Possible Role of IRS-4 in the Origin of Multifocal Hepatocellular Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Patients
2.3. Cell Culture
2.4. Immunohistochemistry (IHC) and Immunocytochemistry (ICC)
2.5. Biochemical and Molecular Biology Methods
2.6. Cellular Adhesion to Collagen Experiments
2.7. Cell Migration Assay
2.8. Statistical Analysis
3. Results
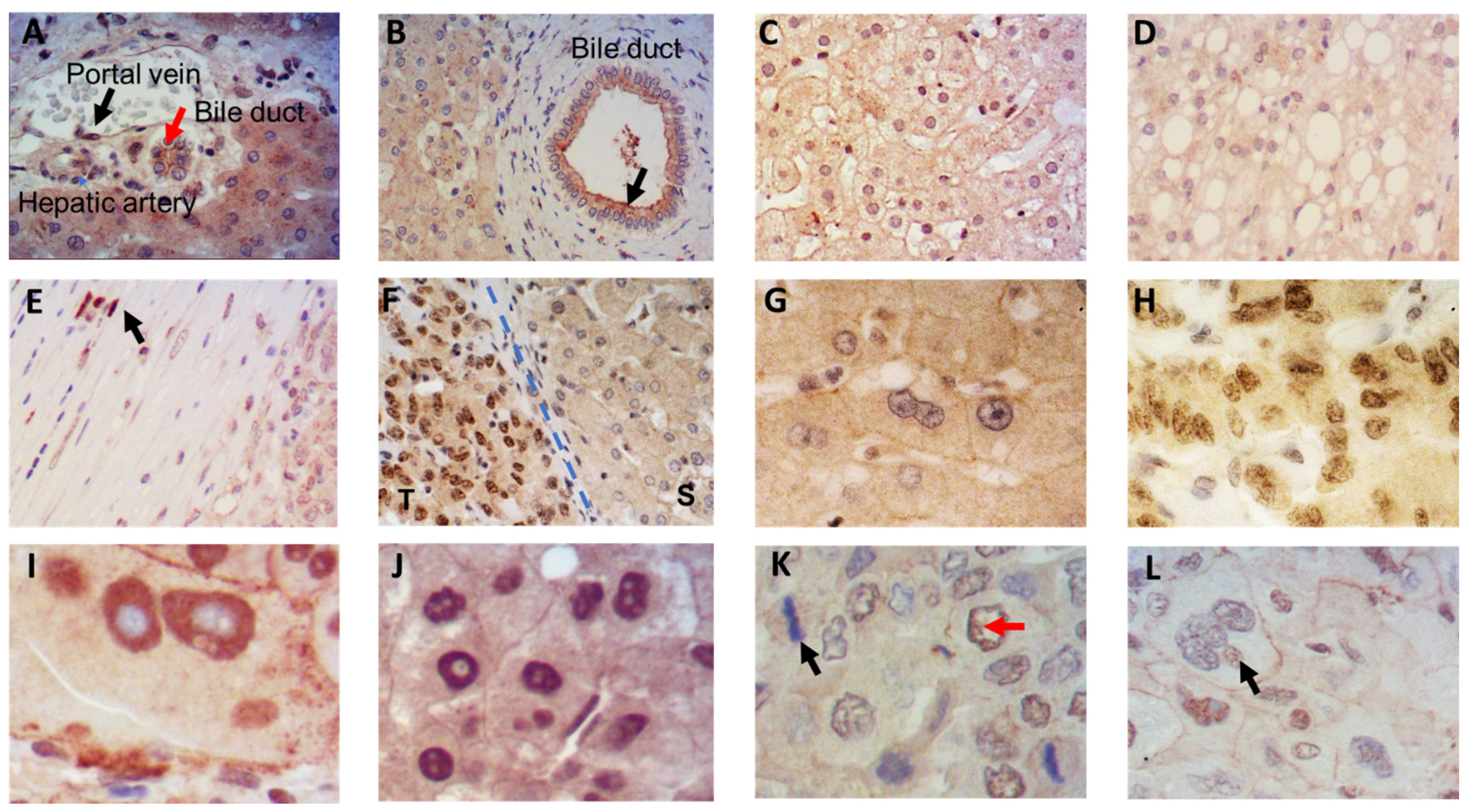
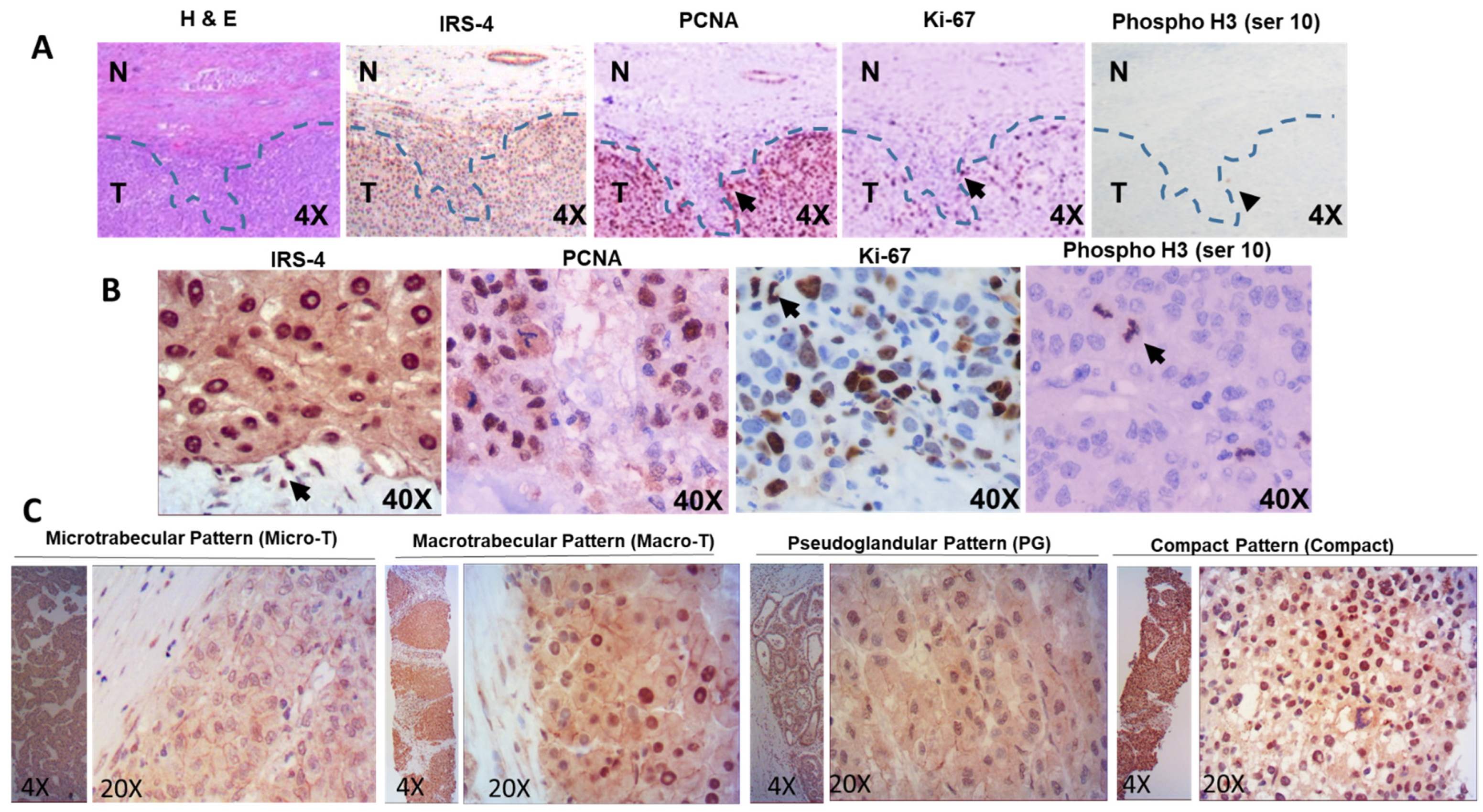
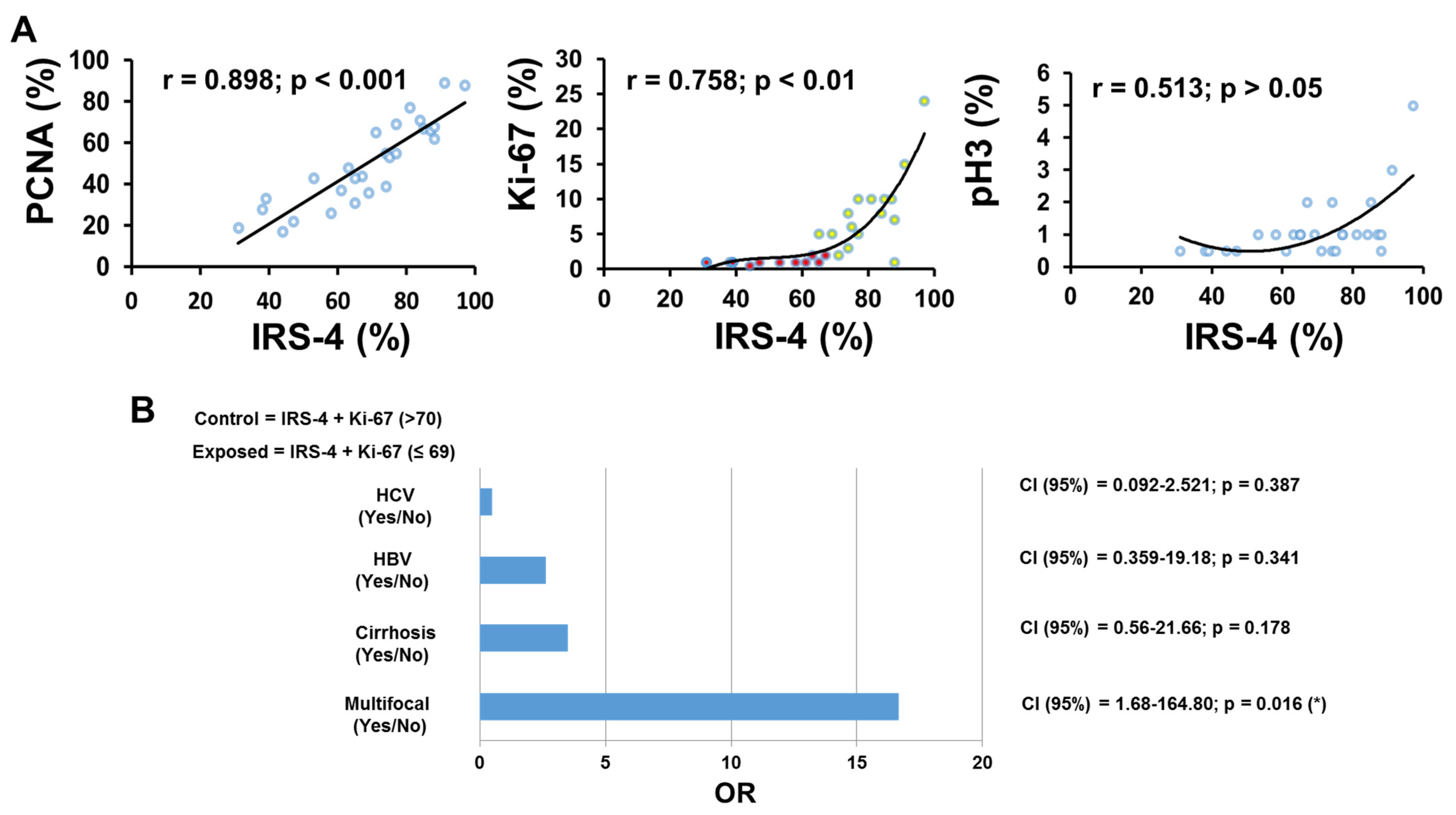
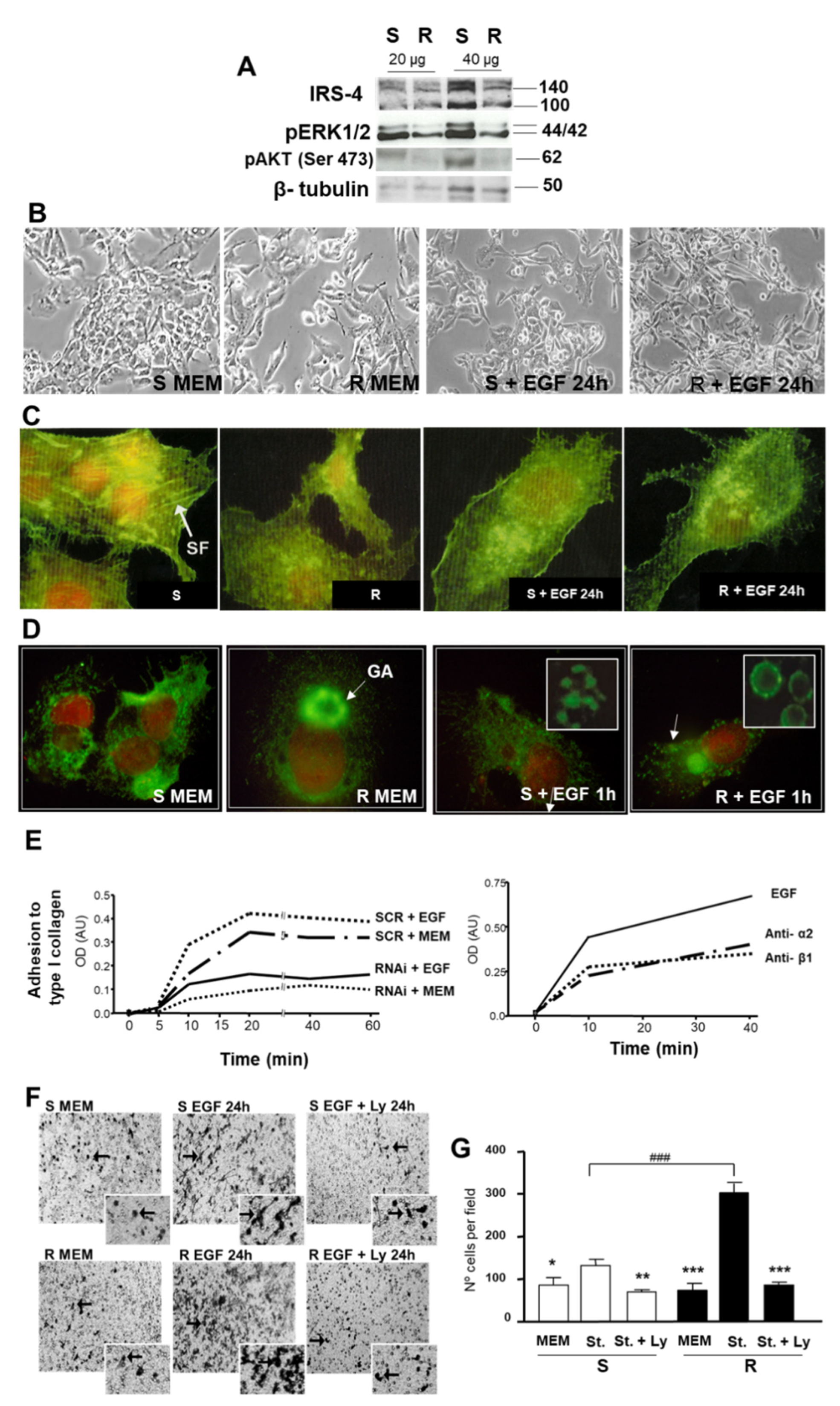
3.1. Localization of IRS-4 in Human HCC
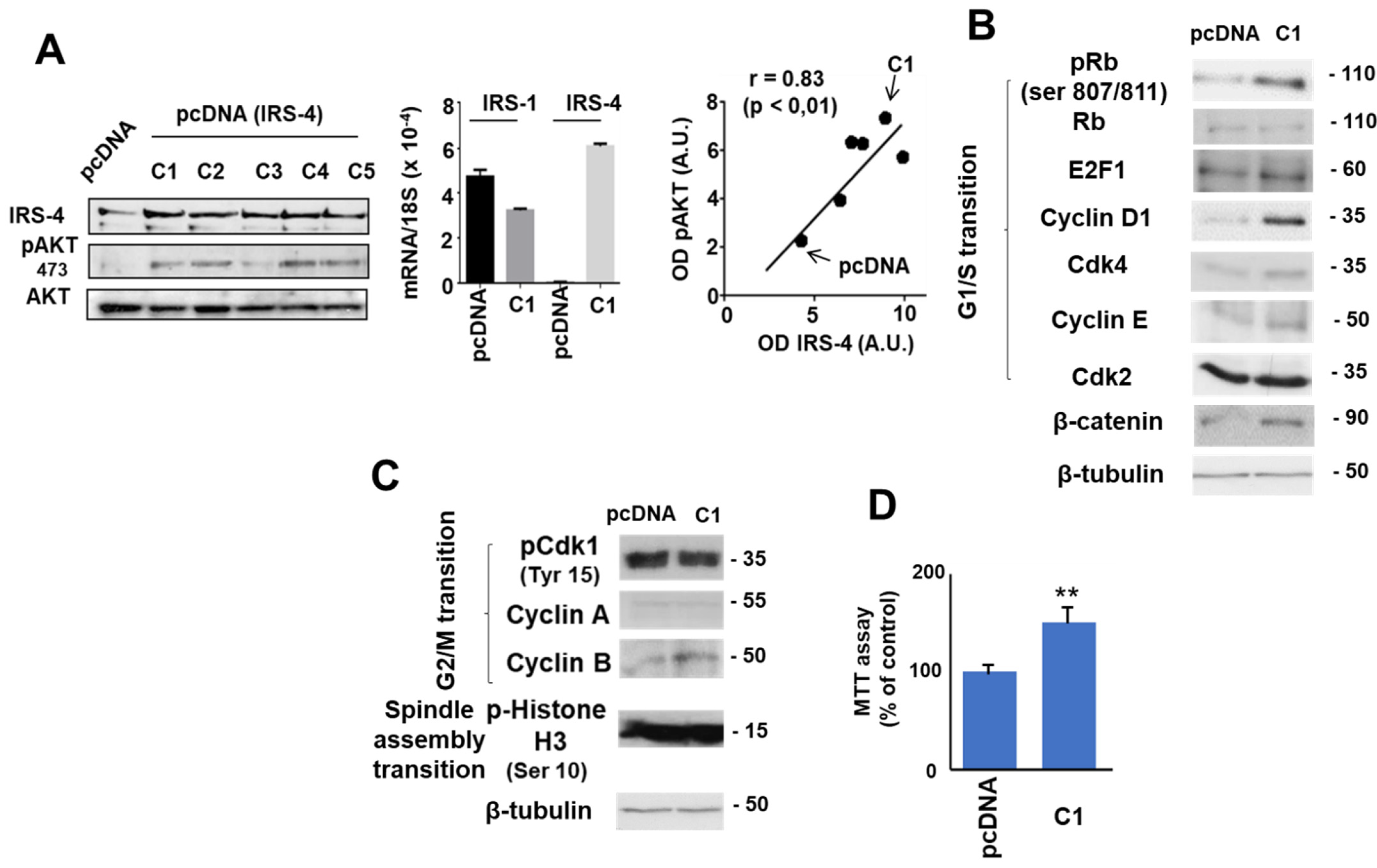
3.2. Role of IRS-4 in the Proliferation of HepG2 Cells
3.3. Study of the Role of IRS-4 Depletion on Cell Morphology, Integrin α2 Distribution, Cell Adhesion, and Migration of HepG2 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singal, A.G.S.; Lampertico, P.; Nahon, P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J. Hepatol. 2020, 72, 250–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fantin, V.R.; Lavan, B.E.; Wang, Q.; Jenkins, N.A.; Gilbert, D.J.; Copeland, N.G.; Keller, S.R.; Lienhard, G.E. Cloning, tissue expression, and chromosomal location of the mouse insulin receptor substrate 4 gene. Endocrinology 1999, 140, 1329–1337. [Google Scholar] [CrossRef] [PubMed]
