Breast and Gut Microbiota Action Mechanisms in Breast Cancer Pathogenesis and Treatment
Abstract
:Simple Summary
Abstract
1. Introduction
2. Approaches to Assess Microbiome: Pros and Cons
3. Microbiota and Breast Cancer
3.1. Mammary Microbiota and Breast Cancer
3.2. Link between Gut Microbiota and Breast Cancer
4. Role of Gut Microbiota in Estrogen Level Regulation
5. Breast Cancer, Microbiota and the Immune System
6. Breast Cancer, Microbiota and Inflammation
7. Breast Cancer, Microbiota and Epigenetic Regulation
8. Diet, Microbiota and Breast Cancer
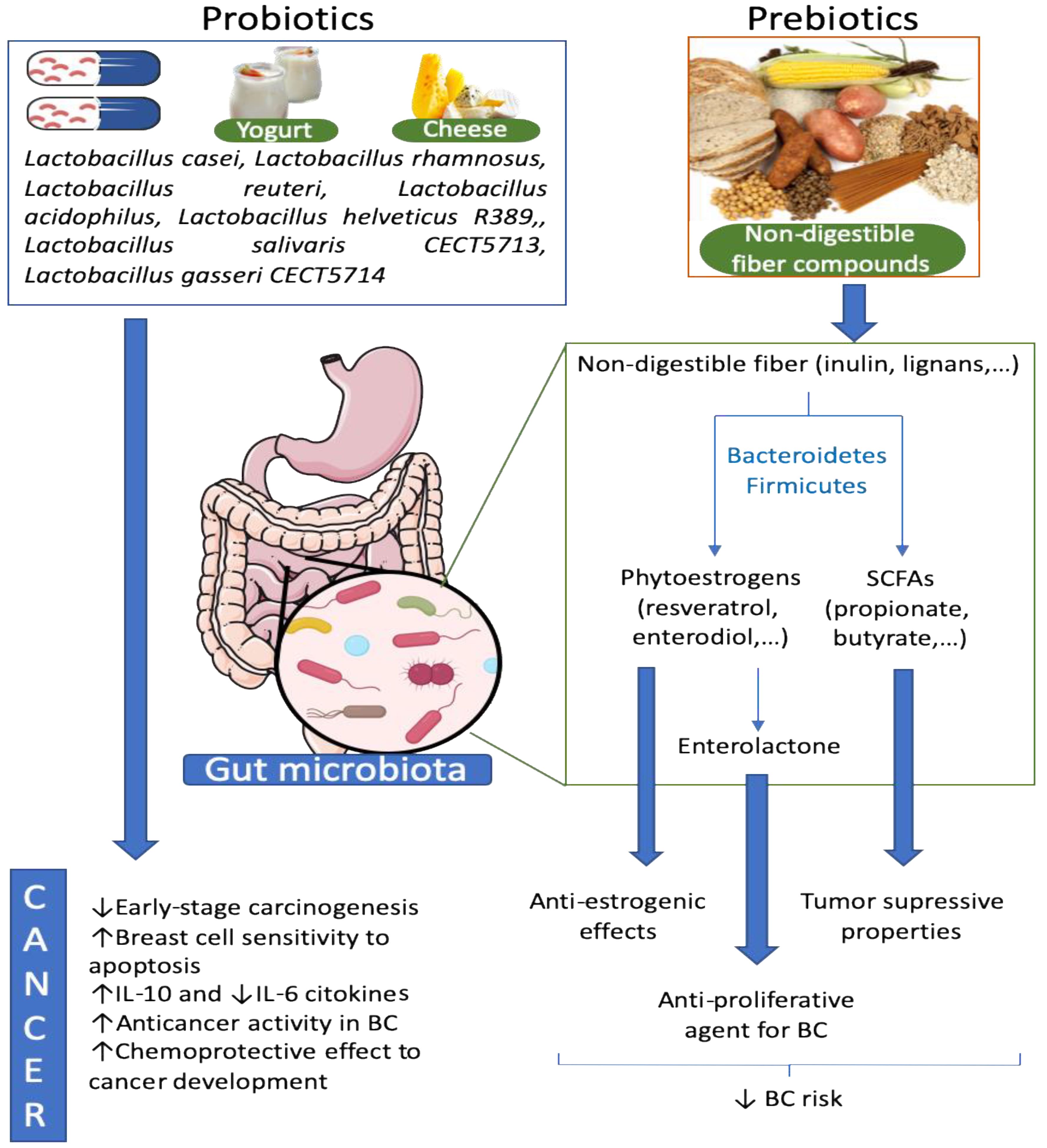
9. Probiotics Effects against Breast Cancer
10. Prebiotics, Microbiota and Breast Cancer
11. Antibiotics, Microbiota and Breast Cancer
12. Microbiota, Anti-Cancer Therapy and Disease Progression
13. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorlie, T.; Tibshirani, R.; Parker, J.; Hastie, T.; Marron, J.S.; Nobel, A.; Deng, S.; Johnsen, H.; Pesich, R.; Geisler, S.; et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. USA 2003, 100, 8418–8423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.; Fan, C.; Oh, D.S.; Marron, J.S.; He, X.; Qaqish, B.F.; Livasy, C.; Carey, L.A.; Reynolds, E.; Dressler, L.; et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genom. 2006, 7, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Murray, Y.; Thun, M.J. Cancer Statistics, 2008. CA Cancer J. Clin. 2008, 58, 71–96. [Google Scholar] [CrossRef] [PubMed]
- Mikó, E.; Kovács, T.; Sebö, E.; Tóth, J.; Csonka, T.; Ujlaki, G.; Sipos, A.; Szabó, J.; Méhes, G.; Bai, P. Microbiome-Microbial Metabolome-Cancer Cell Interactions in Breast Cancer-Familiar, but Unexpected. Cells 2019, 8, 293. [Google Scholar] [CrossRef] [Green Version]
- Jovel, J.; Patterson, J.; Wang, W.; Hotte, N.; O’Keefe, S.; Mitchel, T.; Perry, T.; Kao, D.; Mason, A.L.; Madsen, K.L.; et al. Characterization of the Gut Microbiome Using 16S or Shotgun Metagenomics. Front. Microbiol. 2016, 7, 459. [Google Scholar] [CrossRef] [Green Version]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef] [Green Version]
- Fricker, A.M.; Podlesny, D.; Fricke, W.F. What is new and relevant for sequencing-based microbiome research? A mini-review. J. Adv. Res. 2019, 19, 105–112. [Google Scholar] [CrossRef]
- Shakya, M.; Lo, C.C.; Chain, P.S.G. Advances and Challenges in Metatranscriptomic Analysis. Front. Genet. 2019, 10, 904. [Google Scholar] [CrossRef] [Green Version]
- Earle, K.A.; Billings, G.; Sigal, M.; Lichtman, J.S.; Hans-son, G.C.; Elias, J.E.; Amieva, M.R.; Huang, K.C.; Sonnenburg, J.L. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe 2015, 18, 478–488. [Google Scholar] [CrossRef] [Green Version]
- Lukumbuzya, M.; Schmid, M.; Pjevac, P.; Daims, H.A. Multicolor Fluorescence in situ Hybridization Approach Using an Extended Set of Fluorophores to Visualize Microorganisms. Front. Microbiol. 2019, 10, 1383. [Google Scholar] [CrossRef] [PubMed]
- Willis, J.R.; Gabaldón, T. The Human Oral Microbiome in Health and Disease: From Sequences to Ecosystems. Microorganisms 2020, 8, 308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lacey, J.V.; Kreimer, A.R.; Buys, S.S.; Marcus, P.M.; Chang, S.C.; Leitzmann, M.F.; Hoover, R.N.; Prorok, P.C.; Berg, C.D.; Hartge, P.; et al. Breast cancer epidemiology according to recognized breast cancer risk factors in the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial Cohort. BMC Cancer 2009, 9, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madigan, M.P.; Ziegler, R.G.; Benichou, J.; Byrne, C.; Hoover, R.N. Proportion of breast cancer cases in the United States explained by well-established risk factors. J. Natl. Cancer Inst. 1995, 87, 1681–1685. [Google Scholar] [CrossRef] [Green Version]
- Fernández, M.F.; Reina-Pérez, I.; Astorga, J.M.; Rodríguez-Carrillo, A.; Plaza-Díaz, J.; Fontana, L. Breast Cancer and Its relationship with the microbiota. Int. J. Environ. Res. Public Health 2018, 15, 1747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Gemert, W.A.; Lanting, C.I.; Goldbohm, R.A.; van den Brandt, P.A.; Grooters, H.G.; Kampman, E.; Kiemeney, L.A.L.M.; van Leeuwen, F.E.; Monninkhof, E.M.; de Vries, E.; et al. The proportion of postmenopausal breast cancer cases in the Netherlands attributable to lifestyle-related risk factors. Breast Cancer Res. Treat. 2015, 152, 155–162. [Google Scholar] [CrossRef] [Green Version]
- Peterson, J.; Garges, S.; Giovanni, M.; McInnes, P.; Wang, L.; Schloss, J.A.; Bonazzi, V.; McEwen, J.E.; Wetterstrand, K.A.; Deal, C.; et al. The NIH Human Microbiome Project. Genome Res. 2009, 19, 2317–2323. [Google Scholar] [CrossRef] [Green Version]
- Plottel, C.S.; Blaser, M.J. Microbiome and malignancy. Cell Host Microbe 2011, 10, 324–335. [Google Scholar] [CrossRef] [Green Version]
- Bultman, S.J. Emerging roles of the microbiome in cancer. Carcinogenesis 2014, 35, 249–255. [Google Scholar] [CrossRef] [Green Version]
- Amieva, M.R.; El-Omar, E.M. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology 2008, 134, 306–323. [Google Scholar] [CrossRef] [Green Version]
- Arthur, J.C.; Jobin, C. The complex interplay between inflammation, the microbiota and colorectal cancer. Gut Microbes 2013, 4, 253–258. [Google Scholar] [CrossRef] [Green Version]
- Urbaniak, C.; Cummins, J.; Brackstone, M.; Macklaim, J.M.; Gloor, G.B.; Baban, C.K.; Scott, L.; O’Hanlon, D.M.; Burton, J.P.; Francis, K.P.; et al. Microbiota of human breast tissue. Appl. Environ. Microbiol. 2014, 80, 3007–3014. [Google Scholar] [CrossRef] [Green Version]
- Ramsay, D.T.; Kent, J.C.; Owens, R.A.; Hartmann, P.E. Ultrasound imaging of milk ejection in the breast of lactating women. Pediatrics 2004, 113, 361–367. [Google Scholar] [CrossRef]
- Donnet-Hughes, A.; Perez, P.F.; Doré, J.; Lecrerc, M.; Levenez, F.; Benyacoub, J.; Serrant, P.; Segura-Roggero, I.; Schiffrin, E.J. Potential role of the intestinal microbiota of the mother in neonatal immune education. Proc. Nutr. Soc. 2010, 69, 407–415. [Google Scholar] [CrossRef] [Green Version]
