranjape, T.; Zhu, Y. Genetic and epigenetic association studies suggest a role of microRNA biogenesis gene exportin-5 (XPO5) in breast tumorigenesis. Int. J. Mol. Epidemiol. Genet. 2010, 2, 9–18. [Google Scholar] [PubMed]
- Romero-Córdoba, S.; Salido-Guadarrama, I.; Rodríguez-Dorantes, M.; Hidalgo-Miranda, A. miRNA biogenesis: Biological impact in the development of cancer. Cancer Biol. Ther. 2014, 15, 1444–1455. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Brane, A.C.; Tollefsbol, T.O. MicroRNAs and Epigenetics Strategies to Reverse Breast Cancer. Cells 2019, 8, 1214. [Google Scholar] [CrossRef] [PubMed]
- Prabhu, K.S.; Raza, A.; Karedath, T.; Raza, S.S.; Fathima, H.; Ahmed, E.I.; Kuttikrishnan, S.; Therachiyil, L.; Kulinski, M.; Dermime, S.; et al. Non-Coding RNAs as Regulators and Markers for Targeting of Breast Cancer and Cancer Stem Cells. Cancers 2020, 12, 351. [Google Scholar] [CrossRef]
- Petrović, N.; Davidovic, R.; Bajić, V.; Obradovic, M.M.; Isenovic, R.E. MicroRNA in breast cancer: The association with BRCA1/2. Cancer Biomark. 2017, 19, 119–128. [Google Scholar] [CrossRef]
- Loh, H.-Y.; Norman, B.P.; Lai, K.-S.; Rahman, N.M.A.N.A.; Alitheen, N.B.; Osman, M.A. The Regulatory Role of MicroRNAs in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 4940. [Google Scholar] [CrossRef]
- Liu, H.; Lei, C.; He, Q.; Pan, Z.; Xiao, D.; Tao, Y. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol. Cancer 2018, 17, 1–14. [Google Scholar] [CrossRef]
- Wei, Y.; Li, L.; Wang, N.; Zhang, C.-Y.; Zen, K. Importin 8 Regulates the Transport of Mature MicroRNAs into the Cell Nucleus. J. Biol. Chem. 2014, 289, 10270–10275. [Google Scholar] [CrossRef]
- Byrd, A.E.; Brewer, J.W. Micro (RNA)managing endoplasmic reticulum stress. IUBMB Life 2013, 65, 373–381. [Google Scholar] [CrossRef]
- Macgregor-Das, A.M.; Das, S. A microRNA’s journey to the center of the mitochondria. Am. J. Physiol. Circ. Physiol. 2018, 315, H206–H215. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Giampazolias, E.; Tait, S.W. Mitochondria and the hallmarks of cancer. FEBS J. 2015, 283, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Murri, M.; El Azzouzi, H. MicroRNAs as regulators of mitochondrial dysfunction and obesity. Am. J. Physiol. Circ. Physiol. 2018, 315, H291–H302. [Google Scholar] [CrossRef] [PubMed]
- Galvan, D.L.; Green, N.H.; Danesh, F.R. The hallmarks of mitochondrial dysfunction in chronic kidney disease. Kidney Int. 2017, 92, 1051–1057. [Google Scholar] [CrossRef]
- Wang, W.-X.; Visavadiya, N.P.; Pandya, J.D.; Nelson, P.T.; Sullivan, P.G.; Springer, J.E. Mitochondria-associated microRNAs in rat hippocampus following traumatic brain injury. Exp. Neurol. 2015, 265, 84–93. [Google Scholar] [CrossRef]
- Purohit, S.; Jahagirdar, D.; Kumar, A.; Sharma, N.K. Potential of Taming MicroRNA on Driver Seat to Control Mitochondrial Horses in Breast Carcinoma. MicroRNA 2016, 5, 158–166. [Google Scholar] [CrossRef]
- Bandiera, S.; Rüberg, S.; Girard, M.; Cagnard, N.; Hanein, S.; Chretien, D.; Munnich, A.; Lyonnet, S.; Henrion-Caude, A. Nuclear Outsourcing of RNA Interference Components to Human Mitochondria. PLoS ONE 2011, 6, e20746. [Google Scholar] [CrossRef]
- Li, P.; Jiao, J.; Gao, G.; Prabhakar, B.S. Control of mitochondrial activity by miRNAs. J. Cell. Biochem. 2012, 113, 1104–1110. [Google Scholar] [CrossRef]
- Kim, K.M.; Noh, J.H.; Abdelmohsen, K.; Gorospe, M. Mitochondrial noncoding RNA transport. BMB Rep. 2017, 50, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Barrey, E.; Saint-Auret, G.; Bonnamy, B.; Damas, D.; Boyer, O.; Gidrol, X. Pre-microRNA and Mature microRNA in Human Mitochondria. PLoS ONE 2011, 6, e20220. [Google Scholar] [CrossRef] [PubMed]
- Sripada, L.; Tomar, D.; Singh, R. Mitochondria: One of the destinations of miRNAs. Mitochondrion 2012, 12, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, H.; Das, S. Mitochondrial miRNA (MitomiR): A new player in cardiovascular health. Can. J. Physiol. Pharmacol. 2015, 93, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, N.; Peng, Y.; Tan, Z.; Ciraolo, G.; Wang, D.; Li, R. miRNAs in mtDNA-less cell mitochondria. Cell Death Discov. 2015, 1, 15004. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.-H.; Lee, M.-J.; Galas, D.J.; Wang, K. The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Borralho, P.M.; Steer, C.J.; Rodrigues, C.M.P. Isolation of Mitochondria from Liver and Extraction of Total RNA and Protein: Analyses of MicroRNA and Protein Expressions. In Advanced Structural Safety Studies; Springer Science and Business Media: Berlin, Germany, 2014; Volume 1241, pp. 9–22. [Google Scholar]
- Geiger, J.; Dalgaard, L.T. Isolation and Analysis of Mitochondrial Small RNAs from Rat Liver Tissue and HepG2 Cells. In Advanced Structural Safety Studies; Springer Science and Business Media: Berlin, Germany, 2018; pp. 337–350. [Google Scholar]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef]
