Epstein-Barr Virus Mediated Signaling in Nasopharyngeal Carcinoma Carcinogenesis
Abstract
Simple Summary
Abstract
1. Introduction
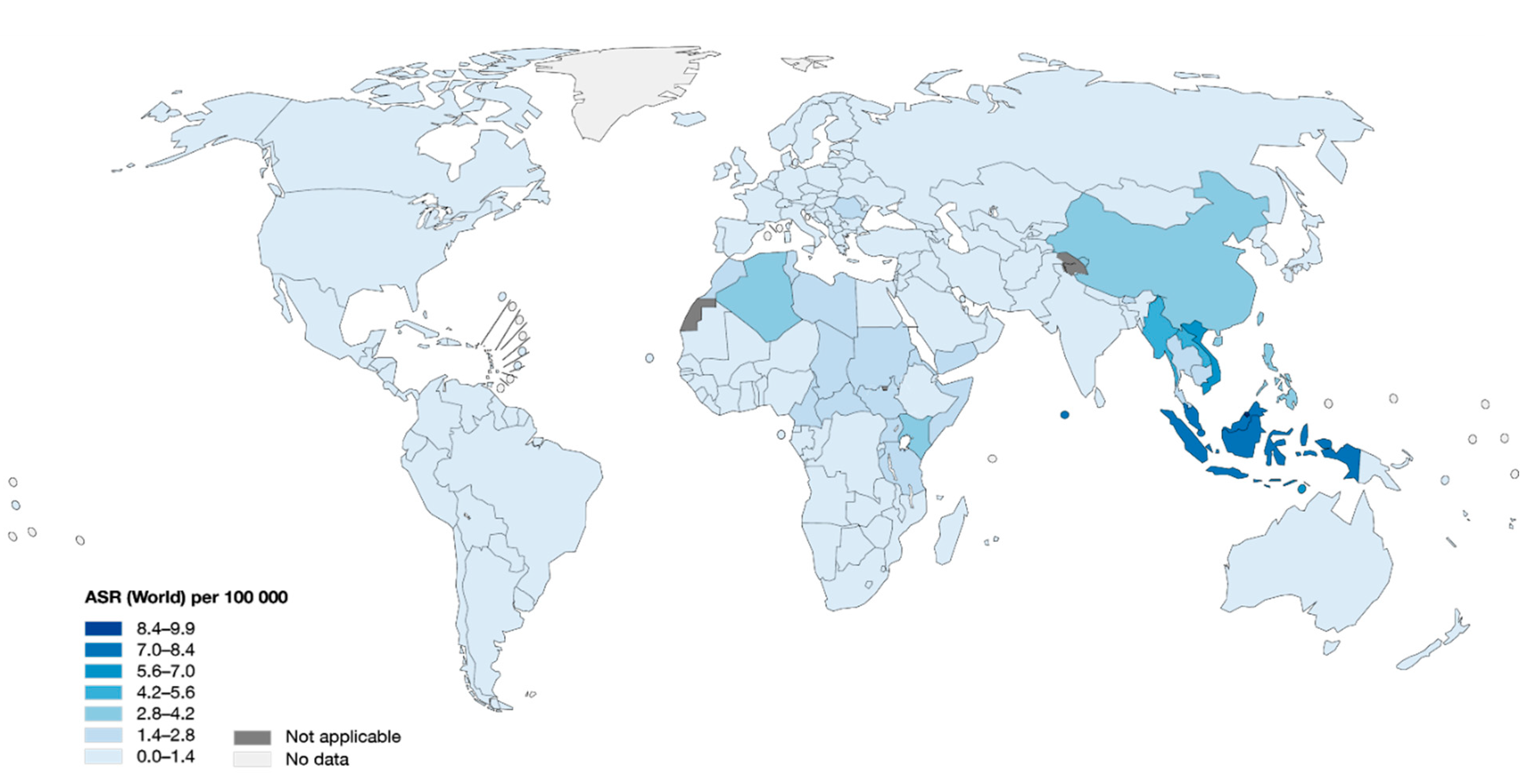
2. Epidemiology of NPC
3. EBV Strains
4. EBV and NPC Carcinogenesis
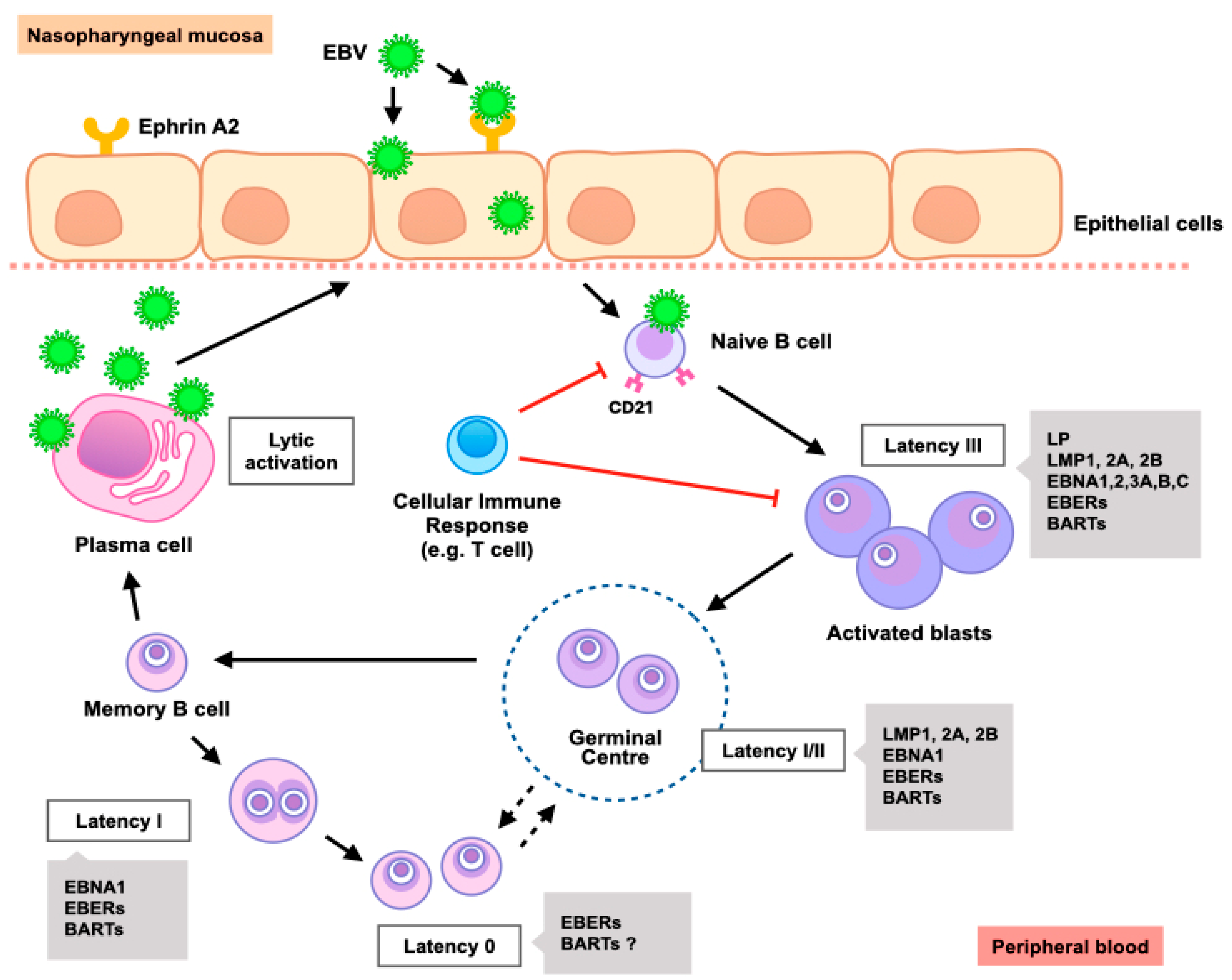
4.1. EBV Life Cycle in Epithelial Cells
4.2. EBV Latency in NPC
4.3. EBV Lytic Infection in NPC
5. EBV-Mediated Signaling
5.1. Wnt/β-Catenin
5.2. JAK/STAT
5.3. PI3K/Akt/mTOR
5.4. EGFR and MAPK
5.5. NF-κB
6. Cancer Hallmarks of EBV-Associated Malignancies
6.1. Immune Evasion
6.2. Metabolic Reprogramming
6.3. Apoptosis
6.4. Metastasis
6.5. Sustaining Proliferative Signal
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Petersson, F. Nasopharyngeal carcinoma: A review. Semin. Diagn. Pathol. 2015, 32, 54–73. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.T.; Adami, H.O. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1765–1777. [Google Scholar] [CrossRef] [PubMed]
- Tsao, S.W.; Tsang, C.M.; Lo, K.W. Epstein-Barr virus infection and nasopharyngeal carcinoma. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2017, 372, 20160270. [Google Scholar] [CrossRef]
- Tulalamba, W.; Janvilisri, T. Nasopharyngeal carcinoma signaling pathway: An update on molecular biomarkers. Int. J. Cell Biol. 2012, 2012, 594681. [Google Scholar] [CrossRef]
- Janvilisri, T. Omics-based identification of biomarkers for nasopharyngeal carcinoma. Dis. Markers 2015, 2015, 762128. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, R.W.; Kutty, M.K.; Dharmalingam, S.K.; Ponnudurai, J.R. Incidence of nasopharyngeal carcinoma in Malaysia, 1968–1977. Br. J. Cancer 1979, 40, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Mak, H.W.; Lee, S.H.; Chee, J.; Tham, I.; Goh, B.C.; Chao, S.S.; Ong, Y.K.; Loh, K.S.; Lim, C.M. Clinical outcome among nasopharyngeal cancer patients in a multi-ethnic society in Singapore. PLoS ONE 2015, 10, e0126108. [Google Scholar] [CrossRef]
- Yu, W.M.; Hussain, S.S.M. Incidence of nasopharyngeal carcinoma in Chinese immigrants, compared with Chinese in China and South East Asia: Review. J. Laryngol. Otol. 2009, 123, 1067–1074. [Google Scholar] [CrossRef]
- Jia, W.H.; Feng, B.J.; Xu, Z.L.; Zhang, X.S.; Huang, P.; Huang, L.X.; Yu, X.J.; Feng, Q.S.; Yao, M.H.; Shugart, Y.Y.; et al. Familial risk and clustering of nasopharyngeal carcinoma in Guangdong, China. Cancer 2004, 101, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.H.; sun Yu, I.T.; Tse, L.A.; Au, J.S.K.; Lau, J.S.M. Tobacco smoking, family history, and the risk of nasopharyngeal carcinoma: A case–referent study in Hong Kong Chinese. Cancer Causes Control. 2015, 26, 913–921. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chang, E.T.; Liu, Q.; Cai, Y.; Zhang, Z.; Chen, G.; Huang, Q.H.; Xie, S.H.; Cao, S.M.; Shao, J.Y.; et al. Quantification of familial risk of nasopharyngeal carcinoma in a high-incidence area. Cancer 2017, 123, 2716–2725. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Zhang, W.; Xie, C.; Wang, B.; Zhang, G.; Zhou, F. Nasopharyngeal carcinoma risk by histologic type in central China: Impact of smoking, alcohol and family history. Int. J. Cancer 2011, 129, 724–732. [Google Scholar] [CrossRef]
- Jia, W.H.; Luo, X.Y.; Feng, B.J.; Ruan, H.L.; Bei, J.X.; Liu, W.S.; Qin, H.D.; Feng, Q.S.; Chen, L.Z.; Yao, S.Y.; et al. Traditional Cantonese diet and nasopharyngeal carcinoma risk: A large-scale case-control study in Guangdong, China. BMC Cancer 2010, 10, 446. [Google Scholar] [CrossRef]
- Guo, X.; Johnson, R.C.; Deng, H.; Liao, J.; Guan, L.; Nelson, G.W.; Tang, M.; Zheng, Y.; De The, G.; O’Brien, S.J.; et al. Evaluation of nonviral risk factors for nasopharyngeal carcinoma in a high-risk population of southern China. Int. J. Cancer 2009, 124, 2942–2947. [Google Scholar] [CrossRef]
- Okekpa, S.I.; Mydin, R.B.S.M.N.; Mangantig, E.; Azmi, N.S.A.; Zahari, S.N.S.; Kaur, G.; Musa, Y. Nasopharyngeal carcinoma (NPC) risk factors: A systematic review and meta-analysis of the association with lifestyle, diets, socioeconomic and sociodemographic in asian region. Asian Pac. J. Cancer Prev. 2019, 20, 3505–3514. [Google Scholar] [CrossRef]
- Shivappa, N.; Hébert, J.R.; Zucchetto, A.; Montella, M.; Libra, M.; Garavello, W.; Rossi, M.; La Vecchia, C.; Serraino, D. Increased risk of nasopharyngeal carcinoma with increasing levels of diet-associated inflammation in an Italian case–control study. Nutr. Cancer 2016, 68, 1123–1130. [Google Scholar] [CrossRef]
- Polesel, J.; Serraino, D.; Negri, E.; Barzan, L.; Vaccher, E.; Montella, M.; Zucchetto, A.; Garavello, W.; Franceschi, S.; La Vecchia, C.; et al. Consumption of fruit, vegetables, and other food groups and the risk of nasopharyngeal carcinoma. Cancer Causes Control. 2013, 24, 1157–1165. [Google Scholar] [CrossRef]
- Turkoz, F.P.; Celenkoglu, G.; Dogu, G.G.; Kalender, M.E.; Coskun, U.; Alkis, N.; Ozkan, M.; Mehmet Turk, H.; Arslan, U.Y. Risk factors of nasopharyngeal carcinoma in Turkey—An epidemiological survey of the anatolian society of medical oncology. Asian Pac. J. Cancer Prev. 2011, 12, 3017–3021. [Google Scholar]
- Liu, Y.T.; Dai, J.J.; Xu, C.H.; Lu, Y.K.; Fan, Y.Y.; Zhang, X.L.; Zhang, C.X.; Chen, Y.M. Greater intake of fruit and vegetables is associated with lower risk of nasopharyngeal carcinoma in Chinese adults: A case-control study. Cancer Causes Control. 2012, 23, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Long, M.; Fu, Z.; Li, P.; Nie, Z. Cigarette smoking and the risk of nasopharyngeal carcinoma: A meta-analysis of epidemiological studies. BMJ Open 2017, 7, 14–17. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.Q.; Qin, H.D.; Ruan, H.L.; Shugart, Y.Y.; Jia, W.H. Quantitative association of tobacco smoking with the risk of nasopharyngeal carcinoma: A comprehensive meta-analysis of studies conducted between 1979 and 2011. Am. J. Epidemiol. 2013, 178, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.H.; Jiang, C.Q.; Ho, S.Y.; Zhang, W.S.; Mai, Z.M.; Xu, L.; Lo, C.M.; Lam, T.H. Smoking and nasopharyngeal carcinoma mortality: A cohort study of 101,823 adults in Guangzhou, China. BMC Cancer 2015, 15, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.X.; Jia, W.H. Familial nasopharyngeal carcinoma. Semin. Cancer Biol. 2002, 12, 443–450. [Google Scholar] [CrossRef]
- Tse, K.P.; Su, W.H.; Chang, K.P.; Tsang, N.M.; Yu, C.J.; Tang, P.; See, L.C.; Hsueh, C.; Yang, M.L.; Hao, S.P.; et al. Genome-wide sssociation study reveals multiple nasopharyngeal carcinoma-associated loci within the HLA region at chromosome 6p21.3. Am. J. Hum. Genet. 2009, 85, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Chung, G.T.Y.; Lui, V.W.Y.; To, K.-F.; Ma, B.B.Y.; Chow, C.; Woo, J.K.S.; Yip, K.Y.; Seo, J.; Hui, E.P.; et al. Exome and genome sequencing of nasopharynx cancer identifies NF-κB pathway activating mutations. Nat. Commun. 2017, 8, 14121. [Google Scholar] [CrossRef] [PubMed]