- Xuan, C.; Shamonki, J.M.; Chung, A.; Donome, M.L.; Chung, M.; Sieling, P.A.; Lee, D.J. Microbial dysbiosis is associated with human breast cancer. PLoS ONE 2014, 9, e83744. [Google Scholar] [CrossRef] [Green Version]
- Yazdi, H.R.; Movafagh, A.; Fallah, F.; Alizadeh Shargh, S.; Mansouri, N.; Heidary Pour, A.; Hashemi, M. Evaluation of Methylobacterium radiotolerance and Sphyngomonas yanoikoaie in sentinel lymph nodes of breast cancer cases. Asian Pac. J. Cancer Prev. 2016, 17, 279–285. [Google Scholar] [CrossRef]
- Wang, H.; Altemus, J.; Niazi, F.; Green, H.; Calhoun, B.C.; Sturgis, C.; Grobmyer, S.R.; Eng, C. Breast tissue, oral and urinary microbiomes in breast cancer. Oncotarget 2017, 8, 88122–88138. [Google Scholar] [CrossRef] [Green Version]
- Thompson, K.J.; Ingle, J.N.; Tang, X.; Chia, N.; Jeraldo, P.R.; Walther-Antonio, M.R.; Kandimalla, K.K.; Johnson, S.; Yao, J.Z.; Harrington, S.C.; et al. A comprehensive analysis of breast cancer microbiota and host gene expression. PLoS ONE 2017, 12, e0188873. [Google Scholar] [CrossRef]
- Meng, S.; Chen, B.; Yang, J.; Wang, J.; Zhu, D.; Meng, Q.; Zhang, L. Study of Microbiomes in Aseptically Collected Samples of Human Breast Tissue Using Needle Biopsy and the Potential Role of in situ Tissue Microbiomes for Promoting Malignancy. Front. Oncol. 2018, 8, 318. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Tian, T.; Wei, Z.; Shih, N.; Feldman, M.D.; Peck, K.N.; DeMichele, A.M.; Alwine, J.C.; Robertson, E.S. Distinct Microbial Signatures Associated with different breast cancer types. Front. Microbiol. 2018, 15, 951. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Wei, Z.; Tan, F.; Peck, K.N.; Shih, N.; Feldman, M.; Rebbeck, T.R.; Alwine, J.C.; Robertson, E.S. Distinct microbiological signatures associated with triple negative breast cancer. Sci. Rep. 2015, 5, 15162. [Google Scholar] [CrossRef] [PubMed]
- Constantini, L.; Magno, S.; Albanese, D.; Donati, C.; Molinari, R.; Filippone, A.; Masetti, R.; Merendino, N. Characterization of human breast tissue microbiota from core needle biopsies through the analysis of multi hypervariable 16S-rRNA gene regions. Sci. Rep. 2018, 8, 16893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urbaniak, C.; Gloor, G.B.; Brackstone, M.; Scott, L.; Tangney, M.; Reid, G. The microbiota of breast tissue and its association with breast cancer. Appl. Environ. Microbiol. 2016, 82, 5039–5048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hieken, T.J.; Chen, J.; Hoskin, T.L.; Walther-Antonio, M.; Johnson, S.; Ramaker, S.; Xiao, J.; Radisky, D.C.; Knutson, K.L.; Kalari, K.R.; et al. The Microbiome of Aseptically Collected Human Breast Tissue in Benign and Malignant Disease. Sci. Rep. 2016, 6, 30751. [Google Scholar] [CrossRef]
- Chan, A.A.; Bashir, M.; Rivas, M.N.; Duvall, K.; Sieling, P.A.; Pieber, T.R.; Vaishampayan, P.A.; Love, S.M.; Lee, D.J. Characterization of the microbiome of nipple aspirate fluid of breast cancer survivors. Sci. Rep. 2016, 6, 28061. [Google Scholar] [CrossRef] [Green Version]
- Chiba, A.; Bawaneh, A.; Velazquez, C.; Clear, K.Y.J.; Wilson, A.S.; Howard-McNatt, M.; Levine, E.A.; Levi-Polyachenko, N.; Yates-Alston, S.A.; Diggle, S.P.; et al. Neoadjuvant Chemotherapy Shifts Breast Tumor Microbiota Populations to Regulate Drug Responsiveness and the Development of Metastasis. Mol. Cancer Res. 2019, 18, 120–129. [Google Scholar] [CrossRef] [Green Version]
- Bard, J.M.; Luu, T.H.; Dravet, F.; Michel, C.; Moyon, T.; Pagniez, A.; Nazih, H.; Bobin-Dubigeon, C. Relationship between intestinal microbiota and clinical characteristics of patients with early stage breast cancer. FASEB J. 2015, 29. [Google Scholar] [CrossRef]
- Luu, T.H.; Michel, C.; Bard, J.M.; Dravet, F.; Nazih, H.; Bobin-Dubigeon, C. Intestinal Proportion of Blautia spp. is associated with clinical stage and histoprognostic grade in patients with early-stage breast cancer. Nutr. Cancer 2017, 69, 267–275. [Google Scholar] [CrossRef]
- Frugé, A.D.; Van der Pol, W.; Rogers, L.Q.; Morrow, C.D.; Tsuruta, Y.; Demark-Wahnefried, W. Fecal Akkermansia muciniphila Is Associated with Body Composition and Microbiota Diversity in Overweight and Obese Women with Breast Cancer Participating in a Presurgical Weight Loss Trial. J. Acad. Nutr. Diet. 2020, 120, 650–659. [Google Scholar] [CrossRef] [Green Version]
- Goedert, J.J.; Jones, G.; Hua, X.; Xu, X.; Yu, G.; Flores, R.; Falk, R.T.; Gail, M.H.; Shi, J.; Ravel, J.; et al. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: A population-based case-control pilot study. J. Natl. Cancer Inst. 2015, 107. [Google Scholar] [CrossRef]
- Zhu, J.; Liao, M.; Yao, Z.; Liang, W.; Li, Q.; Liu, J.; Yang, H.; Ji, Y.; Wei, W.; Tan, A.; et al. Breast cancer in postmenopausal women is associated with an altered gut metagenome. Microbiome 2018, 6, 136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuhrman, B.J.; Feigelson, H.S.; Flores, R.; Gail, M.H.; Xu, X.; Ravel, J.; Goedert, J.J. Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women. J. Clin. Endocrinol. Metab. 2014, 99, 4632–4640. [Google Scholar] [CrossRef] [PubMed]
- Goedert, J.J.; Hua, X.; Bielecka, A.; Okayasu, I.; Milne, G.L.; Jones, G.S.; Fujiwara, M.; Sinha, R.; Wan, Y.; Xu, X.; et al. Postmenopausal breast cancer and oestrogen associations with the IgA-coated and IgA-noncoated faecal microbiota. Br. J. Cancer 2018, 118, 471–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchta Rosean, C.; Bostic, R.R.; Ferey, J.C.M.; Feng, T.Y.; Azar, F.N.; Tung, K.S.; Dozmorov, M.G.; Smirnova, E.; Bos, P.D.; Rutkowski, M.R. Preexisting Commensal Dysbiosis Is a Host-Intrinsic Regulator of Tissue Inflammation and Tumor Cell Dissemination in Hormone Receptor-Positive Breast Cancer. Cancer Res. 2019, 79, 3662–3675. [Google Scholar] [CrossRef] [Green Version]
