Insights into the Role of microRNAs in Colorectal Cancer (CRC) Metabolism
Abstract
:Simple Summary
Abstract
1. Introduction
2. Role of miRNAs in Mitochondrial Metabolism/OXPHOS Metabolism
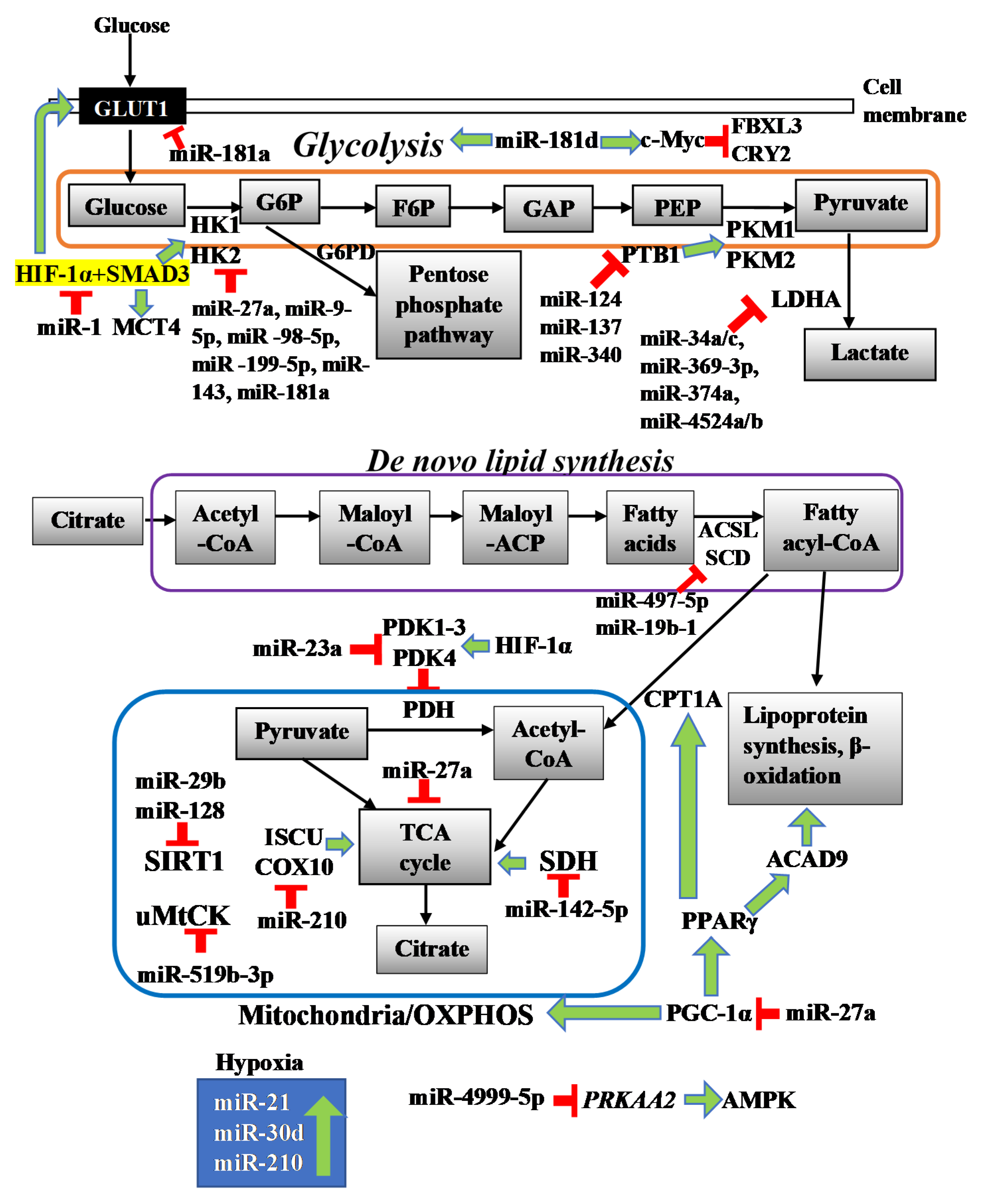
2.1. miR-23a
2.2. miR-519b-3p
2.3. miR-142-5p
2.4. miR-210
2.5. miR-29b and miR-128
2.6. miR-27a
3. Role of miRNAs in Glycolysis
3.1. miR-27a
3.2. miR-143
3.3. miR-9-5p, -98-5p, and -199-5p
3.4. miR-181 Family
3.5. miR-1
3.6. miR-124, miR-137 and miR-340
3.7. miR-4999-5p
4. Role of miRNAs in Lactate Metabolism
miR-34a, miR-34c, miR-369-3p, miR-374a, and miR-4524a/b
5. Role of miRNAs in Lipid Metabolism
5.1. miR-497-5p
5.2. miR-19b-1
6. Hypoxia-Induced Metabolic Reprogramming
miR-21, miR-30d and miR-210
7. Crosstalk between miRNAs and ncRNAs in Metabolic Reprogramming of CRC
8. The Role of miRNAs in the Crosstalk between Metabolism and Liver Metastasis in CRC
9. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2016, 66, 683–691. [Google Scholar] [CrossRef] [Green Version]
- Guinney, J.; Dienstmann, R.; Wang, X.; De Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Fakih, M. Metastatic Colorectal Cancer: Current State and Future Directions. J. Clin. Oncol. 2015, 33, 1809–1824. [Google Scholar] [CrossRef]
- Shen, Y.; Tong, M.; Liang, Q.; Guo, Y.; Sun, H.Q.; Zheng, W.; Ao, L.; Guo, Z.; She, F. Epigenomics alternations and dynamic transcriptional changes in responses to 5-fluorouracil stimulation reveal mechanisms of acquired drug resistance of colorectal cancer cells. Pharm. J. 2017, 18, 23–28. [Google Scholar] [CrossRef] [Green Version]
- Xu, P.; Zhu, Y.; Sun, B.; Xiao, Z. Colorectal cancer characterization and therapeutic target prediction based on microRNA expression profile. Sci. Rep. 2016, 6, 20616. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wang, J. MicroRNAs are important regulators of drug resistance in colorectal cancer. Biol. Chem. 2017, 398, 929–938. [Google Scholar] [CrossRef] [Green Version]
- Tagscherer, K.E.; Fassl, A.; Sinkovic, T.; Richter, J.; Schecher, S.; Macher-Goeppinger, S.; Roth, W. MicroRNA-210 induces apoptosis in colorectal cancer via induction of reactive oxygen. Cancer Cell Int. 2016, 16, 42. [Google Scholar] [CrossRef] [Green Version]
- Pakiet, A.; Kobiela, J.; Stepnowski, P.; Sledzinski, T.; Mika, A. Changes in lipids composition and metabolism in colorectal cancer: A review. Lipids Health Dis. 2019, 18, 29. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, H.; Liu, A.; Fang, C.; Hao, J.; Wang, Z. Lactate dehydrogenase A negatively regulated by miRNAs promotes aerobic glycolysis and is increased in colorectal cancer. Oncotarget 2015, 6, 19456–19468. [Google Scholar] [CrossRef]
- Taniguchi, K.; Sugito, N.; Kumazaki, M.; Shinohara, H.; Yamada, N.; Nakagawa, Y.; Ito, Y.; Otsuki, Y.; Uno, B.; Uchinyama, K.; et al. MicroRNA-124 inhibits cancer cell growth through PTB1/PKM1/PKM2 feedback cascade in colorectal cancer. Cancer Lett. 2015, 363, 17–27. [Google Scholar]
- Dong, H.; Lei, J.; Ding, L.; Wen, Y.; Ju, H.; Zhang, X. MicroRNA: Function, Detection, and Bioanalysis. Chem. Rev. 2013, 113, 6207–6233. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC Couples MicroRNA Biogenesis and Posttranscriptional Gene Silencing. Cell 2005, 123, 631–640. [Google Scholar] [CrossRef] [Green Version]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [Green Version]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar]
- Garofalo, M.; Croce, C.M. MicroRNAs: Master Regulators as Potential Therapeutics in Cancer. Annu. Rev. Pharmacol. Toxicol. 2011, 51, 25–43. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, B. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2008, 19, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Zou, J.; Liu, L.; Wang, Q.; Yin, F.; Yang, Z.; Zhang, W.; Li, L. Downregulation of miR-429 contributes to the development of drug resistance in epithelial ovarian cancer by targeting ZEB1. Am. J. Transl. Res. 2017, 9, 1357–1368. [Google Scholar]