- Escribano, O.; Fernández-Moreno, M.D.; Zueco, J.A.; Menor, C.; Fueyo, J.; Ropero, R.M.; Diaz-Laviada, I.; Román, I.D.; Guijarro, L.G. Insulin receptor substrate-4 signaling in quiescent rat hepatocytes and in regenerating rat liver. Hepatology 2003, 37, 1461–1469. [Google Scholar] [CrossRef] [PubMed]
- Weischenfeldt, J.; Dubash, T.; Drainas, A.P.; Mardin, B.R.; Chen, Y.; Stütz, A.M.; Waszak, S.M.; Bosco, G.; Halvorsen, A.R.; Raeder, B.; et al. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat. Genet. 2017, 49, 65–74. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chen, F.; Fonseca, N.A.; He, Y.; Fujita, M.; Nakagawa, H.; Zhang, Z.; Brazma, A.; PCAWG Transcriptome Working Group; PCAWG Structural Variation Working Group; et al. High-coverage whole-genome analysis of 1220 cancers reveals hundreds of genes deregulated by rearrangement-mediated cis-regulatory alterations. Nat. Commun. 2020, 11, 736. [Google Scholar] [CrossRef] [Green Version]
- Sanmartín-Salinas, P.; Toledo-Lobo, M.V.; Noguerales-Fraguas, F.; Fernández-Contreras, M.E.; Guijarro, L.G. Overexpression of insulin receptor substrate-4 is correlated with clinical staging in colorectal cancer patients. J. Mol. Histol. 2018, 49, 39–49. [Google Scholar] [CrossRef]
- Mehine, M.; Mäkinen, N.; Heinonen, H.R.; Aaltonen, L.A.; Vahteristo, P. Genomics of uterine leiomyomas: Insights from high-throughput sequencing. Fertil. Steril. 2014, 102, 621–629. [Google Scholar] [CrossRef]
- Mertens, F.; Möller, E.; Mandahl, N.; Picci, P.; Perez-Atayde, A.R.; Samson, I.; Sciot, R.; Debiec-Rychter, M. The t(X;6) in subungual exostosis results in transcriptional deregulation of the gene for insulin receptor substrate 4. Int. J. Cancer 2011, 128, 487–491. [Google Scholar] [CrossRef]
- Ikink, G.J.; Boer, M.; Bakker, E.R.; Hilkens, J. IRS4 induces mammary tumorigenesis and confers resistance to HER2-targeted therapy through constitutive PI3K/AKT-pathway hyperactivation. Nat. Commun. 2016, 7, 3567. [Google Scholar] [CrossRef] [Green Version]
- Karrman, K.; Isaksson, M.; Paulsson, K.; Johansson, B. The insulin receptor substrate 4 gene (IRS4) is mutated in paediatric T-cell acute lymphoblastic leukaemia. Br. J. Haematol. 2011, 155, 516–519. [Google Scholar] [CrossRef]