- Horigome, A.; Okubo, R.; Hamazaki, K.; Kinoshita, T.; Katsumata, N.; Uezono, Y.; Xiao, J.Z.; Matsuoka, Y.J. Association between blood omega-3 polyunsaturated fatty acids and the gut microbiota among breast cancer survivors. Benef. Microbes 2019, 10, 751–758. [Google Scholar] [CrossRef]
- Kirkup, B.M.; McKee, A.; Makin, K.A.; Paveley, J.; Caim, S.; Alcon-Giner, C.; Leclaire, C.; Dalby, M.; Le Gall, G.; Andrusaite, A.; et al. Perturbation of the gut microbiota by antibiotics results in accelerated breast tumour growth and metabolic dysregulation. bioRxiv 2019. [Google Scholar] [CrossRef]
- Hoffman, R.M. Development of recombinant methioninase to target the general cancer-specific metabolic defect of methionine dependence: A 40-year odyssey. Expert Opin. Biol. Ther. 2015, 15, 21–31. [Google Scholar] [CrossRef]
- Anderson, W.F.; Rosenberg, P.S.; Prat, A.; Perou, C.M.; Sherman, M.E. How many etiological subtypes of breast cancer: Two, three, four, or more? J. Natl. Cancer Inst. 2014, 106. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [Green Version]
- Flores, R.; Shi, J.; Fuhrman, B.; Xu, X.; Veenstra, T.D.; Gail, M.H.; Gajer, P.; Ravel, J.; Goedert, J.J. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: A cross-sectional study. J. Transl. Med. 2012, 10, 253–258. [Google Scholar] [CrossRef] [Green Version]
- Rea, D.; Coppola, G.; Palma, G.; Barbieri, A.; Luciano, A.; Del Prete, P.; Rossetti, S.; Berretta, M.; Facchini, G.; Perdonà, S.; et al. Microbiota effects on cancer: From risks to therapies. Oncotarget 2018, 9, 17915–17927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dabek, M.; McCrae, S.I.; Stevens, V.J.; Duncan, S.H.; Louis, P. Distribution of beta-glucosidase and beta-glucuronidase activity and of beta-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol. Ecol. 2008, 66, 487–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores, R.; Shi, J.; Gail, M.H.; Gajer, P.; Ravel, J.; Goedert, J.J. Association of fecal microbial diversity and taxonomy with selected enzymatic functions. PLoS ONE 2012, 7, e39745. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Tan, Q.; Fu, Q.; Zhou, Y.; Hu, Y.; Tang, S.; Zhou, Y.; Zhang, J.; Qiu, J.; Lv, Q. Gastrointestinal microbiome and breast cancer: Correlations, mechanisms and potential clinical implications. Breast Cancer 2017, 24, 220–228. [Google Scholar] [CrossRef]
- Savas, P.; Salgado, R.; Denkert, C.; Sotitiou, C.; Darcy, P.K.; Smyth, M.J.; Loi, S. Clinical relevance of host immunity in breast cancer: From TILs to the clinic. Nat. Rev. Clin. Oncol. 2016, 13, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.R.; Provenzano, E.; Dawson, S.J.; Blows, F.M.; Liu, B.; Shah, M.; Earl, H.M.; Poole, C.J.; Hiller, L.; Dunn, J.A.; et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann. Oncol. 2014, 25, 1536–1543. [Google Scholar] [CrossRef]
- Liu, S.; Foulkes, W.D.; Leung, S.; Gao, D.; Lau, S.; Kos, Z.; Nielsen, T.O. Prognostic significance of FOXP3+ tumor infiltrating lymphocytes in breast cancer depends on estrogen receptor and human epidermal growth factor receptor-2 expression status and concurrent cytotoxic T-cell infiltration. Breast Cancer Res. 2014, 16, 432. [Google Scholar] [CrossRef] [Green Version]
- Bates, G.J.; Fox, S.B.; Han, C.; Leek, R.D.; Garcia, J.F.; Harris, A.L.; Banham, A.H. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J. Clin. Oncol. 2006, 24, 5373–5380. [Google Scholar] [CrossRef]
- Gupta, R.; Babb, J.S.; Singh, B.; Chiriboga, L.; Liebes, L.; Adams, S.; Demaria, S. The numbers of FoxP3+ lymphocytes in sentinel lymph nodes of breast cancer patients correlate with primary tumor size but not nodal status. Cancer Investig. 2011, 29, 419–425. [Google Scholar] [CrossRef] [Green Version]
- Curiel, T.J.; Coukos, G.; Zou, L.; Alvarez, X.; Cheng, P.; Mottram, P.; Evdemon-Hogan, M.; Conejo-Cargía, J.R.; Zhang, L.; Burow, M.; et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 2004, 10, 942–949. [Google Scholar] [CrossRef]
- DeNardo, D.G.; Coussens, L.M. Inflammation and breast cancer. Balancing immune response: Crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007, 9, 212. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, L.M.S.; Gomes, S.T.M.; Ishak, R.; Vallinoto, A.C.R. Regulatory T Cell and Forkhead Box Protein 3 as Modulators of Immune Homeostasis. Front. Immunol. 2017, 8, 605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, S.H.; Yu, J. Gut microbiota in colorectal cancer: Mechanisms of action and clinical applications. Nat. Rev. Gastro. Hepat. 2019, 16, 690–704. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Rutkowski, M.R.; Svoronos, N.; Perales-Puchalt, A.; Conejo-Garcia, J.R. The Tumor Macroenvironment: Cancer-Promoting Networks Beyond Tumor Beds. Adv. Cancer Res. 2015, 128, 235–262. [Google Scholar] [CrossRef] [Green Version]
- Wong, D.V.T.; Lima-Júnior, R.C.P.; Carvalho, C.B.M.; Borges, V.F.; Wanderley, C.W.S.; Bem, A.X.C.; Leite, C.A.V.G.; Teixeira, M.A.; Batista, G.L.P.; Silva, R.L.; et al. The Adaptor Protein Myd88 Is a Key Signaling Molecule in the Pathogenesis of Irinotecan-Induced Intestinal Mucositis. PLoS ONE 2015, 10, e0139985. [Google Scholar] [CrossRef]
- Sonis, S.T. A biological approach to mucositis. J. Support Oncol. 2004, 2, 21–32. [Google Scholar]
- Pandey, S.; Singh, S.; Anang, V.; Bhatt, A.N.; Natarajan, K.; Dwarakanath, B.S. Pattern Recognition Receptors in Cancer Progression and Metastasis. Cancer Growth Metastasis 2015, 8, 25–34. [Google Scholar] [CrossRef]
- Bhatelia, K.; Singh, K.; Singh, R. TLRs: Linking inflammation and breast cancer. Cell. Signal. 2014, 26, 2350–2357. [Google Scholar] [CrossRef] [PubMed]