- Serguienko, A.; Grad, I.; Wennerstrøm, A.B.; Meza-Zepeda, L.A.; Thiede, B.; Stratford, E.W.; Myklebost, O.; Munthe, E. Metabolic reprogramming of metastatic breast cancer and melanoma by let-7a microRNA. Oncotarget 2014, 6, 2451–2465. [Google Scholar] [CrossRef] [Green Version]
- Su, Z.; Jiang, G.; Chen, J.; Liu, X.; Zhao, H.; Fang, Z.; He, Y.; Jiang, X.; Xu, G. MicroRNA-429 inhibits cancer cell proliferation and migration by targeting AKT1 in renal cell carcinoma. Mol. Clin. Oncol. 2020, 12, 75–80. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Gao, J.; Huang, Q.; Jin, Y.; Wei, Z. Downregulating microRNA-144 mediates a metabolic shift in lung cancer cells by regulating GLUT1 expression. Oncol. Lett. 2016, 11, 3772–3776. [Google Scholar] [CrossRef] [Green Version]
- Guo, W.; Qiu, Z.; Wang, Z.; Wang, Q.; Tan, N.; Chen, T.; Chen, Z.; Huang, S.; Gu, J.; Li, J.; et al. MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology 2015, 62, 1132–1144. [Google Scholar] [CrossRef]
- Zawacka-Pankau, J.; Grinkevich, V.V.; Hünten, S.; Nikulenkov, F.; Gluch, A.; Li, H.; Enge, M.; Kel, A.; Selivanova, G. Inhibition of Glycolytic Enzymes Mediated by Pharmacologically Activated p53. J. Biol. Chem. 2011, 286, 41600–41615. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.H.; Deng, Z.H.; Hao, H.; Wu, X.L.; Gao, H.; Tang, S.H.; Tang, H. MicroRNA-23a promotes colorectal cancer cell survival by targeting PDK4. Exp. Cell Res. 2018, 373, 171–179. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, M.; Chen, Y.; Li, B. MiR-519b-3p Inhibits the Proliferation and Invasion in Colorectal Cancer via Modulating the uMtCK/Wnt Signaling Pathway. Front. Pharmacol. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Xiao, Z.; Ai, F.; Liu, F.; Chen, X.; Cao, K.; Ren, W.; Zhang, X.; Shu, P.; Zhang, D. miR-142-5p promotes development of colorectal cancer through targeting SDHB and facilitating generation of aerobic glycolysis. Biomed. Pharmacother. 2017, 92, 1119–1127. [Google Scholar] [CrossRef]
- Chen, Z.; Li, Y.; Zhang, H.; Huang, P.; Luthra, R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene 2010, 29, 4362–4368. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Cheng, X.-H. MiR-29b reverses oxaliplatin-resistance in colorectal cancer by targeting SIRT1. Oncotarget 2018, 9, 12304–12315. [Google Scholar] [CrossRef] [Green Version]
- Lian, B.; Yang, D.; Liu, Y.; Shi, G.; Li, J.; Yan, X.; Jin, K.; Liu, X.; Zhao, J.; Shang, W.; et al. miR-128 Targets the SIRT1/ROS/DR5 Pathway to Sensitize Colorectal Cancer to TRAIL-Induced Apoptosis. Cell. Physiol. Biochem. 2018, 49, 2151–2162. [Google Scholar]
- Barisciano, G.; Colangelo, T.; Rosato, V.; Muccillo, L.; Taddei, M.L.; Ippolito, L.; Chiarugi, P.; Galgani, M.; Bruzzaniti, S.; Matarese, G.; et al. miR-27a is a master regulator of metabolic reprogramming and chemoresistance in colorectal cancer. Br. J. Cancer 2020, 122, 1354–1366. [Google Scholar] [CrossRef] [Green Version]
- Gregersen, L.H.; Skanderup, A.J.; Frankel, L.B.; Wen, J.; Krogh, A.; Lund, A.H. MicroRNA-143 down-regulates Hexokinase 2 in colon cancer cells. BMC Cancer 2012, 12, 232. [Google Scholar] [CrossRef] [Green Version]
- Snezhkina, A.V.; Krasnov, G.S.; Zhikrivetskaya, S.O.; Karpova, I.Y.; Fedorova, M.S.; Nyushko, K.M.; Belyakov, M.M.; Gnuchev, N.V.; Sidorov, D.V.; Alekseev, B.Y.; et al. Overexpression of microRNAs miR-9, -98, and -199 Correlates with the Downregulation of HK2 Expression in Colorectal Cancer. Mol. Biol. 2018, 52, 190–199. [Google Scholar] [CrossRef]
- Wei, Z.; Cui, L.; Mei, Z.; Liu, M.; Zhang, D. miR-181a mediates metabolic shift in colon cancer cells via the PTEN/AKT pathway. FEBS Lett. 2014, 588, 1773–1779. [Google Scholar] [CrossRef]
- Guo, X.; Zhu, Y.; Hong, X.; Zhang, M.; Qiu, X.; Wang, Z.; Qi, Z.-Q.; Hong, X. miR-181d and c-myc-mediated inhibition of CRY2 and FBXL3 reprograms metabolism in colorectal cancer. Cell Death Dis. 2017, 8, e2958. [Google Scholar] [CrossRef]