- Hao, P.; Huang, Y.; Peng, J.; Yu, J.; Guo, X.; Bao, F.; Dian, Z.; An, S.; Xu, T.R. IRS4 promotes the progression of non-small cell lung cancer and confers resistance to EGFR-TKI through the activation of PI3K/Akt and Ras-MAPK pathways. Exp. Cell Res. 2021, 21, 112615. [Google Scholar] [CrossRef] [PubMed]
- Sanmartín-Salinas, P.; Guijarro, L.G. Overexpression of IRS-4 Correlates with Procaspase 3 Levels in Tumoural Tissue of Patients with Colorectal Cancer. J. Oncol. 2018, 2018, 3812581. [Google Scholar] [CrossRef] [PubMed]
- Sanmartín-Salinas, P.; Lobo, M.D.V.T.; Noguerales-Fraguas, F.; Londoño, M.T.; Jiménez-Ruiz, A.; Guijarro, L.G. Insulin receptor substrate-4 is overexpressed in colorectal cancer and promotes retinoblastoma-cyclin-dependent kinase activation. J. Gastroenterol. 2018, 53, 932–944. [Google Scholar] [CrossRef] [PubMed]
- Kubota, T.; Kubota, N.; Kadowaki, T. Imbalanced Insulin Actions in Obesity and Type 2 Diabetes: Key Mouse Models of Insulin Signaling Pathway. Cell Metab. 2017, 25, 797–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoxhaj, G.; Dissanayake, K.; MacKintosh, C. Effect of IRS4 levels on PI 3-kinase signalling. PLoS ONE 2013, 8, e73327. