Current Implications of microRNAs in Genome Stability and Stress Responses of Ovarian Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Participation of miRNAs in Pathogenesis and Development of Neoplastic Diseases
3. Aberrant Expression Profiles of miRNAs in Ovarian Cancer
3.1. MiRNAs as Tumor Suppressors
3.2. MiRNAs as Oncogenes
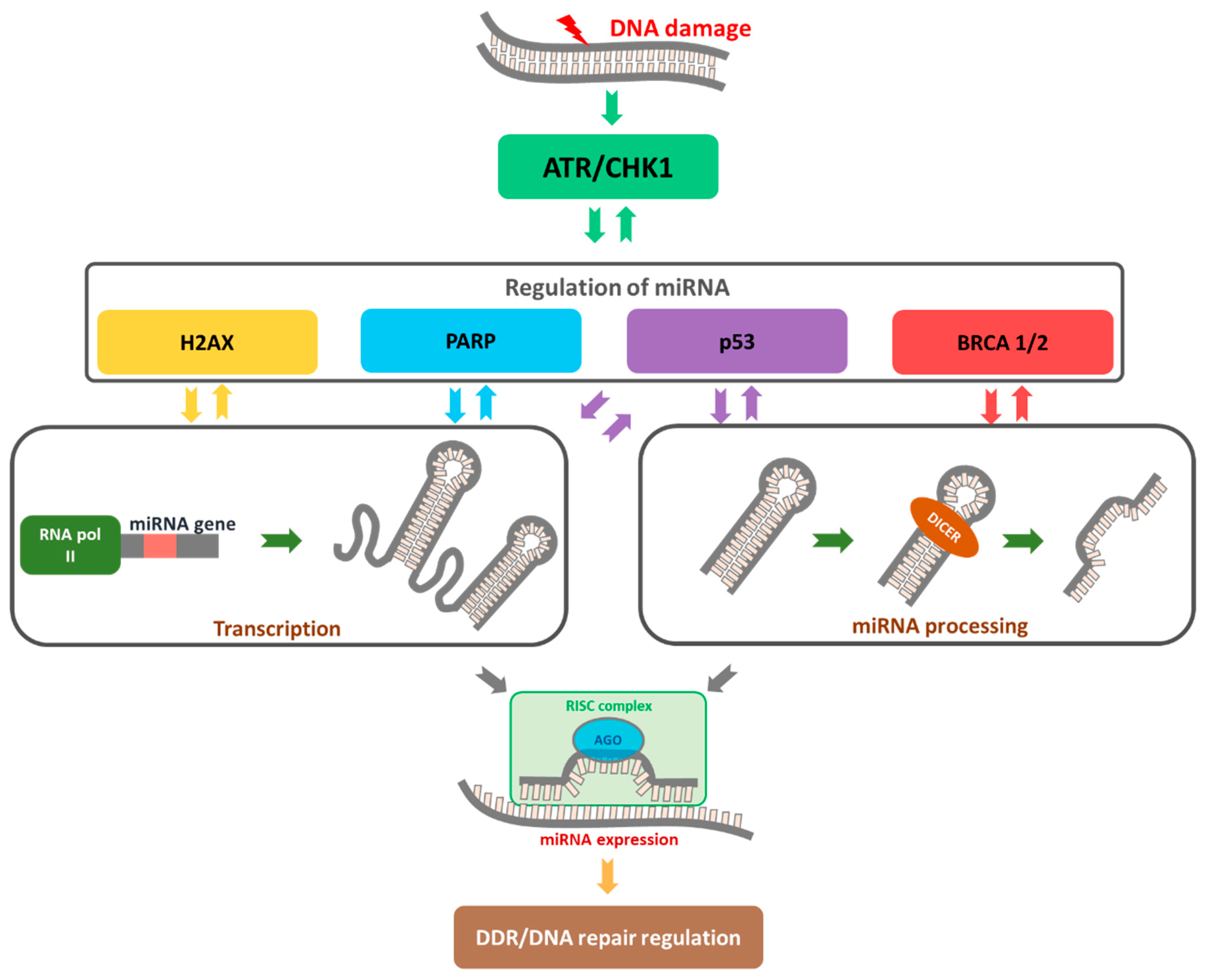
4. MiRNA Functions in Cancer Based on Regulation of DDR
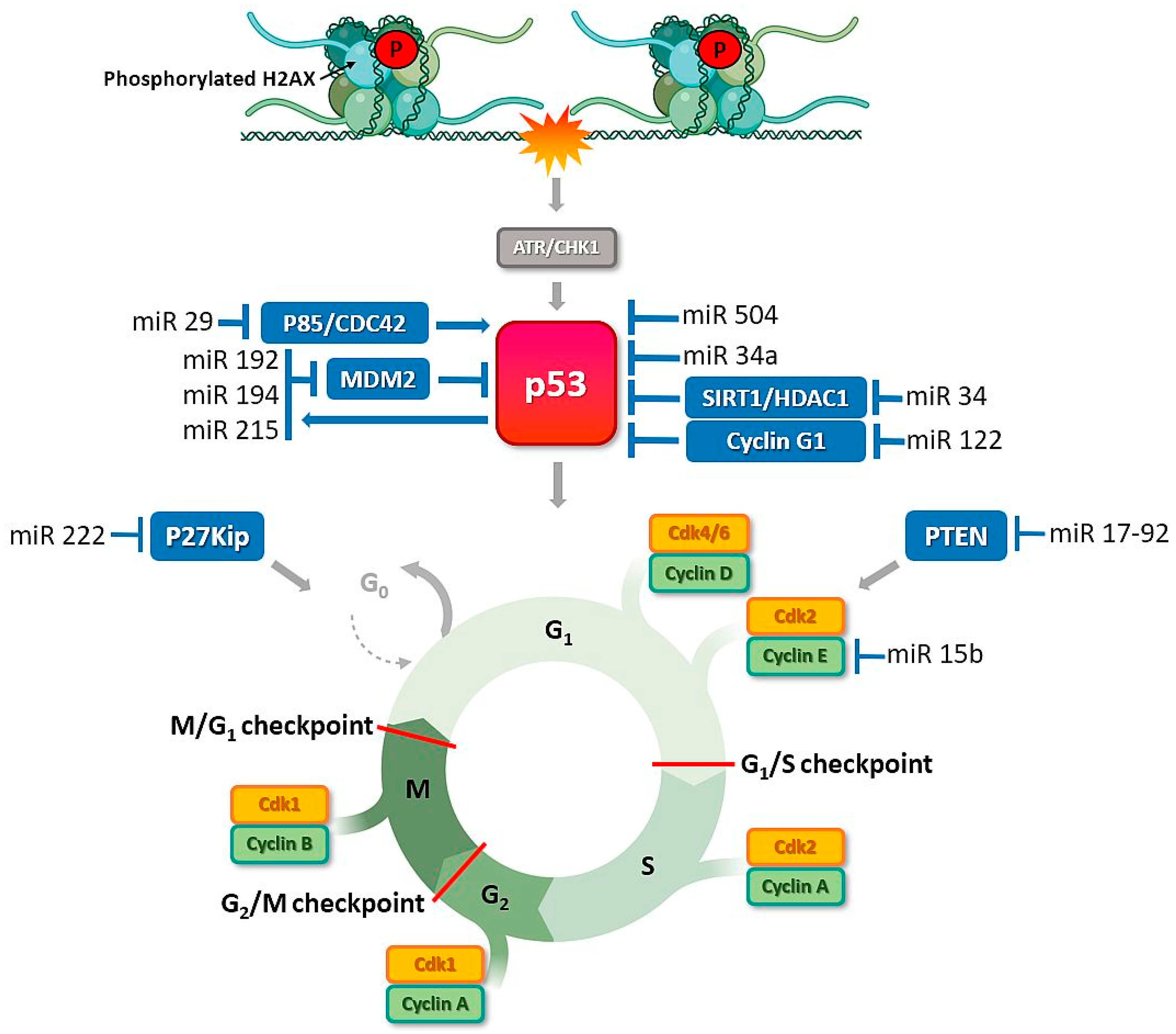
4.1. MiRNAs Are Involved in Cell Cycle Disruption
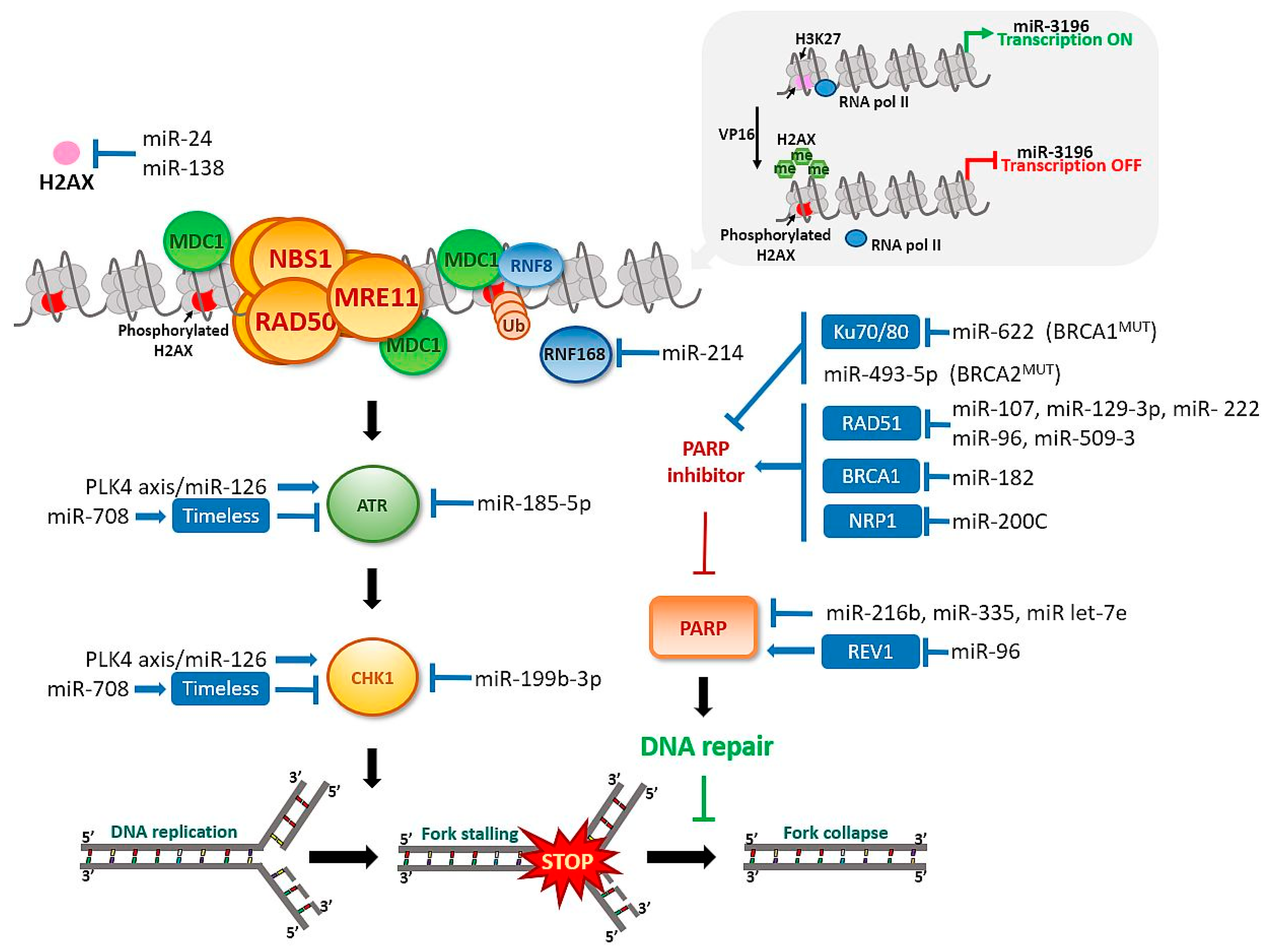
4.2. Functional miRNAs in Activation of the “Response Track” to DNA Damage and the Role of H2AX Histone
4.3. MiRNAs Contributes to DSB DNA Damage Repair System
4.4. MiRNAs Modulate Activity of p53, a Key Protein of the DDR Pathway
5. MiRNAs Associated with DNA Repair Checkpoint Proteins: New Options for Optimizing Ovarian Cancer Therapy
5.1. PARP
5.2. ATR
5.3. CHK1
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ahmed, N.; Kadife, E.; Raza, A.; Short, M.; Jubinsky, P.T.; Kannourakis, G. Ovarian Cancer, Cancer Stem Cells and Current Treatment Strategies: A Potential Role of Magmas in the Current Treatment Methods. Cells 2020, 9, 719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam, M.J.; Roshid, B.; Pervin, S.; Kabir, S.; Chigurupati, S.; Hasan, M.N. A 35 Year Old Bangladeshi Lady with Hereditary Mucinous Ovarian Cancer, Complicated with Omental Metastasis. Mymensingh Med. J. MMJ 2019, 28, 484–489. [Google Scholar]
- Davidson, B.; Tropé, C.G. Ovarian Cancer: Diagnostic, Biological and Prognostic Aspects. Women’s Health 2014, 10, 519–533. [Google Scholar] [CrossRef]
- Bonadio, R.R.D.C.C.; Fogace, R.N.; Miranda, V.C.; Diz, M.D.P.E. Homologous recombination deficiency in ovarian cancer: A review of its epidemiology and management. Clinics 2018, 73, e450s. [Google Scholar] [CrossRef]
- Pujade-Lauraine, E.; Ledermann, J.A.; Selle, F.; Gebski, V.; Penson, R.T.; Oza, A.M.; Korach, J.; Huzarski, T.; Poveda, A.; Pignata, S.; et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017, 18, 1274–1284. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Fu, G.-B.; Tao, Z.; Ouyang, J.; Kong, F.; Jiang, B.-H.; Wan, X.; Chen, K. MiR-497 decreases cisplatin resistance in ovarian cancer cells by targeting mTOR/P70S6K1. Oncotarget 2015, 6, 26457–26471. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Liu, F.; Wang, X.; Ouyang, G. The roles of microRNAs in the regulation of tumor metastasis. Cell Biosci. 2015, 5, 32. [Google Scholar] [CrossRef] [Green Version]
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenet. 2019, 11, 25. [Google Scholar] [CrossRef]
- Han, W.; Cui, H.; Liang, J.; Su, X. Role of MicroRNA-30c in cancer progression. J. Cancer 2020, 11, 2593–2601. [Google Scholar] [CrossRef] [Green Version]
- Hanlon, K.; Rudin, C.E.; Harries, L.W. Investigating the Targets of MIR-15a and MIR-16-1 in Patients with Chronic Lymphocytic Leukemia (CLL). PLoS ONE 2009, 4, e7169. [Google Scholar] [CrossRef] [Green Version]
- Acunzo, M.; Croce, C.M. Downregulation of miR-15a and miR-16-1 at 13q14 in Chronic Lymphocytic Leukemia. Clin. Chem. 2016, 62, 655–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rampazzo, E.; Bojnik, E.; Trentin, L.; Bonaldi, L.; Del Bianco, P.; Frezzato, F.; Visentin, A.; Facco, M.; Semenzato, G.; De Rossi, A. Role of miR-15a/miR-16-1 and the TP53 axis in regulating telomerase expression in chronic lymphocytic leukemia. Haematologica 2017, 102, e253–e256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunter, M.P.; Ismail, N.; Zhang, X.; Aguda, B.D.; Lee, E.J.; Yu, L.; Xiao, T.; Schafer, J.; Lee, M.-L.T.; Schmittgen, T.D.; et al. Detection of microRNA Expression in Human Peripheral Blood Microvesicles. PLoS ONE 2008, 3, e3694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, X.; Takahashi, R.; Hiura, Y.; Hirokawa, G.; Fukushima, Y.; Iwai, N. Plasma miR-208 as a Biomarker of Myocardial Injury. Clin. Chem. 2009, 55, 1944–1949. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xu, S.; Liu, X. MicroRNA profiling of plasma exosomes from patients with ovarian cancer using high-throughput sequencing. Oncol. Lett. 2019, 17, 5601–5607. [Google Scholar] [CrossRef] [Green Version]
- Shapira, I.; Oswald, M.; Lovecchio, J.; Khalili, H.; Menzin, A.; Whyte, J.; Dos Santos, L.; Liang, S.; Bhuiya, T.; Keogh, M.; et al. Circulating biomarkers for detection of ovarian cancer and predicting cancer outcomes. Br. J. Cancer 2014, 110, 976–983. [Google Scholar] [CrossRef] [Green Version]
- Maeda, K.; Sasaki, H.; Ueda, S.; Miyamoto, S.; Terada, S.; Konishi, H.; Kogata, Y.; Ashihara, K.; Fujiwara, S.; Tanaka, Y.; et al. Serum exosomal microRNA-34a as a potential biomarker in epithelial ovarian cancer. J. Ovarian Res. 2020, 13, 47. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, K.; Sawada, K.; Yoshimura, A.; Kinose, Y.; Nakatsuka, E.; Kimura, T. Clinical relevance of circulating cell-free microRNAs in ovarian cancer. Mol. Cancer 2016, 15, 48. [Google Scholar] [CrossRef] [Green Version]