- Bei, J.-X.; Li, Y.; Jia, W.-H.; Feng, B.-J.; Zhou, G.; Chen, L.-Z.; Feng, Q.-S.; Low, H.-Q.; Zhang, H.; He, F.; et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat. Genet. 2010, 42, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Cui, Q.; Feng, Q.-S.; Mo, H.-Y.; Sun, J.; Xia, Y.-F.; Zhang, H.; Foo, J.N.; Guo, Y.-M.; Chen, L.-Z.; Li, M.; et al. An extended genome-wide association study identifies novel susceptibility loci for nasopharyngeal carcinoma. Hum. Mol. Genet. 2016, 25, 3626–3634. [Google Scholar] [CrossRef]
- Yu, G.; Hsu, W.-L.; Coghill, A.E.; Yu, K.J.; Wang, C.-P.; Lou, P.-J.; Liu, Z.; Jones, K.; Vogt, A.; Wang, M.; et al. Whole-Exome Sequencing of nasopharyngeal carcinoma families reveals novel variants potentially involved in nasopharyngeal carcinoma. Sci. Rep. 2019, 9, 9916. [Google Scholar] [CrossRef]
- Yoshizaki, T.; Ito, M.; Murono, S.; Wakisaka, N.; Kondo, S.; Endo, K. Current understanding and management of nasopharyngeal carcinoma. Auris Nasus Larynx 2012, 39, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Carioli, G.; Negri, E.; Kawakita, D.; Garavello, W.; La Vecchia, C.; Malvezzi, M. Global trends in nasopharyngeal cancer mortality since 1970 and predictions for 2020: Focus on low-risk areas. Int. J. Cancer 2017, 140, 2256–2264. [Google Scholar] [CrossRef]
- Tang, L.L.; Chen, W.Q.; Xue, W.Q.; He, Y.Q.; Zheng, R.S.; Zeng, Y.X.; Jia, W.H. Global trends in incidence and mortality of nasopharyngeal carcinoma. Cancer Lett. 2016, 374, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Palser, A.L.; Grayson, N.E.; White, R.E.; Corton, C.; Correia, S.; Ba abdullah, M.M.; Watson, S.J.; Cotten, M.; Arrand, J.R.; Murray, P.G.; et al. Genome diversity of Epstein-Barr Virus from multiple tumor types and normal infection. J. Virol. 2015, 89, 5222–5237. [Google Scholar] [CrossRef] [PubMed]
- Feederle, R.; Klinke, O.; Kutikhin, A.; Poirey, R.; Tsai, M.-H.; Delecluse, H.-J. Epstein-Barr Virus: From the detection of sequence polymorphisms to the recognition of viral types. Curr. Top. Microbiol. Immunol. 2015, 390, 119–148. [Google Scholar]
- Tsai, M.-H.; Lin, X.; Shumilov, A.; Bernhardt, K.; Feederle, R.; Poirey, R.; Kopp-Schneider, A.; Pereira, B.; Almeida, R.; Delecluse, H.-J. The biological properties of different Epstein-Barr virus strains explain their association with various types of cancers. Oncotarget 2017, 8, 10238–10254. [Google Scholar] [CrossRef]
- Kanda, T.; Yajima, M.; Ikuta, K. Epstein-Barr virus strain variation and cancer. Cancer Sci. 2019, 110, 1132–1139. [Google Scholar] [CrossRef]
- Shair, K.H.Y.; Reddy, A.; Cooper, V.S. New insights from elucidating the role of LMP1 in nasopharyngeal carcinoma. Cancers 2018, 10, 86. [Google Scholar] [CrossRef]
- Edwards, R.H.; Seillier-Moiseiwitsch, F.; Raab-Traub, N. Signature amino acid changes in latent membrane protein 1 distinguish Epstein-Barr virus strains. Virology 1999, 261, 79–95. [Google Scholar] [CrossRef]
- Xu, M.; Yao, Y.; Chen, H.; Zhang, S.; Cao, S.M.; Zhang, Z.; Luo, B.; Liu, Z.; Li, Z.; Xiang, T.; et al. Genome sequencing analysis identifies Epstein–Barr virus subtypes associated with high risk of nasopharyngeal carcinoma. Nat. Genet. 2019, 51, 1131–1136. [Google Scholar] [CrossRef]
- Kim, H.; Burassakarn, A.; Kang, Y.; Iizasa, H.; Yoshiyama, H. A single nucleotide polymorphism in the BART promoter region of Epstein-Barr virus isolated from nasopharyngeal cancer cells. Biochem. Biophys. Res. Commun. 2019, 520, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, X.; Chao, Y.; Jia, Y.; Xing, X.; Luo, B. New variations of Epstein-Barr virus-encoded small RNA genes in nasopharyngeal carcinomas, gastric carcinomas, and healthy donors in northern China. J. Med. Virol. 2010, 82, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Correia, S.; Bridges, R.; Wegner, F.; Venturini, C.; Palser, A.; Middeldorp, J.M.; Cohen, J.I.; Lorenzetti, M.A.; Bassano, I.; White, R.E.; et al. Sequence variation of Epstein-Barr Virus: Viral types, geography, codon usage, and diseases. J. Virol. 2018, 92, e01132-18. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.-S.; Li, D.-J.; Liu, Q.-L.; Song, L.-B.; Li, M.-Z.; Zhang, R.-H.; Yu, X.-J.; Wang, H.-M.; Ernberg, I.; Zeng, Y.-X. Genomic sequence analysis of Epstein-Barr virus strain GD1 from a nasopharyngeal carcinoma patient. J. Virol. 2005, 79, 15323–15330. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Fang, X.; Feng, Z.; Guo, Y.-M.; Peng, R.-J.; Liu, T.; Huang, Z.; Feng, Y.; Sun, X.; Xiong, Z.; et al. Direct sequencing and characterization of a clinical isolate of Epstein-Barr Virus from nasopharyngeal carcinoma tissue by using next-generation sequencing technology. J. Virol. 2011, 85, 11291–11299. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.-H.; Raykova, A.; Klinke, O.; Bernhardt, K.; Gärtner, K.; Leung, C.S.; Geletneky, K.; Sertel, S.; Münz, C.; Feederle, R.; et al. Spontaneous lytic replication and epitheliotropism define an Epstein-Barr virus strain found in carcinomas. Cell Rep. 2013, 5, 458–470. [Google Scholar] [CrossRef] [PubMed]
- Kwok, H.; Tong, A.H.Y.; Lin, C.H.; Lok, S.; Farrell, P.J.; Kwong, D.L.W.; Chiang, A.K.S. Genomic sequencing and comparative analysis of Epstein-Barr virus genome isolated from primary nasopharyngeal carcinoma biopsy. PLoS ONE 2012, 7, e36939. [Google Scholar] [CrossRef] [PubMed]
- Tso, K.K.-Y.; Yip, K.Y.-L.; Mak, C.K.-Y.; Chung, G.T.-Y.; Lee, S.-D.; Cheung, S.-T.; To, K.-F.; Lo, K.-W. Complete genomic sequence of Epstein-Barr virus in nasopharyngeal carcinoma cell line C666-1. Infect. Agent. Cancer 2013, 8, 29. [Google Scholar] [CrossRef]
- Arrand, J.R.; Young, L.S.; Tugwood, J.D. Two families of sequences in the small RNA-encoding region of Epstein-Barr virus (EBV) correlate with EBV types A and B. J. Virol. 1989, 63, 983–986. [Google Scholar] [CrossRef]
- Zheng, C.X.; Yan, L.; Nilsson, B.; Eklund, G.; Drettner, B. Epstein-Barr virus infection, salted fish and nasopharyngeal carcinoma: A case-control study in southern. Acta Oncol. 1994, 33, 867–872. [Google Scholar] [CrossRef]
- Zeng, M.-S.; Zeng, Y.-X. Pathogenesis and etiology of nasopharyngeal carcinoma. In Nasopharyngeal Cancer; Springer Nature: London, UK, 2010; pp. 9–25. [Google Scholar]