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, E.P.; Escribano, O.; Chiloeches, A.; Rubio, S.R.; Román, I.D.; Fernández-Moreno, M.D.; Guijarro, L.G. Role of insulin receptor substrate-4 in IGF-I-stimulated HEPG2 proliferation. J. Hepatol. 2007, 46, 1089–1098. [Google Scholar] [CrossRef]
- Cui, A.; Jin, Z.; Gao, Z.; Jin, M.; Zhu, L.; Li, L.; Jin, C.; An, Y. Downregulation of miR-493 promoted melanoma proliferation by suppressing IRS4 expression. Tumour Biol. 2017, 39, 1010428317701640. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Lei, P.; Zeng, W.; Gao, J.; Wu, N. Long Noncoding RNA LINC00173 Promotes the Malignancy of Melanoma by Promoting the Expression of IRS4 through Competitive Binding to microRNA-493. Cancer Manag. Res. 2020, 12, 3131–3144. [Google Scholar] [CrossRef]
- Cantarini, M.C.; de la Monte, S.M.; Pang, M.; Tong, M.; D’Errico, A.; Trevisani, F.; Wands, J.R. Aspartyl-asparagyl beta hydroxylase over-expression in human hepatoma is linked to activation of insulin-like growth factor and notch signaling mechanisms. Hepatology 2006, 44, 446–457. [Google Scholar] [CrossRef]