- Staicu, C.E.; Predescu, D.-V.; Rusu, C.M.; Radu, B.M.; Cretoiu, D.; Suciu, N.; Crețoiu, S.M.; Voinea, S.-C. Role of microRNAs as Clinical Cancer Biomarkers for Ovarian Cancer: A Short Overview. Cells 2020, 9, 169. [Google Scholar] [CrossRef] [Green Version]
- Vaksman, O.; Tropé, C.; Davidson, B.; Reich, R. Exosome-derived miRNAs and ovarian carcinoma progression. Carcinogenesis 2014, 35, 2113–2120. [Google Scholar] [CrossRef]
- Zhou, J.; Gong, G.; Tan, H.; Dai, F.; Zhu, X.; Chen, Y.; Wang, J.; Liu, Y.; Chen, P.; Wu, X.; et al. Urinary microRNA-30a-5p is a potential biomarker for ovarian serous adenocarcinoma. Oncol. Rep. 2015, 33, 2915–2923. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.; Müller, V.; Milde-Langosch, K.; Trillsch, F.; Pantel, K.; Schwarzenbach, H. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget 2016, 7, 16923–16935. [Google Scholar] [CrossRef] [Green Version]
- Kapetanakis, N.-I.; Uzan, C.; Jimenez-Pailhes, A.-S.; Gouy, S.; Bentivegna, E.; Morice, P.; Caron, O.; Gourzones-Dmitriev, C.; Le Teuff, G.; Busson, P. Plasma miR-200b in ovarian carcinoma patients: Distinct pattern of pre/post-treatment variation compared to CA-125 and potential for prediction of progression-free survival. Oncotarget 2015, 6, 36815–36824. [Google Scholar] [CrossRef] [Green Version]
- Chan, S.-H.; Wang, L.-H. Regulation of cancer metastasis by microRNAs. J. Biomed. Sci. 2015, 22, 9. [Google Scholar] [CrossRef] [Green Version]
- Huang, Q.; Gumireddy, K.; Schrier, M.; Le Sage, C.; Nagel, R.; Nair, S.; Egan, D.A.; Li, A.; Huang, G.; Klein-Szanto, A.J.; et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat. Cell Biol. 2008, 10, 202–210. [Google Scholar] [CrossRef]
- Li, N.; Yang, L.; Sun, Y.; Wu, X. MicroRNA-16 inhibits migration and invasion via regulation of the Wnt/β-catenin signaling pathway in ovarian cancer. Oncol. Lett. 2019, 17, 2631–2638. [Google Scholar] [CrossRef] [Green Version]
- Lezina, L.; Purmessur, N.; Antonov, A.V.; Ivanova, T.; Karpova, E.; Krishan, K.; Ivan, M.; Aksenova, V.; Tentler, D.; Garabadgiu, A.V.; et al. miR-16 and miR-26a target checkpoint kinases Wee1 and Chk1 in response to p53 activation by genotoxic stress. Cell Death Dis. 2013, 4, e953. [Google Scholar] [CrossRef]
- Choi, P.-W.; Wong, K.-K. The Functions of MicroRNA-200 Family in Ovarian Cancer: Beyond Epithelial-Mesenchymal Transition. Int. J. Mol. Sci. 2017, 18, 1207. [Google Scholar] [CrossRef] [Green Version]
- Chung, V.Y.; Tan, T.Z.; Tan, M.; Wong, M.K.; Kuay, K.T.; Yang, Z.; Ye, J.; Muller, J.; Koh, C.M.; Guccione, E.; et al. GRHL2-miR-200-ZEB1 maintains the epithelial status of ovarian cancer through transcriptional regulation and histone modification. Sci. Rep. 2016, 6, 19943. [Google Scholar] [CrossRef] [Green Version]
- Shi, C.; Yang, Y.; Zhang, L.; Yu, J.; Qin, S.; Xu, H.; Gao, Y. MiR-200a-3p promoted the malignant behaviors of ovarian cancer cells through regulating PCDH9. OncoTargets Ther. 2019, 12, 8329–8338. [Google Scholar] [CrossRef] [Green Version]
- Brozovic, A.; Duran, G.E.; Wang, Y.C.; Francisco, E.B.; Sikic, B.I. The miR-200 family differentially regulates sensitivity to paclitaxel and carboplatin in human ovarian carcinoma OVCAR-3 and MES-OV cells. Mol. Oncol. 2015, 9, 1678–1693. [Google Scholar] [CrossRef] [PubMed]
- Jiajie, T.; Yanzhou, Y.; Hoi-Hung, A.C.; Zi-Jiang, C.; Wai-Yee, C. Conserved miR-10 family represses proliferation and induces apoptosis in ovarian granulosa cells. Sci. Rep. 2017, 7, 41304. [Google Scholar] [CrossRef] [Green Version]
- Yu, P.-N.; Yan, M.-D.; Lai, H.-C.; Huang, R.-L.; Chou, Y.-C.; Lin, W.-C.; Yeh, L.-T.; Lin, Y.-W. Downregulation ofmiR-29contributes to cisplatin resistance of ovarian cancer cells. Int. J. Cancer 2013, 134, 542–551. [Google Scholar] [CrossRef] [PubMed]
- To, S.K.Y.; Mak, A.S.C.; Fung, Y.M.E.; Che, C.-M.; Li, S.-S.; Deng, W.; Ru, B.; Zhang, J.; Wong, A.S.T. β-catenin downregulates Dicer to promote ovarian cancer metastasis. Oncogene 2017, 36, 5927–5938. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Cai, J.; Cai, L.; Jia, J.; Xie, L.; Zhu, Y.; Huang, B.; Jin, D.; Wang, Z. Let-7e sensitizes epithelial ovarian cancer to cisplatin through repressing DNA double strand break repair. J. Ovarian Res. 2017, 10, 24. [Google Scholar] [CrossRef] [Green Version]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef]
- Biamonte, F.; Santamaria, G.; Sacco, A.; Perrone, F.M.; Di Cello, A.; Battaglia, A.M.; Salatino, A.; Di Vito, A.; Aversa, I.; Venturella, R.; et al. MicroRNA let-7g acts as tumor suppressor and predictive biomarker for chemoresistance in human epithelial ovarian cancer. Sci. Rep. 2019, 9, 5668. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Cao, L.; Wang, Y.; Wang, X.; Liu, N.; You, Y. Regulation of let-7 and its target oncogenes (Review). Oncol. Lett. 2012, 3, 955–960. [Google Scholar] [CrossRef]
- Wei, J.; Zhang, L.; Li, J.; Zhu, S.; Tai, M.; Mason, C.W.; Chapman, J.A.; Reynolds, E.A.; Weiner, C.P.; Zhou, H.H. MicroRNA-205 promotes cell invasion by repressing TCF21 in human ovarian cancer. J. Ovarian Res. 2017, 10, 33. [Google Scholar] [CrossRef]