- Hornig-Do, H.-T.; Günther, G.; Bust, M.; Lehnartz, P.; Bosio, A.; Wiesner, R.J. Isolation of functional pure mitochondria by superparamagnetic microbeads. Anal. Biochem. 2009, 389, 1–5. [Google Scholar] [CrossRef]
- Brown, R.A.M.; Epis, M.R.; Horsham, J.L.; Kabir, T.D.; Richardson, K.L.; Leedman, P.J. Total RNA extraction from tissues for microRNA and target gene expression analysis: Not all kits are created equal. BMC Biotechnol. 2018, 18, 16. [Google Scholar] [CrossRef]
- Yeri, A.; Courtright, A.; Danielson, K.M.; Hutchins, E.D.; Alsop, E.; Carlson, E.; Hsieh, M.; Ziegler, O.; Das, A.; Shah, R.V.; et al. Evaluation of commercially available small RNASeq library preparation kits using low input RNA. BMC Genom. 2018, 19, 331. [Google Scholar] [CrossRef]
- Godoy, P.M.; Bhakta, N.R.; Barczak, A.J.; Cakmak, H.; Fisher, S.; MacKenzie, T.C.; Patel, T.; Price, R.W.; Smith, J.F.; Woodruff, P.G.; et al. Large Differences in Small RNA Composition Between Human Biofluids. Cell Rep. 2018, 25, 1346–1358. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O.; Wind, F.; Negelein, E. The Metabolism of Tumors in The Body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef]
- Potter, M.; Newport, E.; Morten, K.J. The Warburg effect: 80 years on. Biochem. Soc. Trans. 2016, 44, 1499–1505. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The Biology of Cancer: Metabolic Reprogramming Fuels Cell Growth and Proliferation. Cell Metab. 2008, 7, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Courtnay, R.; Ngo, D.C.; Malik, N.; Ververis, K.; Tortorella, S.M.; Karagiannis, T.C. Cancer metabolism and the Warburg effect: The role of HIF-1 and PI3K. Mol. Biol. Rep. 2015, 42, 841–851. [Google Scholar] [CrossRef]
- Ivan, M.; Huang, X. miR-210: Fine-Tuning the Hypoxic Response. Single Mol. Single Cell Seq. 2013, 772, 205–227. [Google Scholar] [CrossRef]
- Ma, X.; Li, C.; Sun, L.; Huang, D.; Li, T.; He, X.; Wu, G.; Yang, Z.; Zhong, X.; Song, L.; et al. Lin28/let-7 axis regulates aerobic glycolysis and cancer progression via PDK1. Nat. Commun. 2014, 5, 5212. [Google Scholar] [CrossRef] [PubMed]
- Serguienko, A.; Grad, I.; Wennerstrøm, A.B.; Meza-Zepeda, L.A.; Thiede, B.; Stratford, E.W.; Myklebost, O.; Munthe, E. Metabolic reprogramming of metastatic breast cancer and melanoma by let-7a microRNA. Oncotarget 2014, 6, 2451–2465. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, Z.; Liu, W.; Li, X. SNHG3 Functions as miRNA Sponge to Promote Breast Cancer Cells Growth Through the Metabolic Reprogramming. Appl. Biochem. Biotechnol. 2020, 191, 1084–1099. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hu, H.; Yan, G.; Wu, T.; Liu, S.; Chen, W.; Ning, Y.; Lu, Z. Long Non-Coding RNA and Breast Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819843889. [Google Scholar] [CrossRef]
- Yuan, Y.; Yao, Y.F.; Hu, S.N.; Gao, J.; Zhang, L.-L. MiR-133a Is Functionally Involved in Doxorubicin-Resistance in Breast Cancer Cells MCF-7 via Its Regulation of the Expression of Uncoupling Protein 2. PLoS ONE 2015, 10, e0129843. [Google Scholar] [CrossRef]
- Baffy, G. Uncoupling protein-2 and cancer. Mitochondrion 2010, 10, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.-A.; Lee, S.; Kwak, M.-K. NFE2L2/NRF2 Activity Is Linked to Mitochondria and AMP-Activated Protein Kinase Signaling in Cancers Through miR-181c/Mitochondria-Encoded Cytochrome c Oxidase Regulation. Antioxid. Redox Signal. 2017, 27, 945–961. [Google Scholar] [CrossRef]
- De La Vega, M.R.; Chapman, E.; Zhang, D.D. NRF2 and the Hallmarks of Cancer. Cancer Cell 2018, 34, 21–43. [Google Scholar] [CrossRef] [PubMed]
- Mihaylova, M.M.; Shaw, R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nature 2011, 13, 1016–1023. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Deng, X.; Gan, X.; Zhang, J.; Ren, G.; Shen, F.; Feng, J.; Cai, W.; Xu, B. Targeting of TLE3 by miR-3677 in human breast cancer promotes cell proliferation, migration and invasion. Oncol. Lett. 2019, 19, 1409–1417. [Google Scholar] [CrossRef] [PubMed]
- Pearson, S.; Loft, A.; Rajbhandari, P.; Simcox, J.; Lee, S.; Tontonoz, P.; Mandrup, S.; Villanueva, C.J. Loss of TLE3 promotes the mitochondrial program in beige adipocytes and improves glucose metabolism. Genes Dev. 2019, 33, 747–762. [Google Scholar] [CrossRef]
- Li, Y.; Li, S.; Zhou, Y.; Meng, X.; Zhang, J.-J.; Xu, D.-P.; Li, H.-B. Melatonin for the prevention and treatment of cancer. Oncotarget 2017, 8, 39896–39921. [Google Scholar] [CrossRef]
- Kubatka, P.; Zubor, P.; Büsselberg, D.; Kwon, T.K.; Adamek, M.; Petrovič, D.; Opatřilová, R.; Gazdikova, K.; Caprnda, M.; Rodrigo, L.; et al. Melatonin and breast cancer: Evidences from preclinical and human studies. Crit. Rev. Oncol. 2018, 122, 133–143. [Google Scholar] [CrossRef]