- Hernández-Breijo, B.; Monserrat, J.; Ramírez-Rubio, S.; Cuevas, E.P.; Vara, D.; Díaz-Laviada, I.; Fernández-Moreno, M.D.; Román, I.D.; Gisbert, J.P.; Guijarro, L.G. Preclinical evaluation of azathioprine plus buthionine sulfoximine in the treatment of human hepatocarcinoma and colon carcinoma. World J. Gastroenterol. 2011, 17, 3899–3911. [Google Scholar] [CrossRef]
- Rubio, C.A. Further studies on the arrest of cell proliferation in tumor cells at the invading front of colonic adenocarcinoma. J. Gastroenterol. Hepatol. 2007, 22, 1877–1881. [Google Scholar] [CrossRef] [PubMed]
- Kohrman, A.Q.; Matus, D.Q. Divide or Conquer: Cell cycle regulation of invasive behaviour. Trends Cell Biol. 2017, 27, 12–25. [Google Scholar] [CrossRef] [Green Version]
- Jung, A.; Schrauder, M.; Oswald, U.; Knoll, C.; Sellberg, P.; Palmqvist, R.; Niedobitek, G.; Brabletz, T.; Kirchner, T. The invasion front of human colorectal adenocarcinomas shows co-localization of nuclear beta-catenin, cyclin D1, and p16INK4A and is a region of low proliferation. Am. J. Pathol. 2001, 159, 1613–1617. [Google Scholar] [CrossRef]