- Xu, W.; Zhang, Z.; Zou, K.; Cheng, Y.; Yang, M.; Chen, H.; Wang, H.; Zhao, J.; Chen, P.; He, L.; et al. MiR-1 suppresses tumor cell proliferation in colorectal cancer by inhibition of Smad3-mediated tumor glycolysis. Cell Death Dis. 2017, 8, e2761. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Zhao, X.; Zhou, Y.; Hu, Y. miR-124, miR-137 and miR-340 regulate colorectal cancer growth via inhibition of the Warburg effect. Oncol. Rep. 2012, 28, 1346–1352. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Hong, Z.; Zhu, J.; Zeng, C.; Tang, Z.; Wang, W.; Huang, H. miR-4999-5p Predicts Colorectal Cancer Survival Outcome and Reprograms Glucose Metabolism by Targeting PRKAA2. OncoTargets Ther. 2020, 13, 1199–1210. [Google Scholar] [CrossRef] [Green Version]
- Gharib, E.; Nasrabadi, P.N.; Zali, M.R. miR-497-5p mediates starvation-induced death in colon cancer cells by targeting acyl-CoA synthetase-5 and modulation of lipid metabolism. J. Cell. Physiol. 2020, 235, 5570–5589. [Google Scholar] [CrossRef]
- Cruz-Gil, S.; Sanchez-Martinez, R.; De Cedrón, M.G.; Martin-Hernandez, R.; Vargas, T.; Molina, S.; Herranz, J.; Davalos, A.; Reglero, G.; Molina, A.R.-D. Targeting the lipid metabolic axisACSL/SCDin colorectal cancer progression by therapeutic miRNAs: miR-19b-1 role. J. Lipid Res. 2017, 59, 14–24. [Google Scholar] [CrossRef] [Green Version]
- Nijhuis, A.; Thompson, H.; Adam, J.; Parker, A.; Gammon, L.; Lewis, A.; Bundy, J.G.; Soga, T.; Jalaly, A.; Propper, D.; et al. Remodelling of microRNAs in colorectal cancer by hypoxia alters metabolism profiles and 5-fluorouracil resistance. Hum. Mol. Genet. 2017, 26, 1552–1564. [Google Scholar] [CrossRef]
- Litwack, G. Chapter 8—Glycolysis and Gluconeogenesis. In Human Biochemistry; Litwack, G., Ed.; Academic Press: Boston, MA, USA, 2018; pp. 183–198. [Google Scholar]
- Li, Q.; Fan, S.; Li, X.; Jin, Y.; He, W.; Zhou, J.; Cen, S.; Yang, Z. Insights into the Phosphoryl Transfer Mechanism of Human Ubiquitous Mitochondrial Creatine Kinase. Sci. Rep. 2016, 6, 38088. [Google Scholar] [CrossRef]
- Gill, A.J. Succinate dehydrogenase (SDH)-deficient neoplasia. Histopathology 2017, 72, 106–116. [Google Scholar] [CrossRef]
- Aldera, A.P.; Govender, D. Gene of the month: SDH. J. Clin. Pathol. 2017, 71, 95–97. [Google Scholar] [CrossRef]
- Tseng, P.-L.; Wu, W.-H.; Hu, T.-H.; Chen, C.-W.; Cheng, H.-C.; Li, C.-F.; Tsai, W.-H.; Tsai, H.-J.; Hsieh, M.-C.; Chuang, J.-H.; et al. Decreased succinate dehydrogenase B in human hepatocellular carcinoma accelerates tumor malignancy by inducing the Warburg effect. Sci. Rep. 2018, 8, 1–16. [Google Scholar] [CrossRef]
- Xiao, Z.; Liu, S.; Ai, F.; Chen, X.; Li, X.; Liu, R.; Ren, W.; Zhang, X.; Shu, P.; Zhang, D. SDHB downregulation facilitates the proliferation and invasion of colorectal cancer through AMPK functions excluding those involved in the modulation of aerobic glycolysis. Exp. Ther. Med. 2017, 15, 864–872. [Google Scholar] [CrossRef] [Green Version]
- Favaro, E.; Ramachandran, A.; McCormick, R.; Gee, H.E.; Blancher, C.; Crosby, M.; Devlin, C.; Blick, C.; Buffa, F.M.; Li, J.-L.; et al. MicroRNA-210 Regulates Mitochondrial Free Radical Response to Hypoxia and Krebs Cycle in Cancer Cells by Targeting Iron Sulfur Cluster Protein ISCU. PLoS ONE 2010, 5, e10345. [Google Scholar] [CrossRef] [Green Version]
- Tang, B.L. Sirt1 and the Mitochondria. Mol. Cells 2016, 39, 87–95. [Google Scholar] [CrossRef] [Green Version]
- Colangelo, T.; Polcaro, G.; Ziccardi, P.; Muccillo, L.; Galgani, M.; Pucci, B.; Milone, M.R.; Budillon, A.; Santopaolo, M.; Mazzoccoli, G.; et al. The miR-27a-calreticulin axis affects drug-induced immunogenic cell death in human colorectal cancer cells. Cell Death Dis. 2016, 7, e2108. [Google Scholar] [CrossRef] [Green Version]
- Colangelo, T.; Polcaro, G.; Ziccardi, P.; Pucci, B.; Muccillo, L.; Galgani, M.; Fucci, A.; Milone, M.R.; Budillon, A.; Santopaolo, M.; et al. Proteomic screening identifies calreticulin as a miR-27a direct target repressing MHC class I cell surface exposure in colorectal cancer. Cell Death Dis. 2016, 7, e2120. [Google Scholar] [CrossRef]