- Wegrzyn, L.R.; Tamimi, R.M.; Rosner, B.A.; Brown, S.B.; Stevens, R.G.; Eliassen, A.H.; Laden, F.; Willett, W.C.; Hankinson, S.E.; Schernhammer, E. Rotating Night-Shift Work and the Risk of Breast Cancer in the Nurses’ Health Studies. Am. J. Epidemiol. 2017, 186, 532–540. [Google Scholar] [CrossRef]
- Jones, E.M.; Schoemaker, M.J.; McFadden, E.C.; Wright, L.B.; Johns, L.E.; Swerdlow, A.J. Night shift work and risk of breast cancer in women: The Generations Study cohort. Br. J. Cancer 2019, 121, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Suofu, Y.; Li, W.; Jean-Alphonse, F.G.; Jia, J.; Khattar, N.K.; Li, J.; Baranov, S.V.; Leronni, D.; Mihalik, A.C.; He, Y.; et al. Dual role of mitochondria in producing melatonin and driving GPCR signaling to block cytochrome c release. Proc. Natl. Acad. Sci. USA 2017, 114, E7997–E8006. [Google Scholar] [CrossRef] [PubMed]
- Proietti, S.; Cucina, A.; Minini, M.; Bizzarri, M. Melatonin, mitochondria, and the cancer cell. Cell. Mol. Life Sci. 2017, 74, 4015–4025. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G. Breast cancer: Occluded role of mitochondria N-acetylserotonin/melatonin ratio in co-ordinating pathophysiology. Biochem. Pharmacol. 2019, 168, 259–268. [Google Scholar] [CrossRef]
- Marques, M.M.C.; Evangelista, A.F.; Macedo, T.; Vieira, R.A.D.C.; Scapulatempo-Neto, C.; Reis, R.M.; Carvalho, A.L.; Da Silva, I.D.C.G. Expression of tumor suppressors miR-195 and let-7a as potential biomarkers of invasive breast cancer. Clinics 2018, 73, 184. [Google Scholar] [CrossRef]
- Singh, R.; Yadav, V.; Kumar, S.; Saini, N. MicroRNA-195 inhibits proliferation, invasion and metastasis in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1. Sci. Rep. 2015, 5, 17454. [Google Scholar] [CrossRef]
- Zhalehjoo, N.; Shakiba, Y.; Panjehpour, M. Gene expression profiles of CYP24A1 and CYP27B1 in malignant and normal breast tissues. Mol. Med. Rep. 2016, 15, 467–473. [Google Scholar] [CrossRef]
- Chajès, V.; Cambot, M.; Moreau, K.; Lenoir, G.M.; Joulin, V. Acetyl-CoA Carboxylase α Is Essential to Breast Cancer Cell Survival. Cancer Res. 2006, 66, 5287–5294. [Google Scholar] [CrossRef]
- Menendez, J.A.; Lupu, R. Fatty acid synthase (FASN) as a therapeutic target in breast cancer. Expert Opin. Ther. Targets 2017, 21, 1001–1016. [Google Scholar] [CrossRef]
- Van Wyhe, R.D.; Rahal, O.M.; Woodward, W.A. Effect of statins on breast cancer recurrence and mortality: A review. Breast Cancer Targets Ther. 2017, 9, 559–565. [Google Scholar] [CrossRef]
- Jeong, S.-Y.; Seol, D.-W. The role of mitochondria in apoptosis. BMB Rep. 2008, 41, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Smaili, S.; Hsu, Y.-T.; Carvalho, A.; Rosenstock, T.R.; Sharpe, J.; Youle, R. Mitochondria, calcium and pro-apoptotic proteins as mediators in cell death signaling. Braz. J. Med. Biol. Res. 2003, 36, 183–190. [Google Scholar] [CrossRef] [PubMed]
- López, J.; Tait, S.W. Mitochondrial apoptosis: Killing cancer using the enemy within. Br. J. Cancer 2015, 112, 957–962. [Google Scholar] [CrossRef] [PubMed]
- Ichim, G.; Lopez, J.; Ahmed, S.U.; Muthalagu, N.; Giampazolias, E.; Delgado, M.E.; Haller, M.; Riley, J.S.; Mason, S.; Athineos, D.; et al. Limited mitochondrial permeabilization causes DNA damage and genomic instability in the absence of cell death. Mol. Cell 2015, 57, 860–872. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, S. A comparative analysis of BCL-2 family. Bioinformation 2019, 15, 299–306. [Google Scholar] [CrossRef]
- Lomonosova, E.; Chinnadurai, G. BH3-only proteins in apoptosis and beyond: An overview. Oncogene 2008, 27, S2–S19. [Google Scholar] [CrossRef]
- Zhou, S.; Huang, Q.; Zheng, S.; Lin, K.; You, J.; Zhang, X. miR-27a regulates the sensitivity of breast cancer cells to cisplatin treatment via BAK-SMAC/DIABLO-XIAP axis. Tumour Biol. 2016, 37, 6837–6845. [Google Scholar] [CrossRef]
- Sharma, P.; Kumar, S. Metformin inhibits human breast cancer cell growth by promoting apoptosis via a ROS-independent pathway involving mitochondrial dysfunction: Pivotal role of superoxide dismutase (SOD). Cell. Oncol. 2018, 41, 637–650. [Google Scholar] [CrossRef]
- Yang, J.; Wu, Y.; Wang, X.; Xu, L.; Zhao, X.; Yang, Y. Chemoresistance is associated with overexpression of HAX-1, inhibition of which resensitizes drug-resistant breast cancer cells to chemotherapy. Tumor Biol. 2017, 39, 1010428317692228. [Google Scholar] [CrossRef]
- Wu, G.; Zhou, W.; Pan, X.; Sun, Y.; Xu, H.; Shi, P.; Li, J.; Gao, L.; Tian, X. miR-100 Reverses Cisplatin Resistance in Breast Cancer by Suppressing HAX-1. Cell. Physiol. Biochem. 2018, 47, 2077–2087. [Google Scholar] [CrossRef]