- Zhu, D.; Huang, X.; Liang, F.; Zhao, L. LncRNA miR503HG interacts with miR-31-5p through multiple ways to regulate cancer cell invasion and migration in ovarian cancer. J. Ovarian Res. 2020, 13, 3. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, F.F.; Jamal, R.; Syafruddin, S.E.; Ab Mutalib, N.S.; Saidin, S.; MdZin, R.R.; Mollah, M.M.H.; Mokhtar, N.M. MicroRNA-200c and microRNA-31 regulate proliferation, colony formation, migration and invasion in serous ovarian cancer. J. Ovarian Res. 2015, 8, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, G.; Cheng, Y. MiR-126-3p inhibits ovarian cancer proliferation and invasion via targeting PLXNB2. Reprod. Biol. 2018, 18, 218–224. [Google Scholar] [CrossRef]
- Xia, X.Y.; Yu, Y.J.; Ye, F.; Peng, G.Y.; Li, Y.J.; Zhou, X.M. MicroRNA-506-3p inhibits proliferation and promotes apoptosis in ovarian cancer cell via targeting SIRT1/AKT/FOXO3a signaling pathway. Neoplasma 2020, 67, 344–353. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Zhang, C.; Zhou, B.; Jiang, D. miR-183 modulated cell proliferation and apoptosis in ovarian cancer through the TGF-β/Smad4 signaling pathway. Int. J. Mol. Med. 2019, 43, 1734–1746. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Li, Y.; Gan, Y.; Tian, R.; Wu, Q.; Shu, G.; Yin, G. Methylation-mediated repression of MiR-424/503 cluster promotes proliferation and migration of ovarian cancer cells through targeting the hub gene KIF23. Cell Cycle 2019, 18, 1601–1618. [Google Scholar] [CrossRef]
- Liao, Y.; Deng, Y.; Liu, J.; Ye, Z.; You, Z.; Yao, S.; He, S. MiR-760 overexpression promotes proliferation in ovarian cancer by downregulation of PHLPP2 expression. Gynecol. Oncol. 2016, 143, 655–663. [Google Scholar] [CrossRef]
- Feng, Y.; Hang, W.; Sang, Z.; Li, S.; Xu, W.; Miao, Y.; Xi, X.; Huang, Q. Identification of exosomal and non-exosomal microRNAs associated with the drug resistance of ovarian cancer. Mol. Med. Rep. 2019, 19, 3376–3392. [Google Scholar] [CrossRef] [Green Version]
- Lv, Y.L.F.; Liu, P.S. MiR-151 promotes ovarian cancer through activation of akt/mTOR signaling pathway by decreasing RhoGDIA. Int. J. Clin. Exp. Pathol. 2016, 9, 11222–11229. [Google Scholar]
- Liu, X.; Yao, B.; Wu, Z. miRNA-199a-5p suppresses proliferation and invasion by directly targeting NF-κB1 in human ovarian cancer cells. Oncol. Lett. 2018, 16, 4543–4550. [Google Scholar] [CrossRef]
- Meng, F.; Henson, R.; Wehbe–Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 Regulates Expression of the PTEN Tumor Suppressor Gene in Human Hepatocellular Cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [CrossRef] [Green Version]
- Misso, G.; Di Martino, M.T.; De Rosa, G.; Farooqi, A.A.; Lombardi, A.; Campani, V.; Zarone, M.R.; Gullà, A.; Tagliaferri, P.; Tassone, P.; et al. Mir-34: A New Weapon Against Cancer? Mol. Ther. Nucleic Acids 2014, 3, e195. [Google Scholar] [CrossRef] [PubMed]
- Frixa, T.; Donzelli, S.; Blandino, G. Oncogenic MicroRNAs: Key Players in Malignant Transformation. Cancers 2015, 7, 2466–2485. [Google Scholar] [CrossRef]
- Kumar, D.; Gorain, M.; Kundu, G.; Kundu, G.C. Therapeutic implications of cellular and molecular biology of cancer stem cells in melanoma. Mol. Cancer 2017, 16, 7. [Google Scholar] [CrossRef] [Green Version]
- Yeh, Y.-M.; Chuang, C.-M.; Chao, K.-C.; Wang, L.-H. MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF-1α. Int. J. Cancer 2013, 133, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; An, Q.; Niu, B.; Lu, X.; Zhang, N.; Cao, X. Role of miR-221/222 in Tumor Development and the Underlying Mechanism. J. Oncol. 2019, 2019, 7252013. [Google Scholar] [CrossRef]
- Amini-Farsani, Z.; Sangtarash, M.H.; Shamsara, M.; Teimori, H. MiR-221/222 promote chemoresistance to cisplatin in ovarian cancer cells by targeting PTEN/PI3K/AKT signaling pathway. Cytotechnology 2018, 70, 203–213. [Google Scholar] [CrossRef]
- Sun, C.; Cao, W.; Qiu, C.; Li, C.; Dongol, S.; Zhang, Z.; Dong, R.; Song, K.; Yang, X.; Zhang, Q.; et al. MiR-509-3 augments the synthetic lethality of PARPi by regulating HR repair in PDX model of HGSOC. J. Hematol. Oncol. 2020, 13, 9. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Li, Q.; Huang, H.; Li, Y.; Li, L.; Hou, W.; You, Z. Overexpression of miRNA-221 promotes cell proliferation by targeting the apoptotic protease activating factor-1 and indicates a poor prognosis in ovarian cancer. Int. J. Oncol. 2017, 50, 1087–1096. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Guo, H.; Lu, S. MiR-335-5p restores cisplatin sensitivity in ovarian cancer cells through targeting BCL2L2. Cancer Med. 2018, 7, 4598–4609. [Google Scholar] [CrossRef] [Green Version]
- Guan, R.; Cai, S.; Sun, M.; Xu, M. Upregulation of miR-520b promotes ovarian cancer growth. Oncol. Lett. 2017, 14, 3155–3161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, M.; Lü, M.; Yao, G.; Tian, H.; Lian, J.; Liu, L.; Liang, M.; Wang, Y.; Sun, F. Transactivation of microRNA-383 by Steroidogenic Factor-1 Promotes Estradiol Release from Mouse Ovarian Granulosa Cells by Targeting RBMS1. Mol. Endocrinol. 2012, 26, 1129–1143. [Google Scholar] [CrossRef] [Green Version]