- Lehn, S.; Tobin, N.P.; Berglund, P.; Nilsson, K.; Sims, A.H.; Jirström, K.; Härkönen, P.; Lamb, R.; Landberg, G. Down-regulation of the oncogene cyclin D1 increases migratory capacity in breast cancer and is linked to unfavorable prognostic features. Am. J. Pathol. 2010, 177, 2886–2897. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, T.; Shinkai, K.; Mukai, M.; Yoshioka, K.; Fujii, Y.; Nakahara, K.; Matsuda, H.; Akedo, H. Cell-cycle-dependent invasion in vitro by rat ascites hepatoma cells. Int. J. Cancer 1995, 63, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Mejlvang, J.; Kriajevska, M.; Vandewalle, C.; Chernova, T.; Sayan, A.E.; Berx, G.; Mellon, J.K.; Tulchinsky, E. Direct repression of cyclin D1 by SIP1 attenuates cell cycle progression in cells undergoing an epithelial mesenchy mal transition. Mol. Biol. Cell 2007, 18, 4615–4624. [Google Scholar] [CrossRef] [Green Version]
- Homma, Y.; Kanno, S.I.; Sasaki, K.; Nishita, M.; Yasui, A.; Asano, T.; Ohashi, K.; Mizuno, K. Insulin receptor substrate-4 binds to Slingshot-1 phosphatase and promotes cofilin dephosphorylation. J. Biol. Chem. 2014, 289, 26302–26313. [Google Scholar] [CrossRef] [Green Version]