- Moser, A.R.; Mattes, E.M.; Dove, W.F.; Lindstrom, M.J.; Haag, J.D.; Gould, M.N. ApcMin, a mutation in the murine Apc gene, predisposes to mammary carcinomas and focal alveolar hyperplasias. Proc. Natl. Acad. Sci. USA 1993, 90, 8977–8981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, V.P.; Poutahidis, T.; Ge, Z.; Nambiar, P.R.; Boussahmain, C.; Wang, Y.Y.; Horwitz, B.H.; Fox, J.G.; Erdman, S.E. Innate immune inflammatory response against enteric bacteria Helicobacter hepaticus induces mammary adenocarcinoma in mice. Cancer Res. 2006, 66, 7395–7400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, V.P.; Poutahidis, T.; Ge, Z.; Nambiar, P.R.; Horwitz, B.H.; Fox, J.G.; Erdman, S.E. Proinflammatory CD4+ CD45RB (hi) lymphocytes promote mammary and intestinal carcinogenesis in Apc(Min/+) mice. Cancer Res. 2006, 66, 57–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toland, A. Aberrant epigenetic regulation in breast cancer. In Patho-Epigenetics of Disease; Miranovitz, J., Niller, H., Eds.; Springer: New York, NY, USA, 2012; pp. 91–122. [Google Scholar] [CrossRef]
- Jeffery, I.B.; O’Toole, P.W. Diet-microbiota interactions and their implications for healthy living. Nutrients 2013, 5, 234–252. [Google Scholar] [CrossRef] [PubMed]
- Canani, R.B.; Costanzo, M.; Leone, L. The epigenetic effects of butyrate: Potential therapeutic implications for clinical practice. Clin. Epigenetics 2012, 4, 4. [Google Scholar] [CrossRef]
- Paul, B.; Barnes, S.; Demark-Wahnefried, W.; Morrow, C.; Salvador, C.; Skibola, C.; Tollefsbol, T.O. Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin. Epigenet. 2015, 7, 112. [Google Scholar] [CrossRef] [Green Version]
- Anand, P.; Kunnumakkara, A.B.; Sundaram, C.; Harikumar, K.B.; Tharakan, S.T.; Lai, O.S.; Sung, B.; Aggarwal, B.B. Cancer is a preventable disease that requires major lifestyle changes. Pharm. Res. 2008, 25, 2097–2116. [Google Scholar] [CrossRef]
- Leeming, E.R.; Johnson, A.J.; Spector, T.D.; Le Roy, C.I. Effect of Diet on the Gut Microbiota: Rethinking Intervention Duration. Nutrients 2019, 11, 2862. [Google Scholar] [CrossRef] [Green Version]
- Klement, R.J.; Pazienza, V. Impact of Different Types of Diet on Gut Microbiota Profiles and Cancer Prevention and Treatment. Medicina 2019, 55, 84. [Google Scholar] [CrossRef] [Green Version]
- Ostan, R.; Lanzarini, C.; Pini, E.; Scurti, M.; Vianello, D.; Bertarelli, C.; Fabbri, C.; Izzi, M.; Palmas, G.; Biondi, F.; et al. Inflammaging and Cancer: A challenge for the mediterranean diet. Nutrients 2015, 7, 2589–2621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Lacoba, R.; Pardo-Garcia, I.; Amo-Saus, E.; Escribano-Sotos, F. Mediterranean diet and health outcomes: A systematic meta-review. Eur. J. Public Health 2018, 28, 955–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shively, C.A.; Register, T.C.; Appt, S.E.; Clarkson, T.B.; Uberseder, B.; Clear, K.Y.J.; Wilson, A.S.; Chiba, A.; Tooze, J.A.; Cook, K.L. Consumption of Mediterranean versus Western Diet Leads to Distinct Mammary Gland Microbiome Populations. Cell Rep. 2018, 25, 47–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pellegrini, M.; Ippolito, M.; Monge, T.; Violi, R.; Cappello, P.; Ferrocino, I.; Cocolin, L.S.; De Francesco, A.; Bo, S.; Finocchiaro, C. Gut microbiota composition after diet and probiotics in overweight breast cancer survivors: A randomized open-label pilot intervention trial. Nutrition 2020, 74, 110749. [Google Scholar] [CrossRef]
- Wieërs, G.; Belkhir, L.; Enaud, R.; Leclercq, S.; Philippart de Foy, J.M.; Dequenne, I.; de Timary, P.; Cani, P.D. How Probiotics Affect the Microbiota. Front. Cell. Infect. Microbiol. 2020, 9, 454. [Google Scholar] [CrossRef] [Green Version]
- Terpou, A.; Papadaki, A.; Lappa, I.K.; Kachrimanidou, V.; Bosnea, L.A.; Kopsahelis, N. Probiotics in Food Systems: Significance and Emerging Strategies Towards Improved Viability and Delivery of Enhanced Beneficial Value. Nutrients 2019, 11, 1591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Moreno de LeBlanc, A.; Matar, C.; Theriault, C.; Perdigon, G. Effects of milk fermented by Lactobacillus helveticus R389 on immune cells associated to mammary glands in normal and a breast cancer model. Immunobiology 2005, 210, 349–358. [Google Scholar] [CrossRef]
- Yazdi, M.H.; Soltan Dallal, M.M.; Hassan, Z.M.; Holakuyee, M.; Agha Amiri, S.; Abolhassani, M.; Mahdavi, M. Oral administration of Lactobacillus acidophilus induces IL-12 production in spleen cell culture of. Br. J. Nutr. 2010, 104, 227–232. [Google Scholar] [CrossRef] [Green Version]
- Lakritz, J.R.; Poutahidis, T.; Levkovich, T.; Varian, B.J.; Ibrahim, Y.M.; Chatzigiagkos, A.; Mirabal, S.; Alm, E.J.; Erdman, S.E. Beneficial bacteria stimulate host immune cells to counteract dietary and genetic predisposition to mammary cancer in mice. Int. J. Cancer 2014, 135, 529–540. [Google Scholar] [CrossRef] [Green Version]
- Kassayová, M.; Bobrov, N.; Strojný, L.; Kisková, T.; Mikeš, J.; Demečková, V.; Orendáš, P.; Bojková, B.; Péč, M.; Kubatka, P.; et al. Preventive effects of probiotic bacteria Lactobacillus plantarum and dietary fiber in chemically-induced mammary carcinogenesis. Anticancer Res. 2014, 34, 4969–4975. [Google Scholar]
- Imani Fooladi, A.A.; Yazdi, M.H.; Pourmand, M.R.; Mirshafiey, A.; Hassan, Z.M.; Azizi, T.; Mahdavi, M.; Soltan Dallal, M.M. Th1 Cytokine Production Induced by Lactobacillus acidophilus in BALB/c Mice Bearing Transplanted Breast Tumor. Jundishapur J. Microbiol. 2015, 8, e17354. [Google Scholar] [CrossRef] [Green Version]
- Hassan, Z.; Mustafa, S.; Rahim, R.A.; Isa, N.M. Anti-breast cancer effects of live, heat-killed and cytoplasmic fractions of Enterococcus faecalis and Staphylococcus hominis isolated from human breast milk. In Vitro Cell. Dev. Biol. Anim. 2016, 52, 337–348. [Google Scholar] [CrossRef]
- Zamberi, N.R.; Abu, N.; Mohamed, N.E.; Nordin, N.; Keong, Y.S.; Beh, B.K.; Zakaria, Z.A.B.; Rahman, N.M.A.N.A.; Alitheen, N.B. The Antimetastatic and Antiangiogenesis Effects of Kefir Water on Murine Breast Cancer Cells. Integr. Cancer Ther. 2016, 15, NP53–NP66. [Google Scholar] [CrossRef] [PubMed]