- Nakayama, I.; Shibazaki, M.; Yashima-Abo, A.; Miura, F.; Sugiyama, T.; Masuda, T.; Maesawa, C. Loss of HOXD10 expression induced by upregulation of miR-10b accelerates the migration and invasion activities of ovarian cancer cells. Int. J. Oncol. 2013, 43, 63–71. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; He, J.; Su, F.; Ding, N.; Hu, W.; Yao, B.; Wang, W.; Zhou, G. Repression of ATR pathway by miR-185 enhances radiation-induced apoptosis and proliferation inhibition. Cell Death Dis. 2013, 4, e699. [Google Scholar] [CrossRef] [Green Version]
- Boele, J.; Persson, H.; Shin, J.W.; Ishizu, Y.; Newie, I.S.; Søkilde, R.; Hawkins, S.M.; Coarfa, C.; Ikeda, K.; Takayama, K.-I.; et al. PAPD5-mediated 3’ adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease. Proc. Natl. Acad. Sci. USA 2014, 111, 11467–11472. [Google Scholar] [CrossRef] [Green Version]
- Bao, J.; Yu, Y.; Chen, J.; He, Y.; Chen, X.; Ren, Z.; Xue, C.; Liu, L.; Hu, Q.; Li, J.; et al. MiR-126 negatively regulates PLK-4 to impact the development of hepatocellular carcinoma via ATR/CHEK1 pathway. Cell Death Dis. 2018, 9, 1045. [Google Scholar] [CrossRef] [Green Version]
- Zou, X.; Zhu, C.; Zhang, L.; Zhang, Y.; Fu, F.; Chen, Y.; Zhou, J. MicroRNA-708 Suppresses Cell Proliferation and Enhances Chemosensitivity of Cervical Cancer Cells to cDDP by Negatively Targeting Timeless. OncoTargets Ther. 2020, 13, 225–235. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.E.; Meghani, K.; Brault, M.-E.; Leclerc, L.; He, Y.; Day, T.A.; Elias, K.M.; Drapkin, R.; Weinstock, D.M.; Dao, F.; et al. Platinum and PARP Inhibitor Resistance Due to Overexpression of MicroRNA-622 in BRCA1-Mutant Ovarian Cancer. Cell Rep. 2016, 14, 429–439. [Google Scholar] [CrossRef] [Green Version]
- Marchini, S.; Cavalieri, D.; Fruscio, R.; Calura, E.; Garavaglia, D.; Nerini, I.F.; Mangioni, C.; Cattoretti, G.; Clivio, L.; Beltrame, L.; et al. Association between miR-200c and the survival of patients with stage I epithelial ovarian cancer: A retrospective study of two independent tumour tissue collections. Lancet Oncol. 2011, 12, 273–285. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, X.; Huang, Y.; Zhang, Q.; Zhou, J.; Zhang, X.; Wang, X. miR-200b and miR-200c co-contribute to the cisplatin sensitivity of ovarian cancer cells by targeting DNA methyltransferases. Oncol. Lett. 2018, 17, 1453–1460. [Google Scholar] [CrossRef] [Green Version]
- Cha, S.Y.; Choi, Y.H.; Hwang, S.; Jeong, J.-Y.; An, H.J. Clinical Impact of microRNAs Associated with Cancer Stem Cells as a Prognostic Factor in Ovarian Carcinoma. J. Cancer 2017, 8, 3538–3547. [Google Scholar] [CrossRef] [Green Version]
- Kan, C.W.S.; Hahn, M.A.; Gard, G.B.; Maidens, J.; Huh, J.Y.; Marsh, D.J.; Howell, V.M. Elevated levels of circulating microRNA-200 family members correlate with serous epithelial ovarian cancer. BMC Cancer 2012, 12, 627. [Google Scholar] [CrossRef] [Green Version]
- Iorio, M.V.; Visone, R.; Di Leva, G.; Donati, V.; Petrocca, F.; Casalini, P.; Taccioli, C.; Volinia, S.; Liu, C.-G.; Alder, H.; et al. MicroRNA Signatures in Human Ovarian Cancer. Cancer Res. 2007, 67, 8699–8707. [Google Scholar] [CrossRef] [Green Version]
- Nam, E.J.; Yoon, H.; Kim, S.W.; Kim, H.; Kim, Y.T.; Kim, J.H.; Kim, S. MicroRNA Expression Profiles in Serous Ovarian Carcinoma. Clin. Cancer Res. 2008, 14, 2690–2695. [Google Scholar] [CrossRef] [Green Version]
- Calura, E.; Fruscio, R.; Paracchini, L.; Bignotti, E.; Ravaggi, A.; Martini, P.; Sales, G.; Beltrame, L.; Clivio, L.; Ceppi, L.; et al. miRNA Landscape in Stage I Epithelial Ovarian Cancer Defines the Histotype Specificities. Clin. Cancer Res. 2013, 19, 4114–4123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elgaaen, B.V.; Olstad, O.K.; Haug, K.B.F.; Brusletto, B.; Sandvik, L.; Staff, A.C.; Gautvik, K.M.; Davidson, B. Global miRNA expression analysis of serous and clear cell ovarian carcinomas identifies differentially expressed miRNAs including miR-200c-3p as a prognostic marker. BMC Cancer 2014, 14, 80. [Google Scholar] [CrossRef] [Green Version]
- Yeom, K.-H.; Lee, Y.; Han, J.; Suh, M.R.; Kim, V.N. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006, 34, 4622–4629. [Google Scholar] [CrossRef] [Green Version]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef] [Green Version]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef] [Green Version]
- Palmero, E.; De Campos, S.G.P.; Campos, M.; De Souza, N.C.N.; Guerreiro, I.D.C.; Carvalho, A.L.; Marques, M.M.C. Mechanisms and role of microRNA deregulation in cancer onset and progression. Genet. Mol. Biol. 2011, 34, 363–370. [Google Scholar] [CrossRef]