- Fernandez-Marcos, P.J.; Auwerx, J. Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis. Am. J. Clin. Nutr. 2011, 93, 884S–890S. [Google Scholar] [CrossRef] [Green Version]
- Pucci, S.; Zonetti, M.J.; Fisco, T.; Polidoro, C.; Bocchinfuso, G.; Palleschi, A.; Novelli, G.; Spagnoli, L.G.; Mazzarelli, P. Carnitine palmitoyl transferase-1A (CPT1A): A new tumor specific target in human breast cancer. Oncotarget 2016, 7, 19982–19996. [Google Scholar] [CrossRef] [Green Version]
- Nouws, J.; Nijtmans, L.G.; Houten, S.M.; Brand, M.V.D.; Huynen, M.A.; Venselaar, H.; Hoefs, S.; Gloerich, J.; Kronick, J.; Hutchin, T.; et al. Acyl-CoA Dehydrogenase 9 Is Required for the Biogenesis of Oxidative Phosphorylation Complex I. Cell Metab. 2010, 12, 283–294. [Google Scholar] [CrossRef] [Green Version]
- Potter, M.; Newport, E.; Morten, K.J. The Warburg effect: 80 years on. Biochem. Soc. Trans. 2016, 44, 1499–1505. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Tan, M.; Cai, Q. The Warburg effect in tumor progression: Mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett. 2014, 356, 156–164. [Google Scholar] [CrossRef] [Green Version]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef] [Green Version]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Zhou, P.; Chen, W.-G.; Li, X.-W. MicroRNA-143 acts as a tumor suppressor by targeting hexokinase 2 in human prostate cancer. Am. J. Cancer Res. 2015, 5, 2056–2063. [Google Scholar]
- Hui, L.; Zhang, J.; Guo, X. MiR-125b-5p suppressed the glycolysis of laryngeal squamous cell carcinoma by down-regulating hexokinase-2. Biomed. Pharmacother. 2018, 103, 1194–1201. [Google Scholar] [CrossRef]
- Bai, J.-W.; Xue, H.-Z.; Zhang, C. Down-regulation of microRNA-143 is associated with colorectal cancer progression. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4682–4687. [Google Scholar]
- Benitez, J.A.; Ma, J.; D’Antonio, M.; Boyer, A.; Camargo, M.F.; Zanca, C.; Kelly, S.; Khodadadi-Jamayran, A.; Jameson, N.M.; Andersen, M.; et al. PTEN regulates glioblastoma oncogenesis through chromatin-associated complexes of DAXX and histone H3.3. Nat. Commun. 2017, 8, 15223. [Google Scholar] [CrossRef]
- Wu, R.-C.; Young, I.-C.; Chen, Y.-F.; Chuang, S.-T.; Toubaji, A.; Wu, M.-Y. Identification of the PTEN-ARID4B-PI3K pathway reveals the dependency on ARID4B by PTEN-deficient prostate cancer. Nat. Commun. 2019, 10, 4332. [Google Scholar] [CrossRef]
- Li, S.; Shen, Y.; Wang, M.; Yang, J.; Lv, M.; Li, P.; Chen, Z.; Yang, J. Loss of PTEN expression in breast cancer: Association with clinicopathological characteristics and prognosis. Oncotarget 2017, 8, 32043–32054. [Google Scholar] [CrossRef] [Green Version]
- Lin, P.-C.; Lin, J.-K.; Lin, H.-H.; Lan, Y.-T.; Lin, C.-C.; Yang, S.-H.; Chen, W.-S.; Liang, W.-Y.; Jiang, J.-K.; Chang, S.-C. A comprehensive analysis of phosphatase and tensin homolog deleted on chromosome 10 (PTEN) loss in colorectal cancer. World J. Surg. Oncol. 2015, 13, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Lien, E.; Lyssiotis, C.A.; Cantley, L.C. Metabolic Reprogramming by the PI3K-Akt-mTOR Pathway in Cancer. Methods Mol. Biol. 2016, 207, 39–72. [Google Scholar] [CrossRef]
- Hoxhaj, G.; Manning, B.D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 2019, 20, 74–88. [Google Scholar] [CrossRef]
- Zhao, Y.; Hu, X.; Liu, Y.; Dong, S.; Wen, Z.; He, W.; Zhang, S.; Huang, Q.; Shi, M. ROS signaling under metabolic stress: Cross-talk between AMPK and AKT pathway. Mol. Cancer 2017, 16, 79. [Google Scholar] [CrossRef] [Green Version]
- Tu, K.; Liu, Z.; Yao, B.; Han, S.; Yang, W. MicroRNA-519a promotes tumor growth by targeting PTEN/PI3K/AKT signaling in hepatocellular carcinoma. Int. J. Oncol. 2015, 48, 965–974. [Google Scholar] [CrossRef] [Green Version]