- Serban, D.E. Gastrointestinal cancers: Influence of gut microbiota, probiotics and prebiotics. Cancer Lett. 2014, 345, 258–270. [Google Scholar] [CrossRef] [PubMed]
- Vivarelli, S.; Falzone, L.; Basile, M.S.; Nicolosi, D.; Genovese, C.; Libra, M.; Salmeri, M. Benefits of using probiotics as adjuvants in anticancer therapy (Review). World Acad. Sci. J. 2019, 1, 125–135. [Google Scholar] [CrossRef]
- Toi, M.; Hirota, S.; Tomotaki, A.; Sato, N.; Hozumi, Y.; Anan, K.; Nagashima, T.; Tokuda, Y.; Masuda, N.; Ohsumi, S.; et al. Probiotic Beverage with Soy Isoflavone Consumption for Breast Cancer Prevention: A Case-Control Study. Curr. Nutr. Food Sci. 2013, 9, 194–200. [Google Scholar] [CrossRef]
- Davani-Davari, D.; Negahdaripour, M.; Karimzadeh, I.; Seifan, M.; Mohkam, M.; Masoumi, S.J.; Berenjian, A.; Ghasemi, Y. Prebiotics: Definition, Types, Sources, Mechanisms, and Clinical Applications. Foods 2019, 8, 92. [Google Scholar] [CrossRef] [Green Version]
- Levy, J.I.; Fabian, M.P.; Peters, J.L. Meta-Analytic Approaches for Multistressor Dose-Response Function Development: Strengths, Limitations, and Case Studies. Risk Anal. 2015, 35, 1040–1049. [Google Scholar] [CrossRef]
- Chen, S.; Chen, Y.; Ma, S.; Zheng, R.; Zhao, P.; Zhang, L.; Liu, Y.; Yu, Q.; Deng, Q.; Zhang, K. Dietary fibre intake and risk of breast cancer: A systematic review and meta-analysis of epidemiological studies. Oncotarget 2016, 7, 80980–80989. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, R.; Rylander-Rudqvist, T.; Ye, W.; Saji, S.; Adlercreutz, H.; Wolk, A. Dietary fiber intake and risk of postmenopausal breast cancer defined by estrogen and progesterone receptor status-a prospective cohort study among Swedish women. Int. J. Cancer 2008, 122, 403–412. [Google Scholar] [CrossRef]
- Zengul, A.G.; Demark-Wahnefried, W.; Barnes, S.; Morrow, C.D.; Bertrand, B.; Berryhil, T.F.; Frugé, A.D. Associations between Dietary Fiber, the Fecal Microbiota and Estrogen Metabolism in Postmenopausal Women with Breast Cancer. Nutr. Cancer. 2020, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Navarro, S.L.; Li, F.; Lampe, J.W. Mechanisms of action of isothiocyanates in cancer chemoprevention: An update. Food Funct. 2011, 2, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Kolaátorovaá, L.; Lapčík, O.; Staárka, L. Phytoestrogens and the intestinal microbiome. J. Physiol. Res. 2018, 67, S401–S408. [Google Scholar] [CrossRef] [PubMed]
- Stojanov, S.; Kreft, S. Gut Microbiota and the Metabolism of Phytoestrogens. Rev. Bras. Farmacogn. 2020, 30, 145–154. [Google Scholar] [CrossRef]
- Zhu, Y.; Kawaguchi, K.; Kiyama, R. Differential and directional estrogenic signaling pathways induced by enterolignans and their precursors. PLoS ONE 2017, 12, e0171390. [Google Scholar] [CrossRef] [Green Version]
- Zaineddin, A.K.; Vrieling, A.; Buck, K.; Becker, S.; Linseisen, J.; Flesh-Janys, D.; Kaaks, R.; Chang-Claude, J. Serum enterolactone and postmenopausal breast cancer risk by estrogen, progesterone and herceptin 2 receptor status. Int. J. Cancer 2012, 130, 1401–1410. [Google Scholar] [CrossRef]
- Mabrok, H.B.; Klopfleisch, R.; Ghanem, K.Z.; Clavel, T.; Blaut, M.; Loh, G. Lignan transformation by gut bacteria lowers tumor burden in a gnotobiotic rat model of breast cancer. Carcinogenesis 2012, 33, 203–208. [Google Scholar] [CrossRef] [Green Version]
- Fink, B.N.; Steck, S.E.; Wolff, M.S.; Britton, J.A.; Kabat, G.C.; Gaudet, M.M.; Abrahamson, P.E.; Bell, P.; Schroeder, J.C.; Teitelbaum, S.L. Dietary flavonoid intake and breast cancer survival among women on Long Island. Cancer Epidemiol. Biomarkers Prev. 2007, 16, 2285–2292. [Google Scholar] [CrossRef] [Green Version]
- Boyapati, S.M.; Shu, X.O.; Ruan, Z.X.; Dai, Q.; Cai, Q.; Gao, Y.T.; Zheng, W. Soyfood intake and breast cancer survival: A followup of the Shanghai Breast Cancer Study. Breast Cancer Res. Treat. 2005, 92, 11–17. [Google Scholar] [CrossRef]
- Velentzis, L.S.; Woodside, J.V.; Cantwell, M.M.; Leathem, A.J.; Keshtgar, M.R. Do phytoestrogens reduce the risk of breast cancer and breast cancer recurrence? What clinicians need to know. Eur. J. Cancer 2008, 44, 1799–1806. [Google Scholar] [CrossRef] [Green Version]
- Swann, R.; Perkins, K.A.; Velentzis, L.S.; Ciria, c.; Dutton, S.J.; Mulligan, A.A.; Woodside, J.V.; Cantwell, M.M.; Leathem, A.J.; Robertson, C.E. The DietCompLyf study: A prospective cohort study of breast cancer survival and phytoestrogen consumption. Maturitas 2013, 75, 232–240. [Google Scholar] [CrossRef] [Green Version]
- Verheus, M.; van Gils, C.H.; Keinan-Boker, L.; Grace, P.B.; Bingham, S.A.; Peeters, P.H. Plasma phytoestrogens and subsequent breast cancer risk. J. Clin. Oncol. 2007, 25, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Holley, D.; Collins, L.B.; Montgomery, S.A.; Whitmore, A.C.; Hillhouse, A.; Curry, K.P.; Renner, S.W.; Greenwalt, A.; Ryan, E.; et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014, 4, 1387–1397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salimi, V.; Shahsavari, Z.; Safizadeh, B.; Hosseini, A.; Khademian, N.; Tavakoli-Yaraki, M. Sodium butyrate promotes apoptosis in breast cancer cells through reactive oxygen species (ROS) formation and mitochondrial impairment. Lipids Health Dis. 2017, 16, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.J.; Kim, J.S. The effect of combined therapy with 5-aza-2′-deoxycylidine, sodium butyrate, and tamoxifen on apoptosis of breast cancer cell lines. J. Clin. Oncol. 2011, 29. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, P.C.; Ma, Y.B.; Fan, R.; Gao, F.F.; Zhang, J.W.; Wei, L. Sodium butyrate-induced apoptosis and ultrastructural changes in MCF-7 breast cancer cells. Ultrastruct. Pathol. 2016, 40, 200–204. [Google Scholar] [CrossRef]
- Puupponen-Pimiä, R.; Aura, A.M.; Oksman-Caldentey, K.M.; Myllärinen, P.; Saarela, M.; Mattila-Sandholm, T.; Poutanen, K. Development of functional ingredients for gut health. Trends Food Sci. Tech. 2002, 13, 3–11. [Google Scholar] [CrossRef]