- Yin, F.; Liu, X.; Li, D.; Wang, Q.; Zhang, W.; Li, L. Tumor suppressor genes associated with drug resistance in ovarian cancer (Review). Oncol. Rep. 2013, 30, 3–10. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Pan, X.; Cobb, G.; Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Schwartz, P.; Scarampi, L.; Rutherford, T.; Canuto, E.M.; Yu, H.; Katsaros, D. MicroRNA let-7a: A potential marker for selection of paclitaxel in ovarian cancer management. Gynecol. Oncol. 2011, 122, 366–371. [Google Scholar] [CrossRef]
- Wang, Y.; Lei, X.; Gao, C.; Xue, Y.; Li, X.; Wang, H.; Feng, Y. MiR-506-3p suppresses the proliferation of ovarian cancer cells by negatively regulating the expression of MTMR6. J. Biosci. 2019, 44, 126. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Mitra, R.; Maulik, U.; Zhang, M.Q. Development of the human cancer microRNA network. Silence 2010, 1, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansson, M.D.; Lund, A.H. MicroRNA and cancer. Mol. Oncol. 2012, 6, 590–610. [Google Scholar] [CrossRef] [PubMed]
- Berchuck, A.; Kohler, M.F.; Bast, R.C. Oncogenes in Ovarian Cancer. Hematol. Clin. N. Am. 1992, 6, 813–827. [Google Scholar] [CrossRef]
- Vescarelli, E.; Gerini, G.; Megiorni, F.; Anastasiadou, E.; Pontecorvi, P.; Solito, L.; De Vitis, C.; Camero, S.; Marchetti, C.; Mancini, R.; et al. MiR-200c sensitizes Olaparib-resistant ovarian cancer cells by targeting Neuropilin 1. J. Exp. Clin. Cancer Res. 2020, 39, 3. [Google Scholar] [CrossRef]
- Duran, G.E.; Wang, Y.C.; Moisan, F.; Francisco, E.B.; Sikic, B.I. Decreased levels of baseline and drug-induced tubulin polymerisation are hallmarks of resistance to taxanes in ovarian cancer cells and are associated with epithelial-to-mesenchymal transition. Br. J. Cancer 2017, 116, 1318–1328. [Google Scholar] [CrossRef] [Green Version]
- Huang, R.-X.; Zhou, P.-K. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Target. Ther. 2020, 5, 60. [Google Scholar] [CrossRef]
- Taylor, R.; Cullen, S.P.; Martin, S. Apoptosis: Controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 2008, 9, 231–241. [Google Scholar] [CrossRef]
- Yousefi, B.; Rahmati, M.; Ahmadi, Y. The roles of p53R2 in cancer progression based on the new function of mutant p53 and cytoplasmic p21. Life Sci. 2014, 99, 14–17. [Google Scholar] [CrossRef]
- Fukuda, T.; Yamagata, K.; Fujiyama, S.; Matsumoto, T.; Koshida, I.; Yoshimura, K.; Mihara, M.; Naitou, M.; Endoh, H.; Nakamura, T.; et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat. Cell Biol. 2007, 9, 604–611. [Google Scholar] [CrossRef]
- Gralewska, P.; Gajek, A.; Marczak, A.; Rogalska, A. Participation of the ATR/CHK1 pathway in replicative stress targeted therapy of high-grade ovarian cancer. J. Hematol. Oncol. 2020, 13, 39. [Google Scholar] [CrossRef] [Green Version]
- Saravi, S.; Katsuta, E.; Jeyaneethi, J.; Amin, H.A.; Kaspar, M.; Takabe, K.; Pados, G.; Drenos, F.; Hall, M.; Karteris, E. H2A Histone Family Member X (H2AX) Is Upregulated in Ovarian Cancer and Demonstrates Utility as a Prognostic Biomarker in Terms of Overall Survival. J. Clin. Med. 2020, 9, 2844. [Google Scholar] [CrossRef] [PubMed]
- Medina, P.P.; Slack, F.J. MicroRNAs and cancer: An overview. Cell Cycle 2008, 7, 2485–2492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Huang, J.-W.; Li, M.; Cavenee, W.K.; Mitchell, P.; Zhou, X.; Tewari, M.; Furnari, F.B.; Taniguchi, T. MicroRNA-138 Modulates DNA Damage Response by Repressing Histone H2AX Expression. Mol. Cancer Res. 2011, 9, 1100–1111. [Google Scholar] [CrossRef] [Green Version]
- Georges, S.A.; Biery, M.C.; Kim, S.-Y.; Schelter, J.M.; Guo, J.; Chang, A.N.; Jackson, A.L.; Carleton, M.O.; Linsley, P.S.; Cleary, M.A.; et al. Coordinated Regulation of Cell Cycle Transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008, 68, 10105–10112. [Google Scholar] [CrossRef] [Green Version]
- Agostini, A.; Brunetti, M.; Davidson, B.; Tropé, C.G.; Eriksson, A.G.Z.; Heim, S.; Panagopoulos, I.; Micci, F. The microRNA miR-192/215 family is upregulated in mucinous ovarian carcinomas. Sci. Rep. 2018, 8, 11069. [Google Scholar] [CrossRef]
- Stucki, M.; Clapperton, J.A.; Mohammad, D.; Yaffe, M.B.; Smerdon, S.J.; Jackson, S.P. MDC1 Directly Binds Phosphorylated Histone H2AX to Regulate Cellular Responses to DNA Double-Strand Breaks. Cell 2005, 123, 1213–1226. [Google Scholar] [CrossRef] [Green Version]
- Xiao, A.; Li, H.; Shechter, D.; Ahn, S.H.; Fabrizio, L.A.; Erdjument-Bromage, H.; Ishibe-Murakami, S.; Wang, B.; Tempst, P.; Hofmann, K.; et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nat. Cell Biol. 2008, 457, 57–62. [Google Scholar] [CrossRef] [Green Version]
- Coster, G.; Goldberg, M. The cellular response to DNA damage: A focus on MDC1 and its interacting proteins. Nucleus 2010, 1, 166–178. [Google Scholar] [CrossRef]
- Chapman, R.; Jackson, S.P. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008, 9, 795–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukas, J.; Lukas, C.; Bartek, J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 2011, 13, 1161–1169. [Google Scholar] [CrossRef]