- Nishikawa, J.; Yoshiyama, H.; Iizasa, H.; Kanehiro, Y.; Nakamura, M.; Nishimura, J.; Saito, M.; Okamoto, T.; Sakai, K.; Suehiro, Y.; et al. Epstein-barr virus in gastric carcinoma. Cancers 2014, 6, 2259–2274. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.M.; Yu, K.J.; Mbulaiteye, S.M.; Hildesheim, A.; Bhatia, K. The extent of genetic diversity of Epstein-Barr virus and its geographic and disease patterns: A need for reappraisal. Virus Res. 2009, 143, 209–221. [Google Scholar] [CrossRef] [PubMed]
- zur Hausen, H.; Schulte-Holthausen, H.; Klein, G.; Henle, W.; Henle, G.; Clifford, P.; Santesson, L. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature 1970, 228, 1056–1058. [Google Scholar] [CrossRef] [PubMed]
- Thorley-Lawson, D.A.; Hawkins, J.B.; Tracy, S.I.; Shapiro, M. The pathogenesis of Epstein–Barr virus persistent infection. Curr. Opin. Virol. 2013, 3, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Hutt-Fletcher, L.M. Epstein-Barr Virus entry. J. Virol. 2007, 81, 7825–7832. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Gu, X.; Nathan, C.A.; Hutt-Fletcher, L. Laser-capture microdissection of oropharyngeal epithelium indicates restriction of Epstein-Barr virus receptor/CD21 mRNA to tonsil epithelial cells. J. Oral Pathol. Med. 2008, 37, 626–633. [Google Scholar] [CrossRef]
- Jiang, R.; Gu, X.; Moore-Medlin, T.N.; Nathan, C.-A.; Hutt-Fletcher, L.M. Oral dysplasia and squamous cell carcinoma: Correlation between increased expression of CD21, Epstein-Barr virus and CK19. Oral Oncol. 2012, 48, 836–841. [Google Scholar] [CrossRef][Green Version]
- Wang, H.-B.; Zhang, H.; Zhang, J.-P.; Li, Y.; Zhao, B.; Feng, G.-K.; Du, Y.; Xiong, D.; Zhong, Q.; Liu, W.-L.; et al. Neuropilin 1 is an entry factor that promotes EBV infection of nasopharyngeal epithelial cells. Nat. Commun. 2015, 6, 6240. [Google Scholar] [CrossRef]
- Xiong, D.; Du, Y.; Wang, H.-B.; Zhao, B.; Zhang, H.; Li, Y.; Hu, L.-J.; Cao, J.-Y.; Zhong, Q.; Liu, W.-L.; et al. Nonmuscle myosin heavy chain IIA mediates Epstein–Barr virus infection of nasopharyngeal epithelial cells. Proc. Natl. Acad. Sci. USA 2015, 112, 11036–11041. [Google Scholar] [CrossRef]
- Chen, J.; Sathiyamoorthy, K.; Zhang, X.; Schaller, S.; Perez White, B.E.; Jardetzky, T.S.; Longnecker, R. Ephrin receptor A2 is a functional entry receptor for Epstein–Barr virus. Nat. Microbiol. 2018, 3, 172–180. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Wang, H.-B.; Zhang, A.; Chen, M.-L.; Fang, Z.-X.; Dong, X.-D.; Li, S.-B.; Du, Y.; Xiong, D.; et al. Ephrin receptor A2 is an epithelial cell receptor for Epstein–Barr virus entry. Nat. Microbiol. 2018, 3, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Temple, R.M.; Zhu, J.; Budgeon, L.; Christensen, N.D.; Meyers, C.; Sample, C.E. Efficient replication of Epstein-Barr virus in stratified epithelium in vitro. Proc. Natl. Acad. Sci. USA 2014, 111, 16544–16549. [Google Scholar] [CrossRef] [PubMed]
- Eichelberg, M.R.; Welch, R.; Guidry, J.T.; Ali, A.; Ohashi, M.; Makielski, K.R.; McChesney, K.; Van Sciver, N.; Lambert, P.F.; Keleș, S.; et al. Epstein-Barr Virus infection promotes epithelial cell growth by attenuating differentiation-dependent exit from the cell cycle. mBio 2019, 10, e01332-19. [Google Scholar] [CrossRef]
- Dittmer, D.P.; Hilscher, C.J.; Gulley, M.L.; Yang, E.V.; Chen, M.; Glaser, R. Multiple pathways for Epstein-Barr virus episome loss from nasopharyngeal carcinoma. Int. J. Cancer 2008, 123, 2105–2112. [Google Scholar] [CrossRef]
- Lin, W.; Yip, Y.L.; Jia, L.; Deng, W.; Zheng, H.; Dai, W.; Ko, J.M.Y.; Lo, K.W.; Chung, G.T.Y.; Yip, K.Y.; et al. Establishment and characterization of new tumor xenografts and cancer cell lines from EBV-positive nasopharyngeal carcinoma. Nat. Commun. 2018, 9, 4663. [Google Scholar] [CrossRef] [PubMed]
- Yip, Y.L.; Li, W.; Deng, W.; Jia, L.; Lo, K.W.; Busson, P.; Vérillaud, B.; Liu, X.; Tsang, C.M.; Lung, M.L.; et al. Establishment of a nasopharyngeal carcinoma cell line capable of undergoing lytic Epstein–Barr virus reactivation. Lab. Investig. 2018, 98, 1093–1104. [Google Scholar] [CrossRef]
- Dyson, P.J.; Farrell, P.J. Chromatin structure of Epstein-Barr Virus. J. Gen. Virol. 1985, 66, 1931–1940. [Google Scholar] [CrossRef]
- Teo, M.C.C.; Soo, K.C. Cancer trends and incidences in Singapore. Jpn. J. Clin. Oncol. 2013, 43, 219–224. [Google Scholar] [CrossRef]
- Raab-Traub, N.; Flynn, K. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell 1986, 47, 883–889. [Google Scholar] [CrossRef]
- Lung, R.W.M.; Tong, J.H.M.; To, K.F. Emerging roles of small Epstein-Barr virus derived non-coding RNAs in epithelial malignancy. Int. J. Mol. Sci. 2013, 14, 17378–17409. [Google Scholar] [CrossRef]
- Canaan, A.; Haviv, I.; Urban, A.E.; Schulz, V.P.; Hartman, S.; Zhang, Z.; Palejev, D.; Deisseroth, A.B.; Lacy, J.; Snyder, M.; et al. EBNA1 regulates cellular gene expression by binding cellular promoters. Proc. Natl. Acad. Sci. USA 2009, 106, 22421–22426. [Google Scholar] [CrossRef] [PubMed]
- Dresang, L.R.; Vereide, D.T.; Sugden, B. Identifying sites bound by Epstein-Barr Virus Nuclear Antigen 1 (EBNA1) in the human genome: Defining a position-weighted matrix to predict sites bound by EBNA1 in viral genomes. J. Virol. 2009, 83, 2930–2940. [Google Scholar] [CrossRef] [PubMed]
- Sugden, B.; Warren, N. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J. Virol. 1989, 63, 2644–2649. [Google Scholar] [CrossRef]
- Allen, M.D.; Young, L.S.; Dawson, C.W. The Epstein-Barr Virus-encoded LMP2A and LMP2B proteins promote epithelial cell spreading and motility. J. Virol. 2005, 79, 1789–1802. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, Z.; Wan, X.; Jiang, R.; Deng, L.; Gao, Y.; Tang, J.; Yang, Y.; Zhao, W.; Yan, X.; Yao, K.; et al. Epstein-Barr Virus-encoded latent membrane protein 2A promotes the epithelial-mesenchymal transition in nasopharyngeal carcinoma via metastatic tumor antigen 1 and mechanistic target of rapamycin signaling induction. J. Virol. 2014, 88, 11872–11885. [Google Scholar] [CrossRef]
- Hoebe, E.K.; Le Large, T.Y.S.; Greijer, A.E.; Middeldorp, J.M. BamHI-A rightward frame 1, an Epstein-Barr virus-encoded oncogene and immune modulator. Rev. Med. Virol. 2013, 23, 367–383. [Google Scholar] [CrossRef] [PubMed]
- Hoebe, E.; Wille, C.; Hagemeier, S.; Kenney, S.; Greijer, A.; Middeldorp, J. Epstein-Barr Virus gene BARF1 expression is regulated by the epithelial differentiation factor ΔNp63α in undifferentiated nasopharyngeal carcinoma. Cancers 2018, 10, 76. [Google Scholar] [CrossRef]
- Eliopoulos, A.G.; Stack, M.; Dawson, C.W.; Kaye, K.M.; Hodgkin, L.; Sihota, S.; Rowe, M.; Young, L.S. Epstein-Barr virus-encoded LMP1 and CD40 mediate IL-6 production in epithelial cells via an NF-κB pathway involving TNF receptor-associated factors. Oncogene 1997, 14, 2899–2916. [Google Scholar] [CrossRef]