- Feng, X.; Jiang, J.; Shi, S.; Xie, H.; Zhou, L.; Zheng, S. Knockdown of miR-25 increases the sensitivity of liver cancer stem cells to TRAIL-induced apoptosis via PTEN/PI3K/Akt/Bad signaling pathway. Int. J. Oncol. 2016, 49, 2600–2610. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Liu, Q.; Tang, J.; Xie, Y.; Xu, X.; Huang, R.; Zhang, Y.; Jin, K.; Sun, B. Alpha1-ACT Functions as a Tumour Suppressor in Hepatocellular Carcinoma by Inhibiting the PI3K/AKT/mTOR Signalling Pathway via Activation of PTEN. Cell. Physiol. Biochem. 2017, 41, 2289–2306. [Google Scholar] [CrossRef]
- Huber, A.-L.; Papp, S.J.; Chan, A.B.; Henriksson, E.; Jordan, S.D.; Kriebs, A.; Nguyen, M.; Wallace, M.; Li, Z.; Metallo, C.M.; et al. CRY2 and FBXL3 Cooperatively Degrade c-MYC. Mol. Cell 2016, 64, 774–789. [Google Scholar] [CrossRef] [Green Version]
- Farrell, A.S.; Sears, R.C. MYC Degradation. Cold Spring Harb. Perspect. Med. 2014, 4, a014365. [Google Scholar] [CrossRef]
- Chen, X.; Han, P.; Zhou, T.; Guo, X.; Song, X.; Li, Y. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci. Rep. 2016, 6, 34985. [Google Scholar] [CrossRef]
- Du, Y.-Y.; Zhao, L.-M.; Chen, L.; Sang, M.-X.; Li, J.; Ma, M.; Liu, J.-F. The tumor-suppressive function of miR-1 by targeting LASP1 and TAGLN2 in esophageal squamous cell carcinoma. J. Gastroenterol. Hepatol. 2016, 31, 384–393. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, Y.; Wang, H.; Zhang, G.; Ding, Y.; Zhao, L. Tumor suppressor miR-1 restrains epithelial-mesenchymal transition and metastasis of colorectal carcinoma via the MAPK and PI3K/AKT pathway. J. Transl. Med. 2014, 12, 244. [Google Scholar] [CrossRef] [Green Version]
- Bronisz, A.; Wang, Y.; Nowicki, M.O.; Peruzzi, P.; Ansari, K.I.; Ogawa, D.; Balaj, L.; De Rienzo, G.; Mineo, M.; Nakano, I.; et al. Extracellular vesicles modulate the glioblastoma microenvironment via a tumor suppression signaling network directed by miR-1. Cancer Res. 2013, 74, 738–750. [Google Scholar] [CrossRef] [Green Version]
- Courtnay, R.; Ngo, D.C.; Malik, N.; Ververis, K.; Tortorella, S.M.; Karagiannis, T.C. Cancer metabolism and the Warburg effect: The role of HIF-1 and PI3K. Mol. Biol. Rep. 2015, 42, 841–851. [Google Scholar] [CrossRef]
- Syed, V. TGF-β Signaling in Cancer. J. Cell. Biochem. 2016, 117, 1279–1287. [Google Scholar]
- Muirhead, H. Isoenzymes of pyruvate kinase. Biochem. Soc. Trans. 1990, 18, 193–196. [Google Scholar] [CrossRef]
- Noguchi, T.; Inoue, H.; Tanaka, T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J. Biol. Chem. 1986, 261, 13807–13812. [Google Scholar]
- Christofk, H.R.; Heiden, M.G.V.; Harris, M.H.; Ramanathan, A.; Gerszten, R.E.; Wei, R.; Fleming, M.D.; Schreiber, S.L.; Cantley, L.C. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 2008, 452, 230–233. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, J.; Manley, J.L. Turning on a fuel switch of cancer: hnRNP proteins regulate alternative splicing of pyruvate kinase mRNA. Cancer Res. 2010, 70, 8977–8980. [Google Scholar] [CrossRef] [Green Version]
- Clower, C.V.; Chatterjee, D.; Wang, Z.; Cantley, L.C.; Vander Heiden, M.G.; Krainer, A.R. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc. Natl. Acad. Sci. USA 2010, 107, 1894–1899. [Google Scholar]
- David, C.J.; Chen, M.; Assanah, M.; Canoll, P.; Manley, J.L. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 2009, 463, 364–368. [Google Scholar] [CrossRef]
- Herzig, S.; Shaw, R.J. AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2017, 19, 121–135. [Google Scholar] [CrossRef] [Green Version]
- Kopsiaftis, S.; Sullivan, K.L.; Garg, I.; Taylor, J.A.; Claffey, K.P. AMPKalpha2 Regulates Bladder Cancer Growth through SKP2-Mediated Degradation of p27. Mol. Cancer Res. 2016, 14, 1182–1194. [Google Scholar]
- Xu, Q.; Wu, N.; Li, X.; Guo, C.; Li, C.; Jiang, B.; Wang, H.; Shi, D. Inhibition of PTP1B blocks pancreatic cancer progression by targeting the PKM2/AMPK/mTOC1 pathway. Cell Death Dis. 2019, 10, 1–15. [Google Scholar]