- Sharma, M.; Arora, I.; Stoll, M.L.; Li, Y.; Morrow, C.D.; Barnes, S.; Berryhill, T.F.; Li, S.; Tollefsbol, T.O. Nutritional combinatorial impact on the gut microbiota and plasma short-chain fatty acids levels in the prevention of mammary cancer in Her2/neu estrogen receptor-negative transgenic mice. bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, G.; Wang, Y.; Zhang, Y.; Wan, X.; Li, J.; Liu, K.; Wang, F.; Liu, K.; Liu, Q.; Yang, C.; et al. Anti-cancer activities of tea epigallocatechin-3-gallate in breast cancer patients under radiotherapy. Curr. Mol. Med. 2012, 12, 163–176. [Google Scholar] [CrossRef] [Green Version]
- Sheng, J.; Shi, W.; Guo, H.; Long, W.; Wang, Y.; Qi, J.; Liu, J.; Xu, Y. The Inhibitory Effect of (-)-Epigallocatechin-3-Gallate on Breast Cancer Progression via Reducing SCUBE2 Methylation and DNMT Activity. Molecules 2019, 24, 2899. [Google Scholar] [CrossRef] [Green Version]
- Kupcinskas, J.; Hold, G.L. Microbiota and cancer therapy. Microb. Health Dis. 2019, 1, e185. [Google Scholar]
- Xu, C.; Ruan, B.; Jiang, Y.; Xue, T.; Wang, Z.; Lu, H.; Wei, M.; Wang, S.; Ye, Z.; Zhai, D.; et al. Antibiotics-induced gut microbiota dysbiosis promotes tumor initiation via affecting APC-Th1 development in mice. Biochem. Biophys. Res. Commun. 2017, 488, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velicer, C.M.; Heckbert, S.R.; Lampe, J.W.; Potter, J.D.; Robertson, C.A.; Taplin, S.H. Antibiotic use in relation to the risk of breast cancer. JAMA 2004, 291, 827–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simin, J.; Tamimi, R.M.; Engstrand, L.; Callens, S.; Brusselaers, N. Antibiotic use and the risk of breast cancer: A systematic review and dose-response meta-analysis. Pharmacol. Res. 2020, 160, 105072. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Douglass, J.; Prasath, V.; Neace, M.; Atrchian, S.; Manjili, M.H.; Shokouhi, S.; Habibi, M. The microbiome and breast cancer: A review. Breast Cancer Res. Treat. 2019, 178, 493–496. [Google Scholar] [CrossRef]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [Green Version]
- Saad, R.; Rizkallah, M.R.; Aziz, R.K. Gut Pharmacomicrobiomics: The tip of an iceberg of complex interactions between drugs and gut-associated microbes. Gut Pathog. 2012, 4, 16. [Google Scholar] [CrossRef] [Green Version]
- Viaud, S.; Daillère, R.; Boneca, I.G.; Lepage, P.; Langella, P.; Chamaillard, M.; Pittet, M.J.; Ghiringhelli, F.; Trinchieri, G.; Goldszmid, R.; et al. Gut microbiome and anticancer immune response: Really hot Sh*t! Cell Death Differ. 2015, 22, 199–214. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Liu, K.X.; Qu, J.M.; Wang, X.D. The changes induced by cyclophosphamide in intestinal barrier and microflora in mice. Eur. J. Pharmacol. 2013, 714, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Salva, S.; Marranzino, G.; Villena, J.; Agüero, G.; Alvarez, S. Probiotic Lactobacillus strains protect against myelosuppression and immunosuppression in cyclophosphamide-treated mice. Int. Immunopharmacol. 2014, 22, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Westman, E.L.; Canova, M.J.; Radhi, I.J.; Koteva, K.; Kireeva, I.; Waglechner, N.; Wright, G.D. Bacterial inactivation of the anticancer drug doxorubicin. Chem. Biol. 2012, 19, 1255–1264. [Google Scholar] [CrossRef] [Green Version]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexander, J.L.; Wilson, I.D.; Teare, J.; Marchesi, J.R.; Nicholson, J.K.; Kinross, J.M. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 356–365. [Google Scholar] [CrossRef]
- Daillère, R.; Vétizou, M.; Waldschmitt, N.; Yamazaki, T.; Isnard, C.; Poirier-Colame, V.; Duong, C.P.M.; Flament, C.; Lepage, P.; Roberti, M.P.; et al. Enterococcus hirae and Barnesiella intestinihominis Facilitate Cyclophosphamide-Induced Therapeutic Immunomodulatory Effects. Immunity 2016, 45, 931–943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, D.J.; Pan, J.; Yu, H.L.; Zheng, G.W.; Xu, J.H. Target-oriented discovery of a new esterase-producing strain Enterobacter sp. ECU1107 for whole cell-catalyzed production of (2S,3R)-3-phenylglycidate as a chiral synthon of Taxol. Appl. Microbiol. Biotechnol. 2013, 97, 6293–6300. [Google Scholar] [CrossRef]
- Oelschlaeger, T.A.; Tall, B.D. Invasion of cultured human epithelial cells by Klebsiella pneumoniae isolated from the urinary tract. Infect. Immun. 1997, 65, 2950–2958. [Google Scholar] [CrossRef] [Green Version]
- Hussein, M.H.; Schneider, E.K.; Elliott, A.G.; Han, M.; Reyes-Ortega, F.; Morris, F.; Blastovich, M.A.T.; Jasim, R.; Currie, B.; Mayo, M.; et al. From Breast Cancer to Antimicrobial: Combating Extremely Resistant Gram-Negative “Superbugs” Using Novel Combinations of Polymyxin B with Selective Estrogen Receptor Modulators. Microb. Drug Resist. 2017, 23, 640–650. [Google Scholar] [CrossRef]
- Gerits, E.; Defraine, V.; Vandamme, K.; De Cremer, K.; De Brucker, K.; Thevissen, K.; Cammue, B.P.A.; Beullens, S.; Fauvart, M.; Verstraeten, N.; et al. Repurposing Toremifene for Treatment of Oral Bacterial Infections. Antimicrob. Agents Chemother. 2017, 61, e01846-16. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, A.C.; Didone, L.; Jobson, J.; Sofia, M.K.; Krysan, D.; Dunman, P.M. Adenylate kinase release as a high-throughput-screening-compatible reporter of bacterial lysis for identification of antibacterial agents. Antimicrob. Agents Chemother. 2013, 57, 26–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luxo, C.; Jurado, A.S.; Custódio, J.B.; Madeira, V.M. Toxic effects of tamoxifen on the growth and respiratory activity of Bacillus stearothermophilus. Toxicol. In Vitro 2001, 15, 303–305. [Google Scholar] [CrossRef] [Green Version]