- Lo, A.K.F.; Huang, D.P.; Lo, K.W.; Chui, Y.L.; Li, H.M.; Pang, J.C.S.; Tsao, S.W. Phenotypic alterations induced by the Hong Kong-prevalent Epstein-Barr virus-encoded LMP1 variant (2117-LMP1) in nasopharyngeal epithelial cells. Int. J. Cancer 2004, 109, 919–925. [Google Scholar] [CrossRef]
- Nawandar, D.M.; Ohashi, M.; Djavadian, R.; Barlow, E.; Makielski, K.; Ali, A.; Lee, D.; Lambert, P.F.; Johannsen, E.; Kenney, S.C. Differentiation-dependent LMP1 expression is required for efficient lytic Epstein-Barr Virus reactivation in epithelial cells. J. Virol. 2017, 91, 1–18. [Google Scholar] [CrossRef]
- Caves, E.A.; Butch, R.M.; Cook, S.A.; Wasil, L.R.; Chen, C.; Di, Y.P.; Lee, N.; Shair, K.H.Y. Latent membrane protein 1 is a novel determinant of Epstein-Barr Virus genome persistence and reactivation. mSphere 2017, 2, e00453-17. [Google Scholar] [CrossRef]
- Liu, M.-T.; Chen, Y.-R.; Chen, S.-C.; Hu, C.-Y.; Lin, C.-S.; Chang, Y.-T.; Wang, W.-B.; Chen, J.-Y. Epstein-Barr virus latent membrane protein 1 induces micronucleus formation, represses DNA repair and enhances sensitivity to DNA-damaging agents in human epithelial cells. Oncogene 2004, 23, 2531–2539. [Google Scholar] [CrossRef]
- Mei, Y.-P.; Zhou, J.-M.; Wang, Y.; Huang, H.; Deng, R.; Feng, G.-K.; Zeng, Y.-X.; Zhu, X.-F. Silencing of LMP1 induces cell cycle arrest and enhances chemosensitivity through inhibition of AKT signaling pathway in EBV-positive nasopharyngeal carcinoma cells. Cell Cycle 2007, 6, 1379–1385. [Google Scholar] [CrossRef]
- Skalsky, R.L.; Cullen, B.R. EBV Noncoding RNAs. Curr. Top. Microbiol. Immunol. 2015, 391, 181–217. [Google Scholar]
- Fan, C.; Tang, Y.; Wang, J.; Xiong, F.; Guo, C.; Wang, Y.; Xiang, B.; Zhou, M.; Li, X.; Wu, X.; et al. The emerging role of Epstein-Barr virus encoded microRNAs in nasopharyngeal carcinoma. J. Cancer 2018, 9, 2852–2864. [Google Scholar] [CrossRef]
- Spence, T.; Bruce, J.; Yip, K.W.; Liu, F.-F. MicroRNAs in nasopharyngeal carcinoma. Chin. Clin. Oncol. 2016, 5, 17. [Google Scholar] [CrossRef]
- Young, L.S.; Dawson, C.W. Epstein-Barr virus and nasopharyngeal carcinoma. Chin. J. Cancer 2014, 33, 581–590. [Google Scholar] [CrossRef]
- Tsang, C.M.; Yip, Y.L.; Lo, K.W.; Deng, W.; To, K.F.; Hau, P.M.; Lau, V.M.Y.; Takada, K.; Lui, V.W.Y.; Lung, M.L.; et al. Cyclin D1 overexpression supports stable EBV infection in nasopharyngeal epithelial cells. Proc. Natl. Acad. Sci. USA 2012, 109, E3473–E3482. [Google Scholar] [CrossRef]
- Hui, A.B.Y.; Or, Y.Y.Y.; Takano, H.; Tsang, R.K.Y.; To, K.F.; Guan, X.Y.; Sham, J.S.T.; Hung, K.W.K.; Lam, C.N.Y.; Van Hasselt, C.A.; et al. Array-based comparative genomic hybridization analysis identified cyclin D1 as a target oncogene at 11q13.3 in nasopharyngeal carcinoma. Cancer Res. 2005, 65, 8125–8133. [Google Scholar] [CrossRef]
- Song, L.B.; Zeng, M.S.; Liao, W.T.; Zhang, L.; Mo, H.Y.; Liu, W.L.; Shao, J.Y.; Wu, Q.L.; Li, M.Z.; Xia, Y.F.; et al. Bmi-1 is a novel molecular marker of nasopharyngeal carcinoma progression and immortalizes primary human nasopharyngeal epithelial cells. Cancer Res. 2006, 66, 6225–6232. [Google Scholar] [CrossRef]
- Yip, Y.L.; Pang, P.S.; Deng, W.; Tsang, C.M.; Zeng, M.; Hau, P.M.; Man, C.; Jin, Y.; Yuen, A.P.W.; Tsao, S.W. Efficient immortalization of primary nasopharyngeal epithelial cells for EBV infection study. PLoS ONE 2013, 8, 1–13. [Google Scholar] [CrossRef][Green Version]
- Murata, T. Regulation of Epstein-Barr virus reactivation from latency. Microbiol. Immunol. 2014, 58, 307–317. [Google Scholar] [CrossRef]
- Hitt, M.M.; Allday, M.J.; Hara, T.; Karran, L.; Jones, M.D.; Busson, P.; Tursz, T.; Ernberg, I.; Griffin, B.E. EBV gene expression in an NPC-related tumour. EMBO J. 1989, 8, 2639–2651. [Google Scholar] [CrossRef]
- Coghill, A.E.; Hsu, W.-L.; Pfeiffer, R.M.; Juwana, H.; Yu, K.J.; Lou, P.-J.; Wang, C.-P.; Chen, J.-Y.; Chen, C.-J.; Middeldorp, J.M.; et al. Epstein-Barr virus serology as a potential screening marker for nasopharyngeal carcinoma among high-risk individuals from multiplex families in Taiwan. Cancer Epidemiol. Biomark. Prev. 2014, 23, 1213–1219. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Q.; Khin, N.S.; Chua, M.L.K. The evolution of Epstein-Barr virus detection in nasopharyngeal carcinoma. Cancer Biol. Med. 2018, 15, 1–5. [Google Scholar]
- Hsu, M.; Wu, S.-Y.; Chang, S.-S.; Su, I.-J.; Tsai, C.-H.; Lai, S.-J.; Shiau, A.-L.; Takada, K.; Chang, Y. Epstein-Barr virus lytic transactivator Zta enhances chemotactic activity through induction of interleukin-8 in nasopharyngeal carcinoma cells. J. Virol. 2008, 82, 3679–3688. [Google Scholar] [CrossRef]
- Pudney, V.A.; Leese, A.M.; Rickinson, A.B.; Hislop, A.D. CD8+ immunodominance among Epstein-Barr virus lytic cycle antigens directly reflects the efficiency of antigen presentation in lytically infected cells. J. Exp. Med. 2005, 201, 349–360. [Google Scholar] [CrossRef]
- Chang, Y.-H.; Lee, C.-P.; Su, M.-T.; Wang, J.-T.; Chen, J.-Y.; Lin, S.-F.; Tsai, C.-H.; Hsieh, M.-J.; Takada, K.; Chen, M.-R. Epstein-Barr virus BGLF4 kinase retards cellular S-phase progression and induces chromosomal abnormality. PLoS ONE 2012, 7, e39217. [Google Scholar] [CrossRef]
- Wu, C.-C.; Liu, M.-T.; Chang, Y.-T.; Fang, C.-Y.; Chou, S.-P.; Liao, H.-W.; Kuo, K.-L.; Hsu, S.-L.; Chen, Y.-R.; Wang, P.-W.; et al. Epstein-Barr virus DNase (BGLF5) induces genomic instability in human epithelial cells. Nucleic Acids Res. 2010, 38, 1932–1949. [Google Scholar] [CrossRef]
- Chiu, S.-H.; Wu, C.-C.; Fang, C.-Y.; Yu, S.-L.; Hsu, H.-Y.; Chow, Y.-H.; Chen, J.-Y. Epstein-Barr virus BALF3 mediates genomic instability and progressive malignancy in nasopharyngeal carcinoma. Oncotarget 2014, 5, 8583–8601. [Google Scholar] [CrossRef]
- Takada, K. Role of EBER and BARF1 in nasopharyngeal carcinoma (NPC) tumorigenesis. Semin. Cancer Biol. 2012, 22, 162–165. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Su, Q.; Liu, H.; Wang, D.; Zhang, W.; Lu, Z.; Chen, Y.; Huang, X.; Li, W.; Zhang, C.; et al. Frizzled7 Promotes Epithelial-to-mesenchymal transition and stemness via activating canonical Wnt/β-catenin pathway in gastric cancer. Int. J. Biol. Sci. 2018, 14, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhu, H.; Gao, Z.; Li, J.; Zhuang, J.; Dong, Y.; Shen, B.; Li, M.; Zhou, H.; Guo, H.; et al. Wnt7a activates canonical Wnt signaling, promotes bladder cancer cell invasion, and is suppressed by miR-370-3p. J. Biol. Chem. 2018, 293, 6693–6706. [Google Scholar] [CrossRef]