- Sun, X.; Li, Y.; Zheng, M.; Zuo, W.; Zheng, W. MicroRNA-223 Increases the Sensitivity of Triple-Negative Breast Cancer Stem Cells to TRAIL-Induced Apoptosis by Targeting HAX-1. PLoS ONE 2016, 11, e0162754. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Hu, Y.; Xu, L.; Fu, Y.; Tu, J.; Zhao, H.; Zhang, S.; Hong, R.; Gu, X. The role of miR-125b-mitochondria-caspase-3 pathway in doxorubicin resistance and therapy in human breast cancer. Tumor Biol. 2015, 36, 7185–7194. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Zhao, X.; Wang, J.; Lv, L.; Wang, C.; Feng, L.; Shen, L.; Ren, W. miR-125b regulates the drug-resistance of breast cancer cells to doxorubicin by targeting HAX-1. Oncol. Lett. 2017, 15, 1621–1629. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Wang, S.; Zhao, Y.; Zhang, Z.; Qin, C.; Yang, X. MiR-519d impedes cisplatin-resistance in breast cancer stem cells by down-regulating the expression of MCL-1. Oncotarget 2017, 8, 22003–22013. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Saini, N. Downregulation of BCL2 by miRNAs augments drug-induced apoptosis—A combined computational and experimental approach. J. Cell Sci. 2012, 125, 1568–1578. [Google Scholar] [CrossRef]
- Kühlbrandt, W. Structure and function of mitochondrial membrane protein complexes. BMC Biol. 2015, 13, 89. [Google Scholar] [CrossRef]
- Bachmann, M.; Pontarin, G.; Szabò, I. The Contribution of Mitochondrial Ion Channels to Cancer Development and Progression. Cell. Physiol. Biochem. 2019, 53, 63–78. [Google Scholar] [CrossRef]
- Choudhury, A.R.; Singh, K.K. Mitochondrial determinants of cancer health disparities. Semin. Cancer Biol. 2017, 47, 125–146. [Google Scholar] [CrossRef]
- Stojkovič, G.; Makarova, A.V.; Wanrooij, P.H.; Forslund, J.; Burgers, P.M.; Wanrooij, S. Oxidative DNA damage stalls the human mitochondrial replisome. Sci. Rep. 2016, 6, 28942. [Google Scholar] [CrossRef]
- Quirós, P.M.; Mottis, A.; Auwerx, J. Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 2016, 17, 213–226. [Google Scholar] [CrossRef]
- Kuznetsov, A.V.; Margreiter, R. Heterogeneity of Mitochondria and Mitochondrial Function within Cells as Another Level of Mitochondrial Complexity. Int. J. Mol. Sci. 2009, 10, 1911–1929. [Google Scholar] [CrossRef] [PubMed]
- Vyas, S.; Zaganjor, E.; Haigis, M.C. Mitochondria and Cancer. Cell 2016, 166, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Luo, X.; Xiao, L.; Tang, M.; Bode, A.M.; Dong, Z.; Cao, Y. The Role of PGC1 in Cancer Metabolism and its Therapeutic Implications. Mol. Cancer Ther. 2016, 15, 774–782. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-Y.; McDonnell, D.P. Molecular Pathways: The Metabolic Regulator Estrogen-Related Receptor as a Therapeutic Target in Cancer. Clin. Cancer Res. 2012, 18, 6089–6095. [Google Scholar] [CrossRef] [PubMed]
- LeBleu, V.S.; O’Connell, J.T.; Herrera, K.N.G.; Wikman, H.; Pantel, K.; Haigis, M.C.; De Carvalho, F.M.; Damascena, A.; Chinen, L.T.D.; Rocha, R.M.; et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nature 2014, 16, 992–1003. [Google Scholar] [CrossRef]
- Cai, F.-F.; Xu, C.; Pan, X.; Cai, L.; Lin, X.-Y.; Chen, S.; Biskup, E. Prognostic value of plasma levels of HIF-1a and PGC-1a in breast cancer. Oncotarget 2016, 7, 77793–77806. [Google Scholar] [CrossRef]
- Anderson, R.M.; Barger, J.L.; Edwards, M.G.; Braun, K.H.; O’Connor, C.E.; Prolla, T.A.; Weindruch, R. Dynamic regulation of PGC-1α localization and turnover implicates mitochondrial adaptation in calorie restriction and the stress response. Aging Cell 2008, 7, 101–111. [Google Scholar] [CrossRef]
- Lou, C.; Xiao, M.; Cheng, S.; Lu, X.; Jia, S.; Ren, Y.; Li, Z. MiR-485-3p and miR-485-5p suppress breast cancer cell metastasis by inhibiting PGC-1α expression. Cell Death Dis. 2016, 7, e2159. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, X.; Liu, J.; Guo, H.; Xu, H.; Zhang, G. PGC-1 alpha interacts with microRNA-217 to functionally regulate breast cancer cell proliferation. Biomed. Pharmacother. 2017, 85, 541–548. [Google Scholar] [CrossRef]
- Fallah, Y.; Brundage, J.; Allegakoen, P.; Shajahan, A.N. MYC-Driven Pathways in Breast Cancer Subtypes. Biomolecules 2017, 7, 53. [Google Scholar] [CrossRef]
- Li, F.; Wang, Y.; Zeller, K.I.; Potter, J.J.; Wonsey, D.R.; O’Donnell, K.A.; Kim, J.-W.; Yustein, J.T.; Lee, L.A.; Dang, C.V. Myc Stimulates Nuclearly Encoded Mitochondrial Genes and Mitochondrial Biogenesis. Mol. Cell. Biol. 2005, 25, 6225–6234. [Google Scholar] [CrossRef] [PubMed]
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Jiang, G.-Q. MiR-4282 inhibits proliferation, invasion and metastasis of human breast cancer by targeting Myc. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8763–8771. [Google Scholar] [PubMed]
- Hardy, L.; Frison, M.; Campanella, M. Breast cancer cells exploit mitophagy to exert therapy resistance. Oncotarget 2018, 9, 14040–14041. [Google Scholar] [CrossRef]
- Daoud, H.; Gruchy, N.; Constans, J.-M.; Moussaoui, E.; Saumureau, S.; Bayou, N.; Amy, M.; Védrine, S.; Vu, P.Y.; Rötig, A.; et al. Haploinsufficiency of the GPD2 gene in a patient with nonsyndromic mental retardation. Qual. Life Res. 2008, 124, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Alkhaja, A.K.; Jans, D.C.; Nikolov, M.; Vukotic, M.; Lytovchenko, O.; Ludewig, F.; Schliebs, W.; Riedel, D.; Urlaub, H.; Jakobs, S.; et al. MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol. Biol. Cell 2012, 23, 247–257. [Google Scholar] [CrossRef]