- Byrd, C.A.; Bornmann, W.; Erdjument-Bromage, H.; Tempst, P.; Pavletich, N.; Rosen, N.; Nathan, F.; Ding, A. Heat shock protein 90 mediates macrophage activation by Taxol and bacterial lipopolysaccharide. Proc. Natl. Acad. Sci. USA 1999, 96, 5645–5650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Routy, B.; Gopalakrishnan, V.; Daillère, R.; Zitvogel, L.; Wargo, J.A.; Kroemer, G. The gut microbiota influences anticancer immunosurveillance and general health. Nat. Rev. Clin. Oncol. 2018, 15, 382–396. [Google Scholar] [CrossRef] [PubMed]
- van Vliet, M.J.; Harmsen, H.J.; de Bont, E.S.; Tissing, W.J. The role of intestinal microbiota in the development and severity of chemotherapy-induced mucositis. PLoS Pathog. 2010, 6, e1000879. [Google Scholar] [CrossRef] [Green Version]

| Study | Microbiome Tissue Related | Cohort | Sample Type | Main Methodology | Most Relevant Results |
|---|---|---|---|---|---|
| Xuan, C. et al., 2014 [25] | Breast | 20 patients ER+ BC | Breast tumor tissue and its paired normal adjacent tissue | Pyrosequencing V4 16S rDNA Pipeline: QIIME | ↑Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes and Verrucomicrobia (96.6%) in breast tissue. ↑Methylobacterium radiotolerans in BC tissue. ↑Sphingomonas yanoikuyae in paired normal tissue. |
| Urbaniak, C. et al., 2014 [22] | Breast | 43 Canadian women (11 with benign tumors, 27 cancerous tumors and 5 healthy individuals) and 38 Irish women (33 women with BC and 5 healthy individuals) | Breast tissue coming from lumpectomies, mastectomies and breast reductions | V6 16S rRNA sequencing (Ion Torrent) Pipeline: UCLUST | ↑Proteobacteria and Firmicutes in breast tissue. ↑Bacillus (11.4%) and Acinetobacter (10%) in Canadian women. ↑Enterobacteriaceae (30.8%) and Staphylococcus (12.7%) in Irish women. ↑Escherichia coli in BC tissue. |
| Yazdi, H.R. et al., 2016 [26] | Breast- Sentinel lymph | 123 sentinel lymph nodes and 123 normal adjacent breast tissue samples | Sentinel lymph nodes and breast tissue | RT-PCR and pyrosequencing | ↑Methylobacterium Radiotolerance in lymph cancer nodes samples compared to normal adjacent samples. |
| Wang, H. et al., 2017 [27] | Breast | 57 women with invasive breast carcinoma and 21 healthy women | Urine Bilateral breast tissue from control patients underwent cosmetic procedures Tumor and ipsilateral adjacent normal breast tissue for cases by mastectomy | V3-V4 16S rRNA sequencing (Illumina) Pipeline: UCLUST | ↓Methylobacterium and ↑Corynebacterium, Staphylococcus, Actinomyces and Propionibacteriaceae in patients with invasive breast carcinoma compared to healthy individuals. |
| Thompson, K.J. et al., 2017 [28] | Breast | 668 tumor tissues (HER2+, ER+, TNC) and 72 normal adjacent tissues from The Cancer Genome Atlas (TCGA) | Breast tumor tissues and normal adjacent tissues | V3-V5 16S rRNA amplified sequencing data | ↑Proteobacteria, Actinobacteria and Firmicutes in breast tissues. ↑Proteobacteria, Mycobacterium fortuitum and Mycobacterium phlei in BC samples. ↑Actinobacteria in normal adjacent tissue. |
| Meng, S. et al., 2018 [29] | Breast | 22 Chinese patients with benign tumor and 72 malignant BC patients | Breast tissue | V1-V2 16S rRNA sequencing (Illumina HiSeq) | ↑Propionicimonas, Micrococcaceae, Caulobacteraceae, Rhodobacteraceae, Nocardioidaceae and Methylobacteriaceae, in BC tissues (ethno-specific) ↓Bacteroidaceae and ↑ Agrococcus are related with malignancy |
| Banerjee, S. et al., 2018 [30] | Breast | 20 normal breast tissue and 148 BC tissue (50 ER or PR+, 34 HER2+, 24 TP and 40 TN) | Breast tissues | Pathochips array | ↑Proteobacteria ↑Actinomyces in the four BC subtypes studied. |
| Banerjee, S. et al., 2015 [31] | Breast | 100 women with triple negative BC (TNBC), 17 matched controls and 20 non-matched controls | Breast tissue. Matched controls were obtained from the adjacent non-cancerous breast tissue of the same patients with BC and non-matched were from different healthy women. | PathoChip array | ↑Brevundimonas diminuta, Arcanobacterium haemolyticum, Peptoniphilus indolicus, Prevotella nigrescens, Propiniobacterium jensenii and Capnocytophaga canimorsus in TNBC. Among virus, ↑ Herpesviridae, Retroviridae, Parapoxviridae, Polyomaviridae, Papillomaviridae in TNBC. |
| Costantini, L. et al., 2018 [32] | Breast | 16 Mediterranean patients with BC (12 samples were collected from core needle biopsies (CNB) and 7 from surgical excision biopsies (SEB); 3 patients were processed with both procedure) | Fresh tumor breast tissue and paired breast healthy tissue | V3 16S-rRNA gene amplicons sequencing (Ion Torrent) | ↑Ralstonia in breast tissue. No significant differences between healthy adjacent breast tissues and BC tissues. |
| Urbaniak, C. et al., 2016 [33] | Breast | 58 women: 13 benign, 45 cancerous tumors and 23 healthy women | Breast tissue | V6 16S rRNA gene sequencing (Illumina MiSeq) Pipeline: QIIME | ↑Bacillus, Enterobacteriaceae, Staphylococcus, Comamondaceae and Bacteroidetes and ↓ Prevotella, Lactococcus, Streptococcus, Corynebacterium and Staphylococcus in BC patients compared to healthy controls. |
| Hieken, T.J. et al., 2016 [34] | Breast | 28 women undergoing non-mastectomy breast surgery: 13 benign breast disease and 15 invasive BC (100% ER/PR+ and 29% HER2+) | Breast tissue and breast skin | V3-V5 16S rDNA hypervariable taq sequencing (Illumina MiSeq) Pipeline: IM-TORNADO | ↑Fusobacterium, Atopobium, Gluconacetobacter, Hydrogenophaga and Lactobacillus in BC tissue compared to healthy breast tissue. |
| Chan, A.A. et al., 2016 [35] | Breast | 25 women with breast ductal cancer and 23 healthy women | Nipple aspirate fluid (NAF) and aerolar breast skin | V4 16S rRNA gene sequencing (Illumina MiSeq) Pipeline: Mothur | ↑Alistipes and ↓ unclassified genus of the Sphingomonadaceae family in NAF from women with BC compared to healthy controls. |