- Stewart, D.J. Wnt signaling pathway in non-small cell lung cancer. J. Natl. Cancer Inst. 2014, 106, djt356. [Google Scholar] [CrossRef]
- Jiang, R.; Niu, X.; Huang, Y.; Wang, X. β-Catenin is important for cancer stem cell generation and tumorigenic activity in nasopharyngeal carcinoma. Acta Biochim. Biophys. Sin. 2016, 48, 229–237. [Google Scholar] [CrossRef]
- Webb, N.; Connolly, G.; Tellam, J.; Yap, A.S.; Khanna, R. Epstein-Barr virus associated modulation of Wnt pathway is not dependent on latent membrane protein-1. PLoS ONE 2008, 3, e3254. [Google Scholar] [CrossRef]
- Yap, L.F.; Ahmad, M.; Zabidi, M.M.A.; Chu, T.L.; Chai, S.J.; Lee, H.M.; Lim, P.V.H.; Wei, W.; Dawson, C.; Teo, S.-H.; et al. Oncogenic effects of WNT5A in Epstein-Barr virus-associated nasopharyngeal carcinoma. Int. J. Oncol. 2014, 44, 1774–1780. [Google Scholar] [CrossRef]
- Chen, H.; Lee, J.M.; Zong, Y.; Borowitz, M.; Ng, M.H.; Ambinder, R.F.; Hayward, S.D. Linkage between STAT regulation and Epstein-Barr virus gene expression in tumors. J. Virol. 2001, 75, 2929–2937. [Google Scholar] [CrossRef]
- Chen, H.; Lee, J.M.; Wang, Y.; Huang, D.P.; Ambinder, R.F.; Hayward, S.D. The Epstein-Barr virus latency BamHI-Q promoter is positively regulated by STATs and Zta interference with JAK/STAT activation leads to loss of BamHI-Q promoter activity. Proc. Natl. Acad. Sci. USA 1999, 96, 9339–9344. [Google Scholar] [CrossRef]
- Liu, Y.-P.; Tan, Y.-N.; Wang, Z.-L.; Zeng, L.; Lu, Z.-X.; Li, L.-L.; Luo, W.; Tang, M.; Cao, Y. Phosphorylation and nuclear translocation of STAT3 regulated by the Epstein-Barr virus latent membrane protein 1 in nasopharyngeal carcinoma. Int. J. Mol. Med. 2008, 21, 153–162. [Google Scholar] [CrossRef][Green Version]
- Kung, C.-P.; Raab-Traub, N. Epstein-Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor through effects on Bcl-3 and STAT3. J. Virol. 2008, 82, 5486–5493. [Google Scholar] [CrossRef]
- Kung, C.-P.; Meckes, D.G.; Raab-Traub, N. Epstein-Barr virus LMP1 activates EGFR, STAT3, and ERK through effects on PKCdelta. J. Virol. 2011, 85, 4399–4408. [Google Scholar] [CrossRef]
- Xu, Y.; Shi, Y.; Yuan, Q.; Liu, X.; Yan, B.; Chen, L.; Tao, Y.; Cao, Y. Epstein-Barr Virus encoded LMP1 regulates cyclin D1 promoter activity by nuclear EGFR and STAT3 in CNE1 cells. J. Exp. Clin. Cancer Res. 2013, 32, 90. [Google Scholar] [CrossRef]
- Wang, Z.; Luo, F.; Li, L.; Yang, L.; Hu, D.; Ma, X.; Lu, Z.; Sun, L.; Cao, Y. STAT3 activation induced by Epstein-Barr virus latent membrane protein1 causes vascular endothelial growth factor expression and cellular invasiveness via JAK3 And ERK signaling. Eur. J. Cancer 2010, 46, 2996–3006. [Google Scholar] [CrossRef]
- Cheng, J.-Z.; Chen, J.-J.; Xue, K.; Wang, Z.-G.; Yu, D. Clinicopathologic and prognostic significance of VEGF, JAK2 and STAT3 in patients with nasopharyngeal carcinoma. Cancer Cell Int. 2018, 18, 110. [Google Scholar] [CrossRef]
- Chen, J. Roles of the PI3K/Akt pathway in Epstein-Barr virus-induced cancers and therapeutic implications. World J. Virol. 2012, 1, 154–161. [Google Scholar] [CrossRef]
- Yang, C.-F.; Yang, G.-D.; Huang, T.-J.; Li, R.; Chu, Q.-Q.; Xu, L.; Wang, M.-S.; Cai, M.-D.; Zhong, L.; Wei, H.-J.; et al. EB-virus latent membrane protein 1 potentiates the stemness of nasopharyngeal carcinoma via preferential activation of PI3K/AKT pathway by a positive feedback loop. Oncogene 2016, 35, 3419–3431. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, J.; Yu, D.; Liu, Y.; Xue, K.; Zhao, X. Role of Epstein-Barr Virus in the Development of Nasopharyngeal Carcinoma. Open Med. 2017, 12, 171–176. [Google Scholar] [CrossRef]
- Jiang, H.; Fan, D.; Zhou, G.; Li, X.; Deng, H. Phosphatidylinositol 3-kinase inhibitor(LY294002) induces apoptosis of human nasopharyngeal carcinoma in vitro and in vivo. J. Exp. Clin. Cancer Res. 2010, 29, 34. [Google Scholar] [CrossRef]
- Chen, J.; Hu, C.-F.; Hou, J.-H.; Shao, Q.; Yan, L.-X.; Zhu, X.-F.; Zeng, Y.-X.; Shao, J.-Y. Epstein-Barr virus encoded latent membrane protein 1 regulates mTOR signaling pathway genes which predict poor prognosis of nasopharyngeal carcinoma. J. Transl. Med. 2010, 8, 30. [Google Scholar] [CrossRef]
- Wang, W.; Wen, Q.; Xu, L.; Xie, G.; Li, J.; Luo, J.; Chu, S.; Shi, L.; Huang, D.; Li, J.; et al. Activation of Akt/mTOR pathway is associated with poor prognosis of nasopharyngeal carcinoma. PLoS ONE 2014, 9, e106098. [Google Scholar] [CrossRef]
- Wang, M.-H.; Sun, R.; Zhou, X.-M.; Zhang, M.-Y.; Lu, J.-B.; Yang, Y.; Zeng, L.-S.; Yang, X.-Z.; Shi, L.; Xiao, R.-W.; et al. Epithelial cell adhesion molecule overexpression regulates epithelial-mesenchymal transition, stemness and metastasis of nasopharyngeal carcinoma cells via the PTEN/AKT/mTOR pathway. Cell Death Dis. 2018, 9, 2. [Google Scholar] [CrossRef]
- Yang, Z.; Lan, H.; Chen, X.; Li, P.; Li, S.; Mo, W.; Tang, A. Molecular alterations of the WWOX gene in nasopharyngeal carcinoma. Neoplasma 2014, 61, 170–176. [Google Scholar] [CrossRef][Green Version]
- Zhang, Z.; Yu, X.; Zhou, Z.; Li, B.; Peng, J.; Wu, X.; Luo, X.; Yang, L. LMP1-positive extracellular vesicles promote radioresistance in nasopharyngeal carcinoma cells through P38 MAPK signaling. Cancer Med. 2019, 8, 6082–6094. [Google Scholar] [CrossRef]
- Ma, B.B.Y.; Poon, T.C.W.; To, K.F.; Zee, B.; Mo, F.K.F.; Chan, C.M.L.; Ho, S.; Teo, P.M.L.; Johnson, P.J.; Chan, A.T.C. Prognostic significance of tumor angiogenesis, Ki 67, p53 oncoprotein, epidermal growth factor receptor and HER2 receptor protein expression in undifferentiated nasopharyngeal carcinoma—A prospective study. Head Neck 2003, 25, 864–872. [Google Scholar] [CrossRef]
- Thornburg, N.J.; Raab-Traub, N. Induction of epidermal growth factor receptor expression by Epstein-Barr virus latent membrane protein 1 C-terminal-activating region 1 is mediated by NF-κB p50 homodimer/Bcl-3 complexes. J. Virol. 2007, 81, 12954–12961. [Google Scholar] [CrossRef]
- Tao, Y.; Song, X.; Tan, Y.; Lin, X.; Zhao, Y.; Zeng, L.; Tang, M.; Li, W.; Wu, Q.; Cao, Y. Nuclear translocation of EGF receptor regulated by Epstein-Barr virus encoded latent membrane protein 1. Sci. China. Ser. C Life Sci. 2004, 47, 258–267. [Google Scholar] [CrossRef]
- Dawson, C.W.; Laverick, L.; Morris, M.A.; Tramoutanis, G.; Young, L.S. Epstein-Barr virus-encoded LMP1 regulates epithelial cell motility and invasion via the ERK-MAPK pathway. J. Virol. 2008, 82, 3654–3664. [Google Scholar] [CrossRef]
- Sung, W.-W.; Chu, Y.-C.; Chen, P.-R.; Liao, M.-H.; Lee, J.-W. Positive regulation of HIF-1A expression by EBV oncoprotein LMP1 in nasopharyngeal carcinoma cells. Cancer Lett. 2016, 382, 21–31. [Google Scholar] [CrossRef]