- Nakada, S. Opposing roles of RNF8/RNF168 and deubiquitinating enzymes in ubiquitination-dependent DNA double-strand break response signaling and DNA-repair pathway choice. J. Radiat. Res. 2016, 57, i33–i40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Qin, B.; Lou, Z. Ubiquitin and ubiquitin-like molecules in DNA double strand break repair. Cell Biosci. 2020, 10, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, B.D.; D’Andrea, A.D. Chromatin Remodeling at DNA Double-Strand Breaks. Cell 2013, 152, 1344–1354. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Yin, H.; Zhang, Y.; Feng, Y.; Yan, Z.; Jiang, X.; Bukhari, I.; Iqbal, F.; Cooke, H.J.; Shi, Q. miR-214-mediated downregulation of RNF8 induces chromosomal instability in ovarian cancer cells. Cell Cycle 2014, 13, 3519–3528. [Google Scholar] [CrossRef] [Green Version]
- Wan, G.; Mathur, R.; Hu, X.; Zhang, X.; Lu, X. miRNA response to DNA damage. Trends Biochem. Sci. 2011, 36, 478–484. [Google Scholar] [CrossRef] [Green Version]
- Lal, A.; Pan, Y.; Navarro, F.; Dykxhoorn, D.M.; Moreau, L.; Meire, E.; Bentwich, Z.; Lieberman, J.; Chowdhury, D. miR-24–mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat. Struct. Mol. Biol. 2009, 16, 492–498. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Zhang, L.; Duan, L.; Lu, C. MicroRNA-3196 is inhibited by H2AX phosphorylation and attenuates lung cancer cell apoptosis by downregulating PUMA. Oncotarget 2016, 7, 77764–77776. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Sun, Y.; Zhang, P.-Y.; Qian, M.; Zhang, H.; Chen, X.; Ma, D.; Xu, Y.; Chen, X.; Tang, K.-F. The Fra-1–miR-134–SDS22 feedback loop amplifies ERK/JNK signaling and reduces chemosensitivity in ovarian cancer cells. Cell Death Dis. 2016, 7, e2384. [Google Scholar] [CrossRef]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef] [Green Version]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nat. Cell Biol. 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [Green Version]
- Dietlein, F.; Thelen, L.; Reinhardt, H.C. Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends Genet. 2014, 30, 326–339. [Google Scholar] [CrossRef] [PubMed]
- Alhmoud, J.F.; Woolley, J.F.; Al Moustafa, A.-E.; Malki, M.I. DNA Damage/Repair Management in Cancers. Cancers 2020, 12, 1050. [Google Scholar] [CrossRef]
- Van Gent, D.C.; Hoeijmakers, J.H.J.; Kanaar, R. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2001, 2, 196–206. [Google Scholar] [CrossRef]
- Lieber, M.R. The Mechanism of Double-Strand DNA Break Repair by the Nonhomologous DNA End-Joining Pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef] [Green Version]
- Brandsma, I.; van Gent, D.C. Pathway choice in DNA double strand break repair: Observations of a balancing act. Genome Integr. 2012, 3, 9. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Wei, C.; Li, J.; Xing, P.; Li, J.; Zheng, S.; Chen, X. Cell cycle-dependent control of homologous recombination. Acta Biochim. Biophys. Sin. 2017, 49, 655–668. [Google Scholar] [CrossRef] [Green Version]
- Northall, S.J.; Ivančić-Baće, I.; Soultanas, P.; Bolt, E.L. Remodeling and Control of Homologous Recombination by DNA Helicases and Translocases that Target Recombinases and Synapsis. Genes 2016, 7, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolinjivadi, A.M.; Sannino, V.; De Antoni, A.; Técher, H.; Baldi, G.; Costanzo, V. Moonlighting at replication forks—A new life for homologous recombination proteins BRCA1, BRCA2 and RAD51. FEBS Lett. 2017, 591, 1083–1100. [Google Scholar] [CrossRef] [PubMed]
- Moskwa, P.; Buffa, F.M.; Pan, Y.; Panchakshari, R.; Gottipati, P.; Muschel, R.J.; Beech, J.; Kulshrestha, R.; Abdelmohsen, K.; Weinstock, D.M.; et al. miR-182-Mediated Downregulation of BRCA1 Impacts DNA Repair and Sensitivity to PARP Inhibitors. Mol. Cell 2011, 41, 210–220. [Google Scholar] [CrossRef]
- Pannunzio, N.R.; Watanabe, G.; Lieber, M.R. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10512–10523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiruvella, K.K.; Liang, Z.; Wilson, T.E. Repair of Double-Strand Breaks by End Joining. Cold Spring Harb. Perspect. Biol. 2013, 5, a012757. [Google Scholar] [CrossRef]
- Yan, D.; Ng, W.L.; Zhang, X.; Wang, P.; Zhang, Z.; Mo, Y.-Y.; Mao, H.; Hao, C.; Olson, J.J.; Curran, W.J.; et al. Targeting DNA-PKcs and ATM with miR-101 Sensitizes Tumors to Radiation. PLoS ONE 2010, 5, e11397. [Google Scholar] [CrossRef]
- Chakraborty, A.; Uechi, T.; Kenmochi, N. Guarding the ‘translation apparatus’: Defective ribosome biogenesis and the p53 signaling pathway. Wiley Interdiscip. Rev. RNA 2011, 2, 507–522. [Google Scholar] [CrossRef]
- Dancy, B.M.; Cole, P.A. Protein Lysine Acetylation by p300/CBP. Chem. Rev. 2015, 115, 2419–2452. [Google Scholar] [CrossRef]
- Hasty, P.; Christy, B.A. p53 as an intervention target for cancer and aging. Pathobiol. Aging Age-Relat. Dis. 2013, 3. [Google Scholar] [CrossRef]
- Chen, X.; Che, T. Roles of MicroRNA in DNA Damage and Repair. In DNA Repair; Kruman, I., Ed.; InTech; ISBN 978-953-307-697-3. Available online: http://www.intechopen.com/books/dnarepair/roles-of-microrna-in-dna-damage-and-repair (accessed on 13 April 2021).