- Panera, N.; Crudele, A.; Romito, I.; Gnani, D.; Alisi, A. Focal Adhesion Kinase: Insight into Molecular Roles and Functions in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2017, 18, 99. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guijarro, L.G.; Sanmartin-Salinas, P.; Pérez-Cuevas, E.; Toledo-Lobo, M.V.; Monserrat, J.; Zoullas, S.; Sáez, M.A.; Álvarez-Mon, M.A.; Bujan, J.; Noguerales-Fraguas, F.; et al. Possible Role of IRS-4 in the Origin of Multifocal Hepatocellular Carcinoma. Cancers 2021, 13, 2560. https://doi.org/10.3390/cancers13112560
Guijarro LG, Sanmartin-Salinas P, Pérez-Cuevas E, Toledo-Lobo MV, Monserrat J, Zoullas S, Sáez MA, Álvarez-Mon MA, Bujan J, Noguerales-Fraguas F, et al. Possible Role of IRS-4 in the Origin of Multifocal Hepatocellular Carcinoma. Cancers. 2021; 13(11):2560. https://doi.org/10.3390/cancers13112560
Chicago/Turabian StyleGuijarro, Luis G., Patricia Sanmartin-Salinas, Eva Pérez-Cuevas, M. Val Toledo-Lobo, Jorge Monserrat, Sofía Zoullas, Miguel A. Sáez, Miguel A. Álvarez-Mon, Julia Bujan, Fernando Noguerales-Fraguas, and et al. 2021. "Possible Role of IRS-4 in the Origin of Multifocal Hepatocellular Carcinoma" Cancers 13, no. 11: 2560. https://doi.org/10.3390/cancers13112560