- Liang, J.; Zheng, S.; Xiao, X.; Wei, J.; Zhang, Z.; Ernberg, I.; Matskova, L.; Huang, G.; Zhou, X. Epstein-Barr virus-encoded LMP2A stimulates migration of nasopharyngeal carcinoma cells via the EGFR/Ca2+/calpain/ITGβ4 axis. Biol. Open 2017, 6, 914–922. [Google Scholar] [CrossRef]
- Zheng, H.; Li, L.L.; Hu, D.S.; Deng, X.Y.; Cao, Y. Role of Epstein-Barr virus encoded latent membrane protein 1 in the carcinogenesis of nasopharyngeal carcinoma. Cell. Mol. Immunol. 2007, 4, 185–196. [Google Scholar] [PubMed]
- Chung, G.T.Y.; Lou, W.P.K.; Chow, C.; To, K.F.; Choy, K.W.; Leung, A.W.C.; Tong, C.Y.K.; Yuen, J.W.F.; Ko, C.W.; Yip, T.T.C.; et al. Constitutive activation of distinct NF-κB signals in EBV-associated nasopharyngeal carcinoma. J. Pathol. 2013, 231, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Dai, W.; Cheung, A.K.L.; Ko, J.M.Y.; Kan, R.; Wong, B.W.Y.; Leong, M.M.L.; Deng, M.; Kwok, T.C.T.; Chan, J.Y.-W.; et al. Whole-exome sequencing identifies multiple loss-of-function mutations of NF-κB pathway regulators in nasopharyngeal carcinoma. Proc. Natl. Acad. Sci. USA 2016, 113, 11283–11288. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Dai, W.; Yu, V.Z.; Tao, L.; Lung, M.L. Cylindromatosis Lysine 63 Deubiquitinase (CYLD) regulates NF-kB signaling pathway and modulates fibroblast and endothelial cells recruitment in nasopharyngeal carcinoma. Cancers 2020, 12, 1924. [Google Scholar] [CrossRef]
- Ersing, I.; Bernhardt, K.; Gewurz, B.E. NF-κB and IRF7 pathway activation by Epstein-Barr virus Latent Membrane Protein 1. Viruses 2013, 5, 1587–1606. [Google Scholar] [CrossRef]
- Wang, L.W.; Jiang, S.; Gewurz, B.E. Epstein-Barr Virus LMP1-mediated oncogenicity. J. Virol. 2017, 91, e01718-16. [Google Scholar] [CrossRef]
- Lo, A.K.F.; Lo, K.W.; Tsao, S.W.; Wong, H.L.; Hui, J.W.Y.; To, K.F.; Hayward, D.S.; Chui, Y.L.; Lau, Y.L.; Takada, K.; et al. Epstein-Barr virus infection alters cellular signal cascades in human nasopharyngeal epithelial cells. Neoplasia 2006, 8, 173–180. [Google Scholar]
- Zhang, J.; Jia, L.; Lin, W.; Yip, Y.L.; Lo, K.W.; Lau, V.M.Y.; Zhu, D.; Tsang, C.M.; Zhou, Y.; Deng, W.; et al. Epstein-Barr Virus-encoded latent membrane protein 1 upregulates glucose transporter 1 transcription via the mTORC1/NF-κB signaling pathways. J. Virol. 2017, 91, e02168-16. [Google Scholar] [CrossRef]
- Johansson, P.; Jansson, A.; Rüetschi, U.; Rymo, L. Nuclear factor-kappaB binds to the Epstein-Barr Virus LMP1 promoter and upregulates its expression. J. Virol. 2009, 83, 1393–1401. [Google Scholar] [CrossRef]
- Devergne, O.; Hatzivassiliou, E.; Izumi, K.M.; Kaye, K.M.; Kleijnen, M.F.; Kieff, E.; Mosialos, G. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: Role in NF-κB activation. Mol. Cell. Biol. 1996, 16, 7098–7108. [Google Scholar] [CrossRef]
- Thornburg, N.J.; Pathmanathan, R.; Raab-Traub, N. Activation of nuclear factor-κB p50 homodimer/Bcl-3 complexes in nasopharyngeal carcinoma. Cancer Res. 2003, 63, 8293–8301. [Google Scholar] [PubMed]
- Luftig, M.; Yasui, T.; Soni, V.; Kang, M.-S.; Jacobson, N.; Cahir-McFarland, E.; Seed, B.; Kieff, E. Epstein-Barr virus latent infection membrane protein 1 TRAF-binding site induces NIK/IKK alpha-dependent noncanonical NF-kappaB activation. Proc. Natl. Acad. Sci. USA 2004, 101, 141–146. [Google Scholar] [CrossRef]
- Verhoeven, R.J.A.; Tong, S.; Zhang, G.; Zong, J.; Chen, Y.; Jin, D.-Y.; Chen, M.-R.; Pan, J.; Chen, H. NF-κB signaling regulates expression of Epstein-Barr Virus BART microRNAs and long noncoding RNAs in nasopharyngeal carcinoma. J. Virol. 2016, 90, 6475–6488. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Chen, Y.; Gong, P.; Cai, L.; Lyu, X.; Jiang, Q.; Wang, J.; Lu, J.; Yao, K.; Liu, K.; et al. Higher methylation intensity induced by EBV LMP1 via NF-κB/DNMT3b signaling contributes to silencing of PTEN gene. Oncotarget 2016, 7, 40025–40037. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, R.J.A.; Tong, S.; Zong, J.; Chen, Y.; Tsao, S.-W.; Pan, J.; Chen, H. NF-κB signaling regulates Epstein-Barr Virus BamHI-Q-driven EBNA1 expression. Cancers 2018, 10, 119. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Fathallah, I.; Parroche, P.; Gruffat, H.; Zannetti, C.; Johansson, H.; Yue, J.; Manet, E.; Tommasino, M.; Sylla, B.S.; Hasan, U.A. EBV latent membrane protein 1 is a negative regulator of TLR9. J. Immunol. 2010, 185, 6439–6447. [Google Scholar] [CrossRef]
- Bentz, G.L.; Shackelford, J.; Pagano, J.S. Epstein-Barr virus latent membrane protein 1 regulates the function of interferon regulatory factor 7 by inducing its sumoylation. J. Virol. 2012, 86, 12251–12261. [Google Scholar] [CrossRef]
- Shah, K.M.; Stewart, S.E.; Wei, W.; Woodman, C.B.J.; O’Neil, J.D.; Dawson, C.W.; Young, L.S. The EBV-encoded latent membrane proteins, LMP2A and LMP2B, limit the actions of interferon by targeting interferon receptors for degradation. Oncogene 2009, 28, 3903–3914. [Google Scholar] [CrossRef]
- Wang, M.; Yu, F.; Wu, W.; Wang, Y.; Ding, H.; Qian, L. Epstein-Barr virus-encoded microRNAs as regulators in host immune responses. Int. J. Biol. Sci. 2018, 14, 565–576. [Google Scholar] [CrossRef]
- Dunn, G.P.; Bruce, A.T.; Ikeda, H.; Old, L.J.; Schreiber, R.D. Cancer immunoediting: From immunosurveillance to tumor escape. Nat. Immunol. 2002, 3, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Qin, Z.; Wang, J.; Zheng, X.; Lu, J.; Zhang, X.; Wei, L.; Peng, Q.; Zheng, Y.; Ou, C.; et al. Epstein-Barr Virus miR-BART6-3p inhibits the RIG-I pathway. J. Innate Immun. 2017, 9, 574–586. [Google Scholar] [CrossRef]
- Hooykaas, M.J.G.; van Gent, M.; Soppe, J.A.; Kruse, E.; Boer, I.G.J.; van Leenen, D.; Groot Koerkamp, M.J.A.; Holstege, F.C.P.; Ressing, M.E.; Wiertz, E.J.H.J.; et al. EBV MicroRNA BART16 suppresses type I IFN signaling. J. Immunol. 2017, 198, 4062–4073. [Google Scholar] [CrossRef] [PubMed]
- Conlon, K.C.; Miljkovic, M.D.; Waldmann, T.A. Cytokines in the treatment of cancer. J. Interferon Cytokine Res. 2019, 39, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Tagawa, T.; Albanese, M.; Bouvet, M.; Moosmann, A.; Mautner, J.; Heissmeyer, V.; Zielinski, C.; Lutter, D.; Hoser, J.; Hastreiter, M.; et al. Epstein-Barr viral miRNAs inhibit antiviral CD4+ T cell responses targeting IL-12 and peptide processing. J. Exp. Med. 2016, 213, 2065–2080. [Google Scholar] [CrossRef]
- Albanese, M.; Tagawa, T.; Bouvet, M.; Maliqi, L.; Lutter, D.; Hoser, J.; Hastreiter, M.; Hayes, M.; Sugden, B.; Martin, L.; et al. Epstein-Barr virus microRNAs reduce immune surveillance by virus-specific CD8+ T cells. Proc. Natl. Acad. Sci. USA 2016, 113, E6467–E6475. [Google Scholar] [CrossRef]