- Jones, M.F.; Lal, A. MicroRNAs, wild-type and mutant p53: More questions than answers. RNA Biol. 2012, 9, 781–791. [Google Scholar] [CrossRef]
- Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci USA 2008, 105, 13421–13426. [Google Scholar] [CrossRef] [Green Version]
- Park, S.-Y.; Lee, J.H.; Ha, M.; Nam, J.-W.; Kim, V.N. miR-29 miRNAs activate p53 by targeting p85α and CDC42. Nat. Struct. Mol. Biol. 2008, 16, 23–29. [Google Scholar] [CrossRef]
- Fornari, F.; Gramantieri, L.; Ferracin, M.; Veronese, A.; Sabbioni, S.; A Calin, G.; Grazi, G.L.; Giovannini, C.; Croce, C.M.; Bolondi, L.; et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 2008, 27, 5651–5661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fornari, F.; Gramantieri, L.; Giovannini, C.; Veronese, A.; Ferracin, M.; Sabbioni, S.; Calin, G.A.; Grazi, G.L.; Croce, C.M.; Tavolari, S.; et al. MiR-122/Cyclin G1 Interaction Modulates p53 Activity and Affects Doxorubicin Sensitivity of Human Hepatocarcinoma Cells. Cancer Res. 2009, 69, 5761–5767. [Google Scholar] [CrossRef] [Green Version]
- Corney, D.C.; Flesken-Nikitin, A.; Godwin, A.K.; Wang, W.; Nikitin, A.Y. MicroRNA-34b and MicroRNA-34c Are Targets of p53 and Cooperate in Control of Cell Proliferation and Adhesion-Independent Growth. Cancer Res. 2007, 67, 8433–8438. [Google Scholar] [CrossRef] [Green Version]
- Raver-Shapira, N.; Marciano, E.; Meiri, E.; Spector, Y.; Rosenfeld, N.; Moskovits, N.; Bentwich, Z.; Oren, M. Transcriptional Activation of miR-34a Contributes to p53-Mediated Apoptosis. Mol. Cell 2007, 26, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.-C.; Wentzel, E.A.; Kent, O.A.; Ramachandran, K.; Mullendore, M.; Lee, K.H.; Feldmann, G.; Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J.; et al. Transactivation of miR-34a by p53 Broadly Influences Gene Expression and Promotes Apoptosis. Mol. Cell 2007, 26, 745–752. [Google Scholar] [CrossRef] [Green Version]
- Asslaber, D.; Piñón, J.D.; Seyfried, I.; Desch, P.; Stöcher, M.; Tinhofer, I.; Egle, A.; Merkel, O.; Greil, R. microRNA-34a expression correlates with MDM2 SNP309 polymorphism and treatment-free survival in chronic lymphocytic leukemia. Blood 2010, 115, 4191–4197. [Google Scholar] [CrossRef] [Green Version]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nat. Cell Biol. 2005, 434, 913–917. [Google Scholar] [CrossRef]
- Meghani, K.; Fuchs, W.; Detappe, A.; Drané, P.; Gogola, E.; Rottenberg, S.; Jonkers, J.; Matulonis, U.; Swisher, E.M.; Konstantinopoulos, P.A.; et al. Multifaceted Impact of MicroRNA 493-5p on Genome-Stabilizing Pathways Induces Platinum and PARP Inhibitor Resistance in BRCA2-Mutated Carcinomas. Cell Rep. 2018, 23, 100–111. [Google Scholar] [CrossRef] [Green Version]
- Neijenhuis, S.; Bajrami, I.; Miller, R.; Lord, C.J.; Ashworth, A. Identification of miRNA modulators to PARP inhibitor response. DNA Repair 2013, 12, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, J.-W.; Calses, P.; Kemp, C.J.; Taniguchi, T. MiR-96 Downregulates REV1 and RAD51 to Promote Cellular Sensitivity to Cisplatin and PARP Inhibition. Cancer Res. 2012, 72, 4037–4046. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Niu, Z.; Lin, X.; Tian, Y. MiR-216b increases cisplatin sensitivity in ovarian cancer cells by targeting PARP1. Cancer Gene Ther. 2017, 24, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Kaur, S.; Volinia, S.; Greshock, J.; Lassus, H.; Hasegawa, K.; Liang, S.; Leminen, A.; Deng, S.; Smith, L.; et al. MicroRNA Microarray Identifies Let-7i as a Novel Biomarker and Therapeutic Target in Human Epithelial Ovarian Cancer. Cancer Res. 2008, 68, 10307–10314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortez, D.; Guntuku, S.; Qin, J.; Elledge, S.J. ATR and ATRIP: Partners in Checkpoint Signaling. Science 2001, 294, 1713–1716. [Google Scholar] [CrossRef]
- Cimprich, K.A.; Cortez, D. ATR: An essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008, 9, 616–627. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.-J.; Tian, R.; You, A.-G.; Wang, J.; Wu, Y.-J.; Wang, W.; Zhou, A.-Y.; Wei, X.-L.; He, Q.-D.; Feng, X.; et al. Expression and significance of DNA methyltransferase in sera of patients with lung cancer. Zhonghua Yi Xue Za Zhi 2013, 93, 3822–3825. [Google Scholar]
- Bartek, J.; Lukas, J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 2003, 3, 421–429. [Google Scholar] [CrossRef] [Green Version]
- Buisson, R.; Boisvert, J.L.; Benes, C.H.; Zou, L. Distinct but Concerted Roles of ATR, DNA-PK, and Chk1 in Countering Replication Stress during S Phase. Mol. Cell 2015, 59, 1011–1024. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Li, Y.; Wang, F.; Wang, X.; Cheng, B.; Ye, F.; Xie, X.; Zhou, C.; Lu, W. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene 2012, 32, 976–987. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yao, L.; Liu, F.; Hong, J.; Chen, L.; Zhang, B.; Zhang, W. Characterization of microRNA expression in serous ovarian carcinoma. Int. J. Mol. Med. 2014, 34, 491–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Qu, J.; Xu, F.; Guo, Y.; Wang, Y.; Yu, H.; Qian, B. MiR-195 suppresses non-small cell lung cancer by targeting CHEK1. Oncotarget 2015, 6, 9445–9456. [Google Scholar] [CrossRef] [Green Version]