- Ruzzenente, B.; Metodiev, M.D.; Wredenberg, A.; Bratic, A.; Park, C.B.; Camara, Y.; Milenkovic, D.; Zickermann, V.; Wibom, R.; Hultenby, K.; et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2011, 31, 443–456. [Google Scholar] [CrossRef]
- Guaragnella, N.; Coyne, L.P.; Chen, X.J.; Giannattasio, S. Mitochondria–cytosol–nucleus crosstalk: Learning from Saccharomyces cerevisiae. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef]
- Sripada, L.; Singh, R.; Lipatova, A.V.; Prajapati, P.; Tomar, D.; Bhatelia, K.; Roy, M.; Godbole, M.M.; Chumakov, P.M. hsa-miR-4485 regulates mitochondrial functions and inhibits the tumorigenicity of breast cancer cells. J. Mol. Med. 2017, 95, 641–651. [Google Scholar] [CrossRef]
- Carden, T.; Singh, B.; Mooga, V.; Bajpai, P.; Singh, K.K. Epigenetic modification of miR-663 controls mitochondria-to-nucleus retrograde signaling and tumor progression. J. Biol. Chem. 2017, 292, 20694–20706. [Google Scholar] [CrossRef]
- Guerra, F.; Arbini, A.A.; Moro, L. Mitochondria and cancer chemoresistance. Biochim. Biophys. Acta (BBA) Gen. Subj. 2017, 1858, 686–699. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Sampath, H. Mitochondrial DNA Integrity: Role in Health and Disease. Cells 2019, 8, 100. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Owens, K.M.; Bajpai, P.; Desouki, M.M.; Srinivasasainagendra, V.; Tiwari, H.K.; Singh, K.K. Mitochondrial DNA Polymerase POLG1 Disease Mutations and Germline Variants Promote Tumorigenic Properties. PLoS ONE 2015, 10, e0139846. [Google Scholar] [CrossRef] [PubMed]
- Trotta, A.P.; Chipuk, J.E. Mitochondrial dynamics as regulators of cancer biology. Cell. Mol. Life Sci. 2017, 74, 1999–2017. [Google Scholar] [CrossRef] [PubMed]
- Westermann, B. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 2010, 11, 872–884. [Google Scholar] [CrossRef] [PubMed]
- Tilokani, L.; Nagashima, S.; Paupe, V.; Prudent, J. Mitochondrial dynamics: Overview of molecular mechanisms. Essays Biochem. 2018, 62, 341–360. [Google Scholar] [CrossRef]
- Purohit, P.K.; Edwards, R.; Tokatlidis, K.; Saini, N. MiR-195 regulates mitochondrial function by targeting mitofusin-2 in breast cancer cells. RNA Biol. 2019, 16, 918–929. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Long, B.; Jiao, J.-Q.; Wang, J.-X.; Liu, J.-P.; Li, Q.; Li, P.-F. miR-484 regulates mitochondrial network through targeting Fis1. Nat. Commun. 2012, 3, 781. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Jeong, S.-Y.; Karbowski, M.; Smith, C.L.; Youle, R.J. Roles of the Mammalian Mitochondrial Fission and Fusion Mediators Fis1, Drp1, and Opa1 in Apoptosis. Mol. Biol. Cell 2004, 15, 5001–5011. [Google Scholar] [CrossRef]
- Losón, O.C.; Song, Z.; Chen, H.; Chan, D.C. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell 2013, 24, 659–667. [Google Scholar] [CrossRef]
- Zearo, S.; Kim, E.; Zhu, Y.; Zhao, J.T.; Sidhu, S.B.; Robinson, B.G.; Soon, P.S. MicroRNA-484 is more highly expressed in serum of early breast cancer patients compared to healthy volunteers. BMC Cancer 2014, 14, 200. [Google Scholar] [CrossRef]
- Balkwill, F.; Mantovani, A.; Balkwill, F. Inflammation and cancer: Back to Virchow? Lancet 2001, 357, 539–545. [Google Scholar] [CrossRef]
- Huang, J.; Xie, Y.; Sun, X.; Zeh, H.J.; Kang, R.; Lotze, M.T.; Tang, D. DAMPs, ageing, and cancer: The ‘DAMP Hypothesis’. Ageing Res. Rev. 2014, 24, 3–16. [Google Scholar] [CrossRef]
- Grazioli, S.; Pugin, J. Mitochondrial Damage-Associated Molecular Patterns: From Inflammatory Signaling to Human Diseases. Front. Immunol. 2018, 9, 832. [Google Scholar] [CrossRef]
- Karsch-Bluman, A.; Feiglin, A.; Arbib, E.; Stern, T.; Shoval, H.; Schwob, O.; Berger, M.; Benny, O. Tissue necrosis and its role in cancer progression. Oncogene 2018, 38, 1920–1935. [Google Scholar] [CrossRef] [PubMed]
- Kapetanovic, R.; Bokil, N.J.; Sweet, M.J. Innate immune perturbations, accumulating DAMPs and inflammasome dysregulation: A ticking time bomb in ageing. Ageing Res. Rev. 2015, 24, 40–53. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Zhou, E.; Wang, Y.; Xu, F.; Zhang, D.; Zhong, D. microRNA-200a Inhibits Cell Proliferation by Targeting Mitochondrial Transcription Factor A in Breast Cancer. DNA Cell Biol. 2014, 33, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Park, J.; Tran, Q.; Kim, D.; Hong, Y.; Cho, H.; Kwon, S.H.; Brazil, D.; Kim, S.-H.; Park, J. Mitochondrial transcription factor A (TFAM) is upregulated in glioma. Mol. Med. Rep. 2017, 15, 3781–3786. [Google Scholar] [CrossRef]
- De Araújo, L.F.; Siena, A.D.D.; Plaça, J.R.; Brotto, D.B.; Barros, I.I.; Muys, B.R.; Biagi, C.A.O.; Peronni, K.C.; Sousa, J.F.; Molfetta, G.A.; et al. Mitochondrial transcription factor A (TFAM) shapes metabolic and invasion gene signatures in melanoma. Sci. Rep. 2018, 8, 14190. [Google Scholar] [CrossRef]