- Nachmani, D.; Stern-Ginossar, N.; Sarid, R.; Mandelboim, O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 2009, 5, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Samanta, M.; Takada, K. Modulation of innate immunity system by Epstein-Barr virus-encoded non-coding RNA and oncogenesis. Cancer Sci. 2010, 101, 29–35. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, Z.; Shu, Y.; Zhou, H.; Wang, H.; Zhang, W. BART miRNAs: An unimaginable force in the development of nasopharyngeal carcinoma. Eur. J. Cancer Prev. 2017, 26, 144–150. [Google Scholar] [CrossRef]
- Ye, Y.; Zhou, Y.; Zhang, L.; Chen, Y.; Lyu, X.; Cai, L.; Lu, Y.; Deng, Y.; Wang, J.; Yao, K.; et al. EBV-miR-BART1 is involved in regulating metabolism-associated genes in nasopharyngeal carcinoma. Biochem. Biophys. Res. Commun. 2013, 436, 19–24. [Google Scholar] [CrossRef]
- Lyu, X.; Wang, J.; Guo, X.; Wu, G.; Jiao, Y.; Faleti, O.D.; Liu, P.; Liu, T.; Long, Y.; Chong, T.; et al. EBV-miR-BART1-5P activates AMPK/mTOR/HIF1 pathway via a PTEN independent manner to promote glycolysis and angiogenesis in nasopharyngeal carcinoma. PLoS Pathog. 2018, 14, e1007484. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zheng, J.; Tang, Y.; Lin, Y.; Wang, L.; Li, Y.; Liu, C.; Wu, D.; Cai, L. EBV encoded miRNA BART8-3p promotes radioresistance in nasopharyngeal carcinoma by regulating ATM/ATR signaling pathway. Biosci. Rep. 2019, 39, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Hu, Z.-Y.; Dong, X.; Tan, Z.; Li, W.; Tang, M.; Chen, L.; Yang, L.; Tao, Y.; Jiang, Y.; et al. Targeting Epstein-Barr virus oncoprotein LMP1-mediated glycolysis sensitizes nasopharyngeal carcinoma to radiation therapy. Oncogene 2014, 33, 4568–4578. [Google Scholar] [CrossRef] [PubMed]
- Daker, M.; Bhuvanendran, S.; Ahmad, M.; Takada, K.; Khoo, A.S.-B. Deregulation of lipid metabolism pathway genes in nasopharyngeal carcinoma cells. Mol. Med. Rep. 2013, 7, 731–741. [Google Scholar] [CrossRef]
- Wong, R.S.Y. Apoptosis in cancer: From pathogenesis to treatment. J. Exp. Clin. Cancer Res. 2011, 30, 87. [Google Scholar] [CrossRef]
- Ozaki, T.; Nakagawara, A. Role of p53 in cell death and human cancers. Cancers 2011, 3, 994–1013. [Google Scholar] [CrossRef]
- Choy, E.Y.-W.; Siu, K.-L.; Kok, K.-H.; Lung, R.W.-M.; Tsang, C.M.; To, K.-F.; Kwong, D.L.-W.; Tsao, S.W.; Jin, D.-Y. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J. Exp. Med. 2008, 205, 2551–2560. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, J.; Wei, L.; Peng, Q.; Gao, Y.; Fu, Y.; Lu, Y.; Qin, Z.; Zhang, X.; Lu, J.; et al. Epstein-Barr Virus microRNA miR-BART5-3p inhibits p53 expression. J. Virol. 2018, 92, e01022-18. [Google Scholar] [CrossRef]
- Li, Z.; Chen, X.; Li, L.; Liu, S.; Yang, L.; Ma, X.; Tang, M.; Bode, A.M.; Dong, Z.; Sun, L.; et al. EBV encoded miR-BHRF1-1 potentiates viral lytic replication by downregulating host p53 in nasopharyngeal carcinoma. Int. J. Biochem. Cell Biol. 2012, 44, 275–279. [Google Scholar] [CrossRef]
- Marquitz, A.R.; Mathur, A.; Nam, C.S.; Raab-Traub, N. The Epstein-Barr Virus BART microRNAs target the pro-apoptotic protein Bim. Virology 2011, 412, 392–400. [Google Scholar] [CrossRef]
- Saridakis, V.; Sheng, Y.; Sarkari, F.; Holowaty, M.N.; Shire, K.; Nguyen, T.; Zhang, R.G.; Liao, J.; Lee, W.; Edwards, A.M.; et al. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol. Cell 2005, 18, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Murakami, M.; Verma, S.C.; Cai, Q.; Haldar, S.; Kaul, R.; Wasik, M.A.; Middeldorp, J.; Robertson, E.S. Epstein-Barr Virus nuclear antigen 1 (EBNA1) confers resistance to apoptosis in EBV-positive B-lymphoma cells through up-regulation of survivin. Virology 2011, 410, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.-D.; Huang, T.-J.; Peng, L.-X.; Yang, C.-F.; Liu, R.-Y.; Huang, H.-B.; Chu, Q.-Q.; Yang, H.-J.; Huang, J.-L.; Zhu, Z.-Y.; et al. Epstein-Barr Virus encoded LMP1 upregulates microRNA-21 to promote the resistance of nasopharyngeal carcinomacells to cisplatin-induced apoptosis by suppressing PDCD4 and Fas-L. PLoS ONE 2013, 8, e78355. [Google Scholar]
- Valastyan, S.; Weinberg, R.A. Tumor metastasis: Molecular insights and evolving paradigms. Cell 2011, 147, 275–292. [Google Scholar] [CrossRef]
- Liao, W.; Tian, M.; Chen, N. Characteristic and novel therapeutic strategies of nasopharyngeal carcinoma with synchronous metastasis. Cancer Manag. Res. 2019, 11, 8431–8442. [Google Scholar] [CrossRef]
- Li, A.C.; Xiao, W.W.; Shen, G.Z.; Wang, L.; Xu, A.A.; Cao, Y.Q.; Huang, S.M.; Lin, C.G.; Han, F.; Deng, X.W.; et al. Distant metastasis risk and patterns of nasopharyngeal carcinoma in the era of IMRT: Long-term results and benefits of chemotherapy. Oncotarget 2015, 6, 24511–24521. [Google Scholar] [CrossRef]
- Yip, T.T.C.; Ngan, R.K.C.; Fong, A.H.W.; Law, S.C.K. Application of circulating plasma/serum EBV DNA in the clinical management of nasopharyngeal carcinoma. Oral Oncol. 2014, 50, 527–538. [Google Scholar] [CrossRef]
- Lan, Y.-Y.; Hsiao, J.-R.; Chang, K.-C.; Chang, J.S.-M.; Chen, C.-W.; Lai, H.-C.; Wu, S.-Y.; Yeh, T.-H.; Chang, F.-H.; Lin, W.-H.; et al. Epstein-Barr Virus latent membrane protein 2A promotes invasion of nasopharyngeal carcinoma cells through ERK/Fra-1-mediated induction of matrix metalloproteinase 9. J. Virol. 2012, 86, 6656–6667. [Google Scholar] [CrossRef]
- Port, R.J.; Pinheiro-Maia, S.; Hu, C.; Arrand, J.R.; Wei, W.; Young, L.S.; Dawson, C.W. Epstein-Barr virus induction of the Hedgehog signalling pathway imposes a stem cell phenotype on human epithelial cells. J. Pathol. 2013, 231, 367–377. [Google Scholar] [CrossRef]
- Cai, L.-M.; Lyu, X.-M.; Luo, W.-R.; Cui, X.-F.; Ye, Y.-F.; Yuan, C.-C.; Peng, Q.-X.; Wu, D.-H.; Liu, T.-F.; Wang, E.; et al. EBV-miR-BART7-3p promotes the EMT and metastasis of nasopharyngeal carcinoma cells by suppressing the tumor suppressor PTEN. Oncogene 2015, 34, 2156–2166. [Google Scholar] [CrossRef]
- Wong, A.M.G.; Kong, K.L.; Tsang, J.W.H.; Kwong, D.L.W.; Guan, X.-Y. Profiling of Epstein-Barr virus-encoded microRNAs in nasopharyngeal carcinoma reveals potential biomarkers and oncomirs. Cancer 2012, 118, 698–710. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Li, W.; Wu, Y.; Wei, F.; Gong, Z.; Bo, H.; Wang, Y.; Li, X.; Xiang, B.; Guo, C.; et al. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis. 2016, 7, e2353. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Zong, J.; Lin, W.; Wang, M.; Xu, Y.; Zhou, R.; Lin, S.; Guo, Q.; Chen, H.; Ye, Y.; et al. EBV-miR-BART8-3p induces epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma cells through activating NF-κB and Erk1/2 pathways. J. Exp. Clin. Cancer Res. 2018, 37, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Zeng, Z.; Gong, Z.; Zhang, W.; Li, X.; He, B.; Song, Y.; Li, Q.; Zeng, Y.; Liao, Q.; et al. EBV-miR-BART10-3p facilitates epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma by targeting BTRC. Oncotarget 2015, 6, 41766–41782. [Google Scholar] [CrossRef]