| Chiba, A. et al., 2019 [36] | Breast | 15 women with BC who were treated with neoadjuvant chemotherapy, 18 women with no prior therapy at time of surgery and 9 women who had tumor recurrence | Snap-frozen breast tumor tissue | V4 16S rRNA amplicon sequencing (Illumina Miseq) Pipelinee: Mothur (v.1.39.5) Microarray for confirmation | ↑Pseudomonas spp. in BC tissue after neoadjuvant chemotherapy. ↓Prevotella in the tumor tissue from non-treated patients. ↑Brevundimonas and Staphylococcus in the primary breast tumors in patients developing distant metastases. |
| Bard, J.M. et al., 2015 [37] | Gut | 32 BC women: Invasive ductal (81%), stage 0 (46,9%), grade II (62,5%), ER/PgR+ (80%), HER2+ (15%) | Fecal samples | PCR detecting 16S rRNA gene specific sequences | ↓Blautia spp. in stage I compare to stage III BC. Significant differences in the abundance of Bifidobacterium Blautia, and F. Prausnitzii between clinical stages. |
| Luu, T.H. et al., 2017 [38] | Gut | 31 women with early-stage BC (ER/PgR+ 90% and HER2+ 15%): 15 stage 0, 7 stage I, 7 stage II and 2 stage III. In total, 8 patients were overweight | Fecal samples | Real-time qPCR targeting specific 16S rRNA sequences | ↑Firmicutes, F. prausnitzii and Blautia spp. in overweight and obese women compared to normal weight patients. ↑Bacteroidetes, Clostridium coccoides cluster, Clostridium leptum cluster, Faecalibacterium prausnitzii, and Blautia spp. in patients with stage II/III BC compared to patients in stage 0/I. |
| Fruge, A.D. et al., 2018 [39] | Gut | 32 women with BC stage 0 to II | Fecal samples | V4 16S rRNA gene sequencing (Illumina Miseq) | ↓Akkermansia muciniphila (AM) in BC patients with elevated body fat. ↑Prevotella and Lactobacillus and ↓ Clostridium, Campylobacter and Helicobacter in patients with high abundance of AM compared to patients with low abundance of AM. |
| Goedert, J.J. et al., 2015 [40] | Gut | 48 postmenopausal women with BC and 48 paired control women | Urine and fecal samples | V3-V4 16S rRNA sequencing (Illumina) Pipeline: QIIME | ↑Clostridiaceae, Faecalibacterium, and Ruminococcaceae and ↓ Dorea and Lachnospiraceae in BC patients compared to controls. |
| Zhu, J. et al., 2018 [41] | Gut | 18 premenopausal BC patients, 25 premenopausal healthy control, 44 postmenopausal BC patients and 46 postmenopausal healthy controls | Fecal samples | Illumina sequencing | ↑Escherichia coli, Citrobacter koseri, Acinetobacter radioresistens, Enterococcus gallinarum, Shewanella putrefaciens, Erwinia amylovora, Actinomyces spp. HPA0247, Salmonella enterica, and Fusobacterium nucleatum and ↓Eubacterium eligens and Roseburia inulinivorans in postmenopausal BC patients. |
| Fuhrman, B.J. et al., 2014 [42] | Gut | 60 healthy postmenopausal women | Urine and fecal samples | Pyrosequencing V1-V2 16S rRNA Amplicons Pipeline: QIIME | ↑Clostridiales and ↓Bacteroides related with the ↑ ratio all estrogen metabolites to parent estrogens. |
| Goedert, J.J. et al., 2018 [43] | Gut | 48 postmenopausal women with BC (11 women stage 0, 25 stage I, 10 stage II, 2 stage III; 88% ER+) and 48 paired control healthy women | Urine and fecal samples | V4 16S rRNA gene amplicon sequencing (Illumina MiSeq). Data available in the Sequence Read Archive under BioProject ID PRJNA383849 | ↓α- diversity and altered microbiota composition of both IgA-positive and IgA-negative fecal microbiota of BC patients. |
| Buchta Rosean, C. et al., 2019 [44] | Gut and mammary Breast (mice) | A mouse model of HR+ mammary cancer (5-to-8- week-old) | Feces and mammary tissue of mice | Flow cytometry Custom multiplex U-PLEX 16S rDNA sequencing (released by University of Maryland Institute for Genome Science) | ↑Commensal dysbiosis. ↑Circulating tumor cells and metastatic dissemination. ↑Early inflammation within the mammary gland. |
| Horigome, A. et al., 2019 [45] | Gut | 124 participants (46% history of chemotherapy) (123 women and 1 man) | Capillary blood and fecal samples | V3-V4 region of the bacterial 16S rRNA gene sequencing (Illumina Miseq) Pipeline: QIIME2 Gas chromatography for Fatty acid composition | ↑Actinobacteria and Bacteroidetes are associated to polyunsaturated fatty acid (PUFAs) in patients previously treated with chemotherapy. ↑Bifidobacterium is associated to PUFAs in participants with no history of chemotherapy |
| Kirkup, B.M. et al., 2019 [46] | Gut (mice) | Female C57BL6 mice | Fecal samples | V1+V2 16S rRNA gene sequencing by Illumina (MiSeq) Nuclear Magnetic Resonance (NMR) spectroscopy | ↓Odoribacter and Anaeotruncus (butyrate producing bacteria genera) and ↑ Bacteroides. ↓Butyrate in feces from BC patients treated with antibiotics and ↑ tumor growth. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laborda-Illanes, A.; Sanchez-Alcoholado, L.; Dominguez-Recio, M.E.; Jimenez-Rodriguez, B.; Lavado, R.; Comino-Méndez, I.; Alba, E.; Queipo-Ortuño, M.I. Breast and Gut Microbiota Action Mechanisms in Breast Cancer Pathogenesis and Treatment. Cancers 2020, 12, 2465. https://doi.org/10.3390/cancers12092465
Laborda-Illanes A, Sanchez-Alcoholado L, Dominguez-Recio ME, Jimenez-Rodriguez B, Lavado R, Comino-Méndez I, Alba E, Queipo-Ortuño MI. Breast and Gut Microbiota Action Mechanisms in Breast Cancer Pathogenesis and Treatment. Cancers. 2020; 12(9):2465. https://doi.org/10.3390/cancers12092465
Chicago/Turabian StyleLaborda-Illanes, Aurora, Lidia Sanchez-Alcoholado, María Emilia Dominguez-Recio, Begoña Jimenez-Rodriguez, Rocío Lavado, Iñaki Comino-Méndez, Emilio Alba, and María Isabel Queipo-Ortuño. 2020. "Breast and Gut Microbiota Action Mechanisms in Breast Cancer Pathogenesis and Treatment" Cancers 12, no. 9: 2465. https://doi.org/10.3390/cancers12092465
APA StyleLaborda-Illanes, A., Sanchez-Alcoholado, L., Dominguez-Recio, M. E., Jimenez-Rodriguez, B., Lavado, R., Comino-Méndez, I., Alba, E., & Queipo-Ortuño, M. I. (2020). Breast and Gut Microbiota Action Mechanisms in Breast Cancer Pathogenesis and Treatment. Cancers, 12(9), 2465. https://doi.org/10.3390/cancers12092465