- Wei, J.-L.; Fang, M.; Fu, Z.-X.; Zhang, S.; Guo, J.-B.; Wang, R.; Lv, Z.-B.; Xiong, Y.-F. Sestrin 2 suppresses cells proliferation through AMPK/mTORC1 pathway activation in colorectal cancer. Oncotarget 2017, 8, 49318–49328. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.; Shi, M.; Xie, D.; Wei, D.; Jia, Z.; Zheng, S.; Gao, Y.; Huang, S.; Xie, K. FOXM1 promotes the warburg effect and pancreatic cancer progression via transactivation of LDHA expression. Clin. Cancer Res. 2014, 20, 2595–2606. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Xiong, Y.; Qiao, T.; Li, X.; Jia, L.-T.; Han, Y. Lactate dehydrogenase A: A key player in carcinogenesis and potential target in cancer therapy. Cancer Med. 2018, 7, 6124–6136. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; Zhou, F.; Li, N.; Li, Q.; Wang, L. FOXM1-LDHA signaling promoted gastric cancer glycolytic phenotype and progression. Int. J. Clin. Exp. Pathol. 2015, 8, 6756–6763. [Google Scholar]
- Li, J.; Zhu, S.; Tong, J.; Hao, H.; Yang, J.; Liu, Z.; Wang, Y. Suppression of lactate dehydrogenase A compromises tumor progression by downregulation of the Warburg effect in glioblastoma. NeuroReport 2016, 27, 110–115. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.; Cui, J.; Du, J.; Wei, D.; Jia, Z.; Zhang, J.; Zhu, Z.; Gao, Y.; Xie, K. A novel KLF4/LDHA signaling pathway regulates aerobic glycolysis in and progression of pancreatic cancer. Clin. Cancer Res. 2014, 20, 4370–4380. [Google Scholar]
- Vargas, T.; Moreno-Rubio, J.; Herranz, J.; Cejas, P.; Molina, S.; González-Vallinas, M.; Mendiola, M.; Burgos, E.; Aguayo, C.; Custodio, A.B.; et al. ColoLipidGene: Signature of lipid metabolism-related genes to predict prognosis in stage-II colon cancer patients. Oncotarget 2015, 6, 7348–7363. [Google Scholar] [CrossRef] [Green Version]
- Schlaepfer, I.R.; Rider, L.; Rodrigues, L.U.; Gijón, M.A.; Pac, C.T.; Romero, L.; Cimic, A.; Sirintrapun, S.J.; Glode, L.M.; Eckel, R.H.; et al. Lipid catabolism via CPT1 as a therapeutic target for prostate cancer. Mol. Cancer Ther. 2014, 13, 2361–2371. [Google Scholar] [CrossRef] [Green Version]
- Ricciardi, M.R.; Mirabilii, S.; Allegretti, M.; Licchetta, R.; Calarco, A.; Torrisi, M.R.; Foà, R.; Nicolai, R.; Peluso, G.; Tafuri, A. Targeting the leukemia cell metabolism by the CPT1a inhibition: Functional preclinical effects in leukemias. Blood 2015, 126, 1925–1929. [Google Scholar] [CrossRef] [Green Version]
- Yue, S.; Li, J.; Lee, S.Y.; Lee, H.J.; Shao, T.; Song, B.; Cheng, L.; Masterson, T.A.; Liu, X.; Ratliff, T.L.; et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 2014, 19, 393–406. [Google Scholar] [CrossRef] [Green Version]
- Daniëls, V.W.; Smans, K.; Royaux, I.; Chypre, M.; Swinnen, J.V.; Zaidi, N. Cancer Cells Differentially Activate and Thrive on De Novo Lipid Synthesis Pathways in a Low-Lipid Environment. PLoS ONE 2014, 9, e106913. [Google Scholar] [CrossRef]
- Lopes-Marques, M.; Cunha, I.; Reis-Henriques, M.; Santos, M.; Castro, L.F.C. Diversity and history of the long-chain acyl-CoA synthetase (Acsl) gene family in vertebrates. BMC Evol. Biol. 2013, 13, 271. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Martinez, R.; Cruz-Gil, S.; García-Álvarez, M.S.; Reglero, G.; Molina, A.R.-D. Complementary ACSL isoforms contribute to a non-Warburg advantageous energetic status characterizing invasive colon cancer cells. Sci. Rep. 2017, 7, 11143. [Google Scholar] [CrossRef]
- Chen, W.-C.; Wang, C.-Y.; Hung, Y.-H.; Weng, T.-Y.; Yen, M.-C.; Lai, M.-D. Systematic Analysis of Gene Expression Alterations and Clinical Outcomes for Long-Chain Acyl-Coenzyme A Synthetase Family in Cancer. PLoS ONE 2016, 11, e0155660. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Kuang, D.; Zhao, X.; Chen, D.; Wang, X.; Yang, Q.; Wan, J.; Zhu, Y.; Wang, Y.; Zhang, S.; et al. miR-497-5p inhibits cell proliferation and invasion by targeting KCa3.1 in angiosarcoma. Oncotarget 2016, 7, 58148–58161. [Google Scholar] [CrossRef] [Green Version]