- Xu, Y.J.; Zhou, R.; Zong, J.F.; Lin, W.S.; Tong, S.; Guo, Q.J.; Lin, C.; Lin, S.J.; Chen, Y.X.; Chen, M.R.; et al. Epstein-Barr virus-coded miR-BART13 promotes nasopharyngeal carcinoma cell growth and metastasis via targeting of the NKIRAS2/NF-κB pathway. Cancer Lett. 2019, 447, 33–40. [Google Scholar] [CrossRef]
- Pećina-Slaus, N. Tumor suppressor gene E-cadherin and its role in normal and malignant cells. Cancer Cell Int. 2003, 3, 17. [Google Scholar] [CrossRef]
- Onder, T.T.; Gupta, P.B.; Mani, S.A.; Yang, J.; Lander, E.S.; Weinberg, R.A. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008, 68, 3645–3654. [Google Scholar] [CrossRef]
- Hsu, C.-Y.; Yi, Y.-H.; Chang, K.-P.; Chang, Y.-S.; Chen, S.-J.; Chen, H.-C. The Epstein-Barr virus-encoded microRNA miR-BART9 promotes tumor metastasis by targeting E-cadherin in nasopharyngeal carcinoma. PLoS Pathog. 2014, 10, e1003974. [Google Scholar] [CrossRef]
- Leão, R.; Apolónio, J.D.; Lee, D.; Figueiredo, A.; Tabori, U.; Castelo-Branco, P. Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: Clinical impacts in cancer. J. Biomed. Sci. 2018, 25, 1–12. [Google Scholar] [CrossRef]
- Kawagoe, J.; Ohmichi, M.; Takahashi, T.; Ohshima, C.; Mabuchi, S.; Takahashi, K.; Igarashi, H.; Mori-Abe, A.; Saitoh, M.; Du, B.; et al. Raloxifene inhibits estrogen-induced up-regulation of telomerase activity in a human breast cancer cell line. J. Biol. Chem. 2003, 278, 43363–43372. [Google Scholar] [CrossRef]
- Akiyama, M.; Hideshima, T.; Hayashi, T.; Tai, Y.T.; Mitsiades, C.S.; Mitsiades, N.; Chauhan, D.; Richardson, P.; Munshi, N.C.; Anderson, K.C. Nuclear factor-κB p65 mediates tumor necrosis factor α-induced nuclear translocation of telomerase reverse transcriptase protein. Cancer Res. 2003, 63, 18–21. [Google Scholar] [PubMed]
- Ding, L.; Li, L.L.; Yang, J.; Yong, G.T.; Ye, M.; Shi, Y.; Tang, M.; Yi, W.; Xiao, L.L.; Jian, P.G.; et al. Epstein-Barr virus encoded latent membrane protein 1 modulates nuclear translocation of telomerase reverse transcriptase protein by activating nuclear factor-κB p65 in human nasopharyngeal carcinoma cells. Int. J. Biochem. Cell Biol. 2005, 37, 1881–1889. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xu, Z.; Liu, L.; Luo, X.; Lu, J.; Sun, L.; Cao, Y. Targeting EBV-LMP1 DNAzyme enhances radiosensitivity of nasopharyngeal carcinoma cells by inhibiting telomerase activity. Cancer Biol. Ther. 2014, 15, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Deng, X.; Deng, L.; Gu, H.; Fan, W.; Cao, Y. Telomerase activation by Epstein-Barr virus latent membrane protein 1 is associated with c-Myc expression in human nasopharyngeal epithelial cells. J. Exp. Clin. Cancer Res. 2004, 23, 495–506. [Google Scholar] [PubMed]
- Li, H.M.; Man, C.; Jin, Y.; Deng, W.; Yip, Y.L.; Feng, H.C.; Cheung, Y.C.; Lo, K.W.; Meltzer, P.S.; Wu, Z.G.; et al. Molecular and cytogenetic changes involved in the immortalization of nasopharyngeal epithelial cells by telomerase. Int. J. Cancer 2006, 119, 1567–1576. [Google Scholar] [CrossRef]
- Zhu, D.D.; Zhang, J.; Deng, W.; Yip, Y.L.; Lung, H.L.; Tsang, C.M.; Law, W.T.; Yang, J.; Lau, V.M.Y.; Shuen, W.H.; et al. Significance of NF-κB activation in immortalization of nasopharyngeal epithelial cells. Int. J. Cancer 2016, 138, 1175–1185. [Google Scholar] [CrossRef]
- Yip, Y.-L.; Tsang, C.-M.; Deng, W.; Cheung, P.-Y.; Jin, Y.; Cheung, A.L.; Lung, M.L.; Tsao, S.-W. Expression of Epstein-Barr Virus-encoded LMP1 and hTERT extends the life span and immortalizes primary cultures of nasopharyngeal epithelial cells. J. Med. Virol. 2010, 82, 1711–1723. [Google Scholar] [CrossRef]
- Liu, Y.; Gong, P.; Zhou, N.; Zhang, J.; He, C.; Wang, S.; Peng, H. Insufficient PINX1 expression stimulates telomerase activation by direct inhibition of EBV LMP1-NF-κB axis during nasopharyngeal carcinoma development. Biochem. Biophys. Res. Commun. 2019, 514, 127–133. [Google Scholar] [CrossRef]
- Ding, L.; Li, L.; Yang, J.; Zhou, S.; Li, W.; Tang, M.; Shi, Y.; Yi, W.; Cao, Y. Latent membrane protein 1 encoded by Epstein–Barr Virus induces telomerase activity via p16INK4A/Rb/E2F1 and JNK signaling pathways. J. Med. Virol. 2007, 79, 1153–1163. [Google Scholar] [CrossRef]


| EBV Genomes | Sources | GenBank Accession Number | LMP1 Strains | References |
|---|---|---|---|---|
| GD1 | Saliva of NPC patients | AY961628 | China 1 | [44] |
| GD2 | Biopsy of NPC patients | HQ020558 | China 1 | [45] |
| M81 | LCL isolated from NPC patients | KF373730 | China 1 | [46] |
| HKNPC1 | NPC patient | JQ009376 | China 1 | [47] |
| HNKPC2 | NPC patient | MH590513 | China 1 | [47] |
| HNKPC3 | NPC patient | MH590514 | China 1 | [47] |
| HNKPC4 | NPC patient | MH590515 | China 1 | [47] |
| HNKPC5 | NPC patient | MH590516 | China 1 | [47] |
| HNKPC6 | NPC patient | MH590517 | China 1 | [47] |
| HNKPC7 | NPC patient | MH590518 | China 1 | [47] |
| HNKPC8 | NPC patient | MH590519 | China 1 | [47] |
| HNKPC9 | NPC patient | MH590520 | China 1 | [47] |
| C666-1 | NPC cell lines | KC617875 | China 1 | [48] |
| M-ABA | LCL isolated from NPC patients | LN827527 | B95.8 | [49] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Richardo, T.; Prattapong, P.; Ngernsombat, C.; Wisetyaningsih, N.; Iizasa, H.; Yoshiyama, H.; Janvilisri, T. Epstein-Barr Virus Mediated Signaling in Nasopharyngeal Carcinoma Carcinogenesis. Cancers 2020, 12, 2441. https://doi.org/10.3390/cancers12092441
Richardo T, Prattapong P, Ngernsombat C, Wisetyaningsih N, Iizasa H, Yoshiyama H, Janvilisri T. Epstein-Barr Virus Mediated Signaling in Nasopharyngeal Carcinoma Carcinogenesis. Cancers. 2020; 12(9):2441. https://doi.org/10.3390/cancers12092441
Chicago/Turabian StyleRichardo, Timmy, Pongphol Prattapong, Chawalit Ngernsombat, Nurulfitri Wisetyaningsih, Hisashi Iizasa, Hironori Yoshiyama, and Tavan Janvilisri. 2020. "Epstein-Barr Virus Mediated Signaling in Nasopharyngeal Carcinoma Carcinogenesis" Cancers 12, no. 9: 2441. https://doi.org/10.3390/cancers12092441
APA StyleRichardo, T., Prattapong, P., Ngernsombat, C., Wisetyaningsih, N., Iizasa, H., Yoshiyama, H., & Janvilisri, T. (2020). Epstein-Barr Virus Mediated Signaling in Nasopharyngeal Carcinoma Carcinogenesis. Cancers, 12(9), 2441. https://doi.org/10.3390/cancers12092441