- Yao, J.; Xu, F.; Zhang, D.; Yi, W.; Chen, X.; Chen, G.; Zhou, E. TP73-AS1 promotes breast cancer cell proliferation through miR-200a-mediated TFAM inhibition. J. Cell. Biochem. 2017, 119, 680–690. [Google Scholar] [CrossRef] [PubMed]
- Duarte, F.; Palmeira, C.M.; Rolo, A.P. The Role of microRNAs in Mitochondria: Small Players Acting Wide. Genes 2014, 5, 865–886. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, C.; Zhu, S.; Cao, L.; Kroemer, G.; Zeh, H.; Tang, D.; Kang, R. TFAM is a novel mediator of immunogenic cancer cell death. OncoImmunology 2018, 7, e1431086. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Zhou, S.; Zheng, M.; Deng, X.; Yi, Y.; Huang, T. MiR-199a-3p enhances breast cancer cell sensitivity to cisplatin by downregulating TFAM (TFAM). Biomed. Pharmacother. 2017, 88, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Bonneau, E.; Neveu, B.; Kostantin, E.; Tsongalis, G.; De Guire, V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC 2019, 30, 114–127. [Google Scholar] [PubMed]
- Mishra, S.; Yadav, T.; Rani, V.; Information, P.E.K.F.C. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit. Rev. Oncol. 2016, 98, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Rincon, A.L.; Martinez-Archundia, M.; Martinez-Ruiz, G.U.; Hu, Y.-J.; Tonda, A. Automatic discovery of 100-miRNA signature for cancer classification using ensemble feature selection. BMC Bioinform. 2019, 20, 480. [Google Scholar] [CrossRef]
- Xin, H.; Li, X.; Yang, B.; Zhang, L.; Han, Z.; Han, C. Blood-based multiple-microRNA assay displays a better diagnostic performance than single-microRNA assay in the diagnosis of breast tumor. Tumor Biol. 2014, 35, 12635–12643. [Google Scholar] [CrossRef]
- Joyce, D.P.; Kerin, M.J.; Dwyer, R.M. Exosome-encapsulated microRNAs as circulating biomarkers for breast cancer. Int. J. Cancer 2016, 139, 1443–1448. [Google Scholar] [CrossRef]
- Bertoli, G.; Cava, C.; Castiglioni, I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics 2015, 5, 1122–1143. [Google Scholar] [CrossRef]
- Adhami, M.; Haghdoost, A.A.; Sadeghi, B.; Afshar, R.M. Candidate miRNAs in human breast cancer biomarkers: A systematic review. Breast Cancer 2017, 25, 198–205. [Google Scholar] [CrossRef]
- He, K.; Li, W.-X.; Guan, D.; Gong, M.; Ye, S.; Fang, Z.; Huang, J.-F.; Lu, A. Regulatory network reconstruction of five essential microRNAs for survival analysis in breast cancer by integrating miRNA and mRNA expression datasets. Funct. Integr. Genom. 2019, 19, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Braga, E.A.; Burdennyy, A.M.; Pronina, I.V.; Filippova, E.A.; Kazubskaya, T.P.; Fridman, M.V.; Khodyrev, D.S.; Karpukhin, A.V.; Loginov, V.I.; Kushlinskii, N.E. System of Markers Based on the Methylation of a Group of Proapoptotic Genes in Combination with MicroRNA in the Diagnosis of Breast Cancer. Bull. Exp. Biol. Med. 2020, 168, 366–370. [Google Scholar] [CrossRef] [PubMed]
- Dastmalchi, N.; Safaralizadeh, R.; Baradaran, B.; Hosseinpourfeizi, M.; Baghbanzadeh, A. An update review of deregulated tumor suppressive microRNAs and their contribution in various molecular subtypes of breast cancer. Gene 2020, 729, 144301. [Google Scholar] [CrossRef] [PubMed]
- Haakensen, V.D.; Nygaard, V.; Greger, L.; Aure, M.R.; Fromm, B.; Bukholm, I.R.K.; Lüders, T.; Chin, S.-F.; Git, A.; Caldas, C.; et al. Subtype-specific micro-RNA expression signatures in breast cancer progression. Int. J. Cancer 2016, 139, 1117–1128. [Google Scholar] [CrossRef]
- Ozawa, P.M.M.; Vieira, E.; Lemos, D.D.S.; Souza, I.L.M.; Zanata, S.; Pankievicz, V.C.; Tuleski, T.R.; De Souza, E.M.; Wowk, P.F.; Urban, C.; et al. Identification of miRNAs Enriched in Extracellular Vesicles Derived from Serum Samples of Breast Cancer Patients. Biomolecules 2020, 10, 150. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Ding, M.; Lin, J. Three-microRNA expression signature predicts survival in triple-negative breast cancer. Oncol. Lett. 2020, 19, 301–308. [Google Scholar] [CrossRef]
- Stevic, I.; Müller, V.; Weber, K.; Fasching, P.A.; Karn, T.; Marmé, F.; Schem, C.; Stickeler, E.; Denkert, C.; Van Mackelenbergh, M.; et al. Specific microRNA signatures in exosomes of triple-negative and HER2-positive breast cancer patients undergoing neoadjuvant therapy within the GeparSixto trial. BMC Med. 2018, 16, 179. [Google Scholar] [CrossRef]
- Tripathi, R.; Aier, I.; Chakraborty, P.; Varadwaj, P.K. Unravelling the role of long non-coding RNA-LINC01087 in breast cancer. Non-coding RNA Res. 2020, 5, 1–10. [Google Scholar] [CrossRef]
- Wang, X.; Song, C.; Zhou, X.; Han, X.; Li, J.; Wang, Z.; Shang, H.; Liu, Y.; Cao, H. Mitochondria Associated MicroRNA Expression Profiling of Heart Failure. BioMed Res. Int. 2017, 2017, 1–10. [Google Scholar] [CrossRef]