- Ruan, W.-D.; Wang, P.; Feng, S.; Xue, Y.; Zhang, B. MicroRNA-497 inhibits cell proliferation, migration, and invasion by targeting AMOT in human osteosarcoma cells. OncoTargets Ther. 2016, 9, 303–313. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Meng, X.; Huang, Y.; Lv, Z.; Liu, J.; Wang, G.; Meng, W.; Xue, S.; Zhang, Q.; Zhang, P.; et al. MicroRNA-497 inhibits thyroid cancer tumor growth and invasion by suppressing BDNF. Oncotarget 2016, 8, 2825–2834. [Google Scholar] [CrossRef]
- Chai, L.; Kang, X.-J.; Sun, Z.-Z.; Zeng, M.-F.; Yu, S.-R.; Ding, Y.; Liang, J.-Q.; Li, T.-T.; Zhao, J. MiR-497-5p, miR-195-5p and miR-455-3p function as tumor suppressors by targeting hTERT in melanoma A375 cells. Cancer Manag. Res. 2018, 10, 989–1003. [Google Scholar] [CrossRef] [Green Version]
- Dobrzyń, P. [Stearoyl-CoA desaturase in the control of metabolic homeostasis]. Postępy Biochem. 2012, 58, 166–174. [Google Scholar]
- Sánchez-Martínez, R.; Cruz-Gil, S.; De Cedrón, M.G.; Alvarez-Fernández, M.; Vargas, T.; Molina, S.; García, B.; Herranz, J.; Moreno-Rubio, J.; Reglero, G.; et al. A link between lipid metabolism and epithelial-mesenchymal transition provides a target for colon cancer therapy. Oncotarget 2015, 6, 38719–38736. [Google Scholar] [CrossRef] [Green Version]
- Qu, A.; Du, L.; Yang, Y.; Liu, H.; Li, J.; Wang, L.; Liu, Y.; Dong, Z.; Zhang, X.; Jiang, X.; et al. Hypoxia-Inducible MiR-210 Is an Independent Prognostic Factor and Contributes to Metastasis in Colorectal Cancer. PLoS ONE 2014, 9, e90952. [Google Scholar] [CrossRef] [Green Version]
- Chan, Y.C.; Banerjee, J.; Choi, S.Y.; Sen, C.K. miR-210: The master hypoxamir. Microcirculation 2012, 19, 215–223. [Google Scholar] [CrossRef] [Green Version]
- Yao, R.-W.; Wang, Y.; Chen, L.-L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Zhang, Y.; Liu, L.; Yang, W.; Zhang, Q. HNF1A-AS1 Regulates Cell Migration, Invasion and Glycolysis via Modulating miR-124/MYO6 in Colorectal Cancer Cells. OncoTargets Ther. 2020, 13, 1507–1518. [Google Scholar]
- Wang, D.; Zhu, L.; Liao, M.; Zeng, T.; Zhuo, W.; Yang, S.; Wu, W. MYO6 knockdown inhibits the growth and induces the apoptosis of prostate cancer cells by decreasing the phosphorylation of ERK1/2 and PRAS40. Oncol. Rep. 2016, 36, 1285–1292. [Google Scholar]
- You, W.; Tan, G.; Sheng, N.; Gong, J.; Yan, J.; Chen, D.; Zhang, H.; Wang, Z. Downregulation of myosin VI reduced cell growth and increased apoptosis in human colorectal cancer. Acta Biochim. Biophys. Sin. 2016, 48, 430–436. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Wang, B.; Zhu, W.; Yang, Z. Lentivirus-Mediated Knockdown of Myosin VI Inhibits Cell Proliferation of Breast Cancer Cell. Cancer Biother. Radiopharm. 2015, 30, 330–335. [Google Scholar] [CrossRef]
- Wang, Z.; Ying, M.; Wu, Q.; Wang, R.; Li, Y. Overexpression of myosin VI regulates gastric cancer cell progression. Gene 2016, 593, 100–109. [Google Scholar] [CrossRef] [Green Version]
- Salzman, J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. 2016, 32, 309–316. [Google Scholar] [CrossRef] [Green Version]
- Abu, N.; Jamal, R. Circular RNAs as Promising Biomarkers: A Mini-Review. Front. Physiol. 2016, 7, 355. [Google Scholar] [CrossRef]
- Dong, Y.; He, D.; Peng, Z.; Peng, W.; Shi, W.; Wang, J.; Li, B.; Zhang, C.; Duan, C. Circular RNAs in cancer: An emerging key player. J. Hematol. Oncol. 2017, 10, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Yang, L.; Chen, L.-L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.J.; Zhang, Y.H.; Qin, X.J.; Wang, Y.X.; Fu, J. Circular RNA circDENND4C facilitates proliferation, migration and glycolysis of colorectal cancer cells through miR-760/GLUT1 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2387–2400. [Google Scholar]
- Wang, X.; Zhang, H.; Yang, H.; Bai, M.; Ning, T.; Deng, T.; Liu, R.; Fan, Q.; Zhu, K.; Li, J.; et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol. Oncol. 2020, 14, 539–555. [Google Scholar] [CrossRef]