- Su, B.-B.; Zhou, S.-W.; Gan, C.-B.; Zhang, X.-N. MiR-330-5p regulates tyrosinase and PDIA3 expression and suppresses cell proliferation and invasion in cutaneous malignant melanoma. J. Surg. Res. 2016, 203, 434–440. [Google Scholar] [CrossRef]
- Lee, K.-H.; Chen, Y.-L.; Yeh, S.-D.; Hsiao, M.; Lin, J.-T.; Goan, Y.-G.; Lu, P.-J. MicroRNA-330 acts as tumor suppressor and induces apoptosis of prostate cancer cells through E2F1-mediated suppression of Akt phosphorylation. Oncogene 2009, 28, 3360–3370. [Google Scholar] [CrossRef] [Green Version]
- Bibby, B.A.S.; Reynolds, J.V.; Maher, S.G. MicroRNA-330-5p as a Putative Modulator of Neoadjuvant Chemoradiotherapy Sensitivity in Oesophageal Adenocarcinoma. PLoS ONE 2015, 10, e0134180. [Google Scholar] [CrossRef] [Green Version]
- Kong, R.; Liu, W.; Guo, Y.; Feng, J.; Cheng, C.; Zhang, X.; Ma, Y.; Li, S.; Jiang, J.; Zhang, J.; et al. Inhibition of NOB1 by microRNA-330-5p overexpression represses cell growth of non-small cell lung cancer. Oncol. Rep. 2017, 38, 2572–2580. [Google Scholar] [CrossRef]
- Shao, S.; Tian, J.; Zhang, H.; Wang, S. LncRNA myocardial infarction-associated transcript promotes cell proliferation and inhibits cell apoptosis by targeting miR-330-5p in epithelial ovarian cancer cells. Arch. Med. Sci. 2018, 14, 1263–1270. [Google Scholar] [CrossRef] [PubMed]
- Gong, D.; Feng, P.-C.; Ke, X.-F.; Kuang, H.-L.; Pan, L.-L.; Ye, Q.; Wu, J.-B. Silencing Long Non-coding RNA LINC01224 Inhibits Hepatocellular Carcinoma Progression via MicroRNA-330-5p-Induced Inhibition of CHEK1. Mol. Ther. Nucleic Acids 2020, 19, 482–497. [Google Scholar] [CrossRef]
- Xie, Y.; Wei, R.-R.; Huang, G.-L.; Zhang, M.-Y.; Yuan, Y.-F.; Wang, H.-Y. Checkpoint kinase 1 is negatively regulated by miR-497 in hepatocellular carcinoma. Med. Oncol. 2014, 31, 844. [Google Scholar] [CrossRef]
- Wei, L.; He, Y.; Bi, S.; Li, X.; Zhang, J.; Zhang, S. miRNA-199b-3p suppresses growth and progression of ovarian cancer via the CHK1/E-cadherin/EMT signaling pathway by targeting ZEB1. Oncol. Rep. 2020, 45, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; James, J.; Annunziata, C.M. Topotecan synergizes with CHEK1 (CHK1) inhibitor to induce apoptosis in ovarian cancer cells. BMC Cancer 2015, 15, 196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Wang, G.; Zhang, X.; Wu, D.; Yang, L.; Wang, G.; Hao, D. The expression of miRNAs is associated with tumour genome instability and predicts the outcome of ovarian cancer patients treated with platinum agents. Sci. Rep. 2017, 7, 14736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, P.; Roela, R.A.; Lopez, R.V.M.; Estevez-Diz, M.D.P. The prognostic role of microRNA in epithelial ovarian cancer: A systematic review of literature with an overall survival meta-analysis. Oncotarget 2020, 11, 1085–1095. [Google Scholar] [CrossRef] [PubMed]


| MiRNAs as Oncogenes | Ref | MiRNAs as Suppressors | Ref |
|---|---|---|---|
| miR-138 | [26,27] | miR-16 | [28,29] |
| miR-200 a, a-3p, b, c | [30,31,32,33] | miR-10a, 10b | [34] |
| miR-141 | [30,33] | miR-29 | [35,36] |
| miR-429 | [30,33] | miR-let-7 | [37,38,39,40] |
| miR-205 | [41] | miR-31, 31-5p | [42,43] |
| miR-126-3p | [44] | miR-506-3p | [45] |
| miR-183 | [46] | miR-424-5p | [47] |
| miR-760 | [48,49] | miR-503-5p | [47] |
| miR-151 | [50] | miR-199a-5p | [51] |
| miR-21-5p | [52] | miR-34 | [53] |
| miR-106a | [54] | miR-340-5p | [55] |
| miR-195 | [54] | miR-138 | [56] |
| miR-222 | [57,58] | miR-509-3 | [59] |
| miR-221 | [57,58,60] | miR-335-5p | [61] |
| miR-520b | [62] | miR-383 | [63] |
| miR-10b | [64] | miR-185 | [65] |
| miR-21 | [66] | miR-126 | [67] |
| miR-17-92 | [66] | miR-708 | [68] |
| miR-622 | [69] | miR-200c | [18,70,71] |
| miR-424-5p | [72] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gajek, A.; Gralewska, P.; Marczak, A.; Rogalska, A. Current Implications of microRNAs in Genome Stability and Stress Responses of Ovarian Cancer. Cancers 2021, 13, 2690. https://doi.org/10.3390/cancers13112690
Gajek A, Gralewska P, Marczak A, Rogalska A. Current Implications of microRNAs in Genome Stability and Stress Responses of Ovarian Cancer. Cancers. 2021; 13(11):2690. https://doi.org/10.3390/cancers13112690
Chicago/Turabian StyleGajek, Arkadiusz, Patrycja Gralewska, Agnieszka Marczak, and Aneta Rogalska. 2021. "Current Implications of microRNAs in Genome Stability and Stress Responses of Ovarian Cancer" Cancers 13, no. 11: 2690. https://doi.org/10.3390/cancers13112690
APA StyleGajek, A., Gralewska, P., Marczak, A., & Rogalska, A. (2021). Current Implications of microRNAs in Genome Stability and Stress Responses of Ovarian Cancer. Cancers, 13(11), 2690. https://doi.org/10.3390/cancers13112690