- He, C.; Zheng, S.; Luo, Y.; Wang, B. Exosome Theranostics: Biology and Translational Medicine. Theranostics 2018, 8, 237–255. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Gilam, A.; Conde, J.; Weissglas-Volkov, D.; Oliva, N.; Friedman, E.; Artzi, N.; Shomron, N. Local microRNA delivery targets Palladin and prevents metastatic breast cancer. Nat. Commun. 2016, 7, 12868. [Google Scholar] [CrossRef] [PubMed]
- Yi, W.; Tu, M.-J.; Liu, Z.; Zhang, C.; Batra, N.; Yu, A.-X.; Yu, A.-M. Bioengineered miR-328-3p modulates GLUT1-mediated glucose uptake and metabolism to exert synergistic antiproliferative effects with chemotherapeutics. Acta Pharm. Sin. B 2020, 10, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Ahir, M.; Bhattacharya, S.; Karmakar, S.; Mukhopadhyay, A.; Mukherjee, S.; Ghosh, S.; Chattopadhyay, S.; Patra, P.; Adhikary, A. Tailored-CuO-nanowire decorated with folic acid mediated coupling of the mitochondrial-ROS generation and miR425-PTEN axis in furnishing potent anti-cancer activity in human triple negative breast carcinoma cells. Biomaterials 2016, 76, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, K.; Arun, A.; Singh, S.; Manohar, M.; Chuttani, K.; Konwar, R.; Dwivedi, A.; Soni, R.; Singh, A.K.; Mishra, A.K.; et al. Bivalent Approach for Homodimeric Estradiol Based Ligand: Synthesis and Evaluation for Targeted Theranosis of ER(+) Breast Carcinomas. Bioconjugate Chem. 2016, 27, 961–972. [Google Scholar] [CrossRef]
- Kardani, A.; Yaghoobi, H.; Alibakhshi, A.; Khatami, M. Inhibition of miR-155 in MCF-7 breast cancer cell line by gold nanoparticles functionalized with antagomir and AS1411 aptamer. J. Cell. Physiol. 2020, 235, 6887–6895. [Google Scholar] [CrossRef]


| MicroRNA | Expression | Target | Implications | Potential Use | Reference |
|---|---|---|---|---|---|
| miR-1 | Downregulated | Mitochondrial (MINOS-1, LRPPRC) Cytosol (GP-2) | Mitophagy evasion by BCSCs | Therapeutic target: Inducing mitophagy of stem cells In vitro and In vivo | Zhang et al., 2019 [28] |
| miR-7 miR-375 miR-451 | Downregulated | Cytosolic 14-3-3ζ, protein | Increased NAS/melatonin ratio: Modified melatonin metabolism and cell survival | Therapeutic and prevention target in vitro | Anderson 2019 [86] |
| Let-7 | Downregulated | Mitochondrial PDK-1 Apoptotic proteins | Metabolic reprogramming Increased sensitivity to doxorubicin in TNBC | Therapeutic target Causing mitochondrial ROS production. In combination with doxorubicin, increases sensitivity to the drug. Biomarker of invasiveness | Serguienko et al., 2018 [70] Marques et al., 2018 [87] |
| miR-21 | Upregulated | Mitochondrial apoptotic proteins | Apoptosis evasion | Therapeutic target: Downregulation by metformin may be correlated with the suppression of cell proliferation and migration Early diagnosis Survival prediction TNBC specific prognosis biomarker | Sharma & Kumar 2018 [100] Adhami et al., 2018 [162] He et al., 2019 [163] Wu et al., 2020 [168] |
| miR-24-2 miR-365-2 | Downregulated | Mitochondrial Bcl-2 protein | Apoptosis evasion Etoposide resistance | Therapeutic target: Negative regulator of BCL2, reducing levels of BCL2 protein and increasing apoptosis | Singh & Saini 2012 [107] |
| miR-27a | Upregulated | Mitochondrial proapoptotic protein (BAK) | Apoptosis evasion Increased proliferation and metastasis | Therapeutic target: Knockdown increases sensitivity of T-47D cells to cisplatin | Zhou et al., 2016 [99] |
| miR-100 | Downregulated | Mitochondrial antiapoptotic protein HAX-1 | Apoptosis evasion Cisplatin resistance | Therapeutic target: HAX-1 increases chemosensitivity of breast cancer | Wu et al., 2018 [102] |
| miR-125b | Downregulated | Mitochondrial apoptotic proteins | Apoptosis evasion Doxorubicin resistance | Coadjuvant therapy: miR-125b in combination with doxorubicin leads to loss of mitochondrial membrane potential Diagnosis | Xie et al., 2015 [104] Hu et al., 2018 [105] Braga et al., 2020 [164] |
| miR-133a | Downregulated | Mitochondrial UCP-2 | Metabolic reprogramming Doxorubicin resistance | Therapeutic target: miR133a/UCP-2 signaling for chemotherapy resistant | Yuan et al., 2015 [73] |
| miR-155 | Upregulated | Mitochondrial apoptotic proteins | Apoptosis evasion | Therapeutic target: miR-155 may be downregulated by metformin, which allows suppression of cell proliferation and migration Pathologic complete response Nanomedicine target: Gold Nanoparticle-Aptamer-anti-miR-155 development | Sharma & Kumar 2018 [100] Stevic et al., 2018 [169] Kardani et al., 2020 [178] |
| miR-181c | Upregulated | Mitochondrial MTC01 Cytosolic AMPK | Metabolic reprogramming Cell signalling pathways | Therapeutic target: Potential target to inhibit both NFE2L2/NRF2 and AMPKα in breast cancer cells | Jung et al., 2017 [75] |