- Horibe, S.; Tanahashi, T.; Kawauchi, S.; Murakami, Y.; Rikitake, Y. Mechanism of recipient cell-dependent differences in exosome uptake. BMC Cancer 2018, 18, 47. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Investig. 2016, 126, 1208–1215. [Google Scholar] [CrossRef]
- Hon, K.W.; Abu, N.; Ab Mutalib, N.-S.; Jamal, R. Exosomes as Potential Biomarkers and Targeted Therapy in Colorectal Cancer: A Mini-Review. Front. Pharmacol. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Valderrama-Treviño, A.I.; Barrera-Mera, B.; Ceballos-Villalva, J.C.; Montalvo-Javé, E.E. Hepatic Metastasis from Colorectal Cancer. Eur. J. Hepato-Gastroenterol. 2017, 7, 166–175. [Google Scholar] [CrossRef]
- Sahu, S.S.; Dey, S.; Nabinger, S.C.; Jiang, G.; Bates, A.M.; Tanaka, H.; Liu, Y.; Kota, J. The Role and Therapeutic Potential of miRNAs in Colorectal Liver Metastasis. Sci. Rep. 2019, 9, 15803–15810. [Google Scholar] [CrossRef] [Green Version]
- Ji, D.; Chen, Z.; Li, M.; Zhan, T.; Yao, Y.; Zhang, Z.; Xi, J.; Yan, L.; Gu, J. MicroRNA-181a promotes tumor growth and liver metastasis in colorectal cancer by targeting the tumor suppressor WIF-1. Mol. Cancer 2014, 13, 86. [Google Scholar] [CrossRef] [Green Version]
| MiRNA | Up/Down-Regulation | Function | Target Gene/Pathway | Reference |
|---|---|---|---|---|
| miR-23a | Upregulation | Activate PDH in OXPHOS for ATP production | PDK4 | [25] |
| miR-519b-3p | Downregulation | Promote OXPHOS metabolism and cell proliferation | uMtCK/Wnt signalling | [26] |
| miR-142-5p | Upregulation | Promote aerobic glycolysis and Warburg effect | SDH | [27] |
| miR-210 | Upregulation | Increase ROS production and suppress mitochondrial respiration | ISCU, COX 10 | [8,28] |
| miR-29b | Upregulation | Promote ROS generation and apoptosis | SIRT1, Caspase 9, 7 and 3 | [29] |
| miR-128 | Upregulation | Promote ROS generation and apoptosis | SIRT1 | [30] |
| miR-27a | Upregulation | Suppress mitochondrial respiration | PGC-1α, PPARγ, CPT1A and ACAD9 | [31] |
| Facilitate glycolysis | HK1, HK2 | |||
| miR-143 | Downregulation | Promote aerobic glycolysis | HK2 | [32] |
| miR-9-5p, -98-5p, and -199-5p | Upregulation | Facilitate aerobic glycolysis | HK2 | [33] |
| miR-181a | Upregulation | Increase glucose uptake and lactate production | GLUT1 and HK2 via PTEN/AKT pathway | [34] |
| miR-181d | Upregulation | Promote aerobic glycolysis | c-Myc, CRY2, FBXL3 | [35] |
| miR-1 | Downregulation | Promote aerobic glycolysis | HIF-1α and SMAD3 | [36] |
| miR-124, miR-137 and miR-340 | Upregulation | Inhibit aerobic glycolysis | PTB1/PKM1/PKM2 cascade | [11,37] |
| miR-4999-5p | Upregulation | Increase glucose uptake and lactate production | PRKAA2 | [38] |
| miR-34a, miR-34c, miR-369-3p, miR-374a, and miR-4524a/b | Upregulation | Suppress glycolysis and lactate production | LDHA | [10] |
| miR-497-5p | Downregulation | Promote lipid metabolism | ACSL5 | [39] |
| miR-19b-1 | Upregulation | Inhibit de novo lipogenesis | ACSL/SCD | [40] |
| miR-21, miR-30d and miR-210 | Upregulation | Potential biomarker for hypoxia | - | [41] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wai Hon, K.; Zainal Abidin, S.A.; Othman, I.; Naidu, R. Insights into the Role of microRNAs in Colorectal Cancer (CRC) Metabolism. Cancers 2020, 12, 2462. https://doi.org/10.3390/cancers12092462
Wai Hon K, Zainal Abidin SA, Othman I, Naidu R. Insights into the Role of microRNAs in Colorectal Cancer (CRC) Metabolism. Cancers. 2020; 12(9):2462. https://doi.org/10.3390/cancers12092462
Chicago/Turabian StyleWai Hon, Kha, Syafiq Asnawi Zainal Abidin, Iekhsan Othman, and Rakesh Naidu. 2020. "Insights into the Role of microRNAs in Colorectal Cancer (CRC) Metabolism" Cancers 12, no. 9: 2462. https://doi.org/10.3390/cancers12092462
APA StyleWai Hon, K., Zainal Abidin, S. A., Othman, I., & Naidu, R. (2020). Insights into the Role of microRNAs in Colorectal Cancer (CRC) Metabolism. Cancers, 12(9), 2462. https://doi.org/10.3390/cancers12092462