| miR-195 | Downregulated | Mitochondrial CYP27B1, Bcl-2 MTN-2 Cytosolic lipogenesis enzymes | Metabolic reprogramming Apoptosis evasion Modified mitochondrial dynamics Etoposide resistance | Therapeutic target: miR-195 targets BCL inducing apoptosis. It also reduces proliferation, invasion, and migration. Prognosis Early diagnosis | Singh et al., 2015 [88] Singh & Saini 2012 [107] Purohit et al., 2019 [139] Adhami et al., 2018 [162] |
| miR-199a-3p miR-200a | Upregulated | Mitochondrial TFAM | Mitochondrial genome instability Immune interactions Cisplatin resistance | Therapeutic target: BC cells growth and mitochondrial DNA copy number regulations | Yao et al., 2014 [149] Yao et al., 2018 [152] Fan et al., 2017 [155] |
| miR-210 | Upregulated (by HIF-1α) | Mitochondrial and cytosolic targets | Metabolic reprogramming DNA damage Apoptosis evasion Angiogenesis Cell cycle alterations | Therapeutic target: overexpression makes cells more susceptibleto killing by 3-bromo-pyruvate Prognosis Early diagnosis | Ivan & Huang 2014 [68] Adhami et al., 2018 [162] |
| miR-217 miR-485-3p miR-485-5p | Downregulated | PGC-1α | Mitochondrial biogenesis Increased proliferation Invasion and metastasis | Therapeutic target: In vitro, overexpression of miR-485-3p and miR-485-5p suppressed mitochondrial respiration and potential for cell migration and invasión. In vivo, they inhibited spontaneous metastasis of BC cells. | Lou et al., 2016 [120] Zhang et al., 2017 [121] |
| miR-223 | Downregulated | Mitochondrial HAX-1 | Apoptosis evasion by TRAIL in TNBC | Therapeutic target: Overexpression increases sensitivity of TNBCSCs to TRAIL-induced apoptosis | Sun et al., 2016 [103] |
| miR-330-5p | Upregulated (by SNHG3) | Mitochondrial and cytosolic metabolic enzymes: PKM2 | Metabolic reprogramming | Therapeutic target: When increased, SNHG3 knockdown in CAF-secreted exosomes suppressed glycolysis metabolism and cell proliferation | Li et al., 2020 [71] |
| miR-484 | Upregulated | Mitochondrial Fis-1 | Mitochondrial dynamics Apoptosis evasion Cancer development | Therapeutic target: In vivo models with high or low levels of miR-484 correlates with reduced or enhanced mitochondrial fission, apoptosis, and myocardial infarction | Wang et al., 2012 [140] |
| miR-519d | Downregulated | Mitochondrial antiapoptotic protein MCL-1 | BCSC sensitivity to cisplatin reduced | Coadjuvant therapy: Induced expression of miR-519d in T-47D-cancer stem cells increased their sensitivity to cisplatin via apoptosis | Xie et al., 2017 [106] |
| miR-663 | Downregulated (by ROS) | Mitochondrial ETC components | Mitochondrion-nucleus signalling; increased proliferation, tumorigenesis | Therapeutic target: antimir-663 increased in vitro cellular proliferation and promoted tumor development in vivo | Carden et al., 2017 [132] |
| miR-3677 | Upregulated | TLE3 | Metabolic reprogramming | Therapeutic target: Silencing TLE3 may suppress proliferation and migration in BC cells. | Peng et al., 2020 [78] |
| miR-4282 | Downregulated | Nuclear c-Myc | c-Myc driven pathways; glutaminolysis chemotherapy resistance | Therapeutic target: In silico and in vitro analyses showed that Myc might be the target of miR-4282 in BC | Zhao et al., 2018 [125] |
| miR-4485 | Downregulated | Mitochondrial pre-rRNA 16 S ETC complex I Others | Abnormal ATP production and mitochondrial RNA processing; ROS regulation; apoptosis evasion | Therapeutic target: In vitro miR-4485 downregulated glycolytic pathway genes and decreased proliferation of BC cels. In vivo model, expression of this miR decreased the tumorigenicity | Sripada et al., 2017 [131] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortega, M.A.; Fraile-Martínez, O.; Guijarro, L.G.; Casanova, C.; Coca, S.; Álvarez-Mon, M.; Buján, J.; García-Honduvilla, N.; Asúnsolo, Á. The Regulatory Role of Mitochondrial MicroRNAs (MitomiRs) in Breast Cancer: Translational Implications Present and Future. Cancers 2020, 12, 2443. https://doi.org/10.3390/cancers12092443
Ortega MA, Fraile-Martínez O, Guijarro LG, Casanova C, Coca S, Álvarez-Mon M, Buján J, García-Honduvilla N, Asúnsolo Á. The Regulatory Role of Mitochondrial MicroRNAs (MitomiRs) in Breast Cancer: Translational Implications Present and Future. Cancers. 2020; 12(9):2443. https://doi.org/10.3390/cancers12092443
Chicago/Turabian StyleOrtega, Miguel A., Oscar Fraile-Martínez, Luis G. Guijarro, Carlos Casanova, Santiago Coca, Melchor Álvarez-Mon, Julia Buján, Natalio García-Honduvilla, and Ángel Asúnsolo. 2020. "The Regulatory Role of Mitochondrial MicroRNAs (MitomiRs) in Breast Cancer: Translational Implications Present and Future" Cancers 12, no. 9: 2443. https://doi.org/10.3390/cancers12092443
APA StyleOrtega, M. A., Fraile-Martínez, O., Guijarro, L. G., Casanova, C., Coca, S., Álvarez-Mon, M., Buján, J., García-Honduvilla, N., & Asúnsolo, Á. (2020). The Regulatory Role of Mitochondrial MicroRNAs (MitomiRs) in Breast Cancer: Translational Implications Present and Future. Cancers, 12(9), 2443. https://doi.org/10.3390/cancers12092443










