Tracking the Antibody Immunome in Sporadic Colorectal Cancer by Using Antigen Self-Assembled Protein Arrays
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Nucleic Acid Programmable Protein Array (NAPPArray) Performance and Serum Screening Conditions
2.3. Identification of Differential aAb Serum Profiles Across the Analyzed Samples
2.4. Functional In Silico Analysis
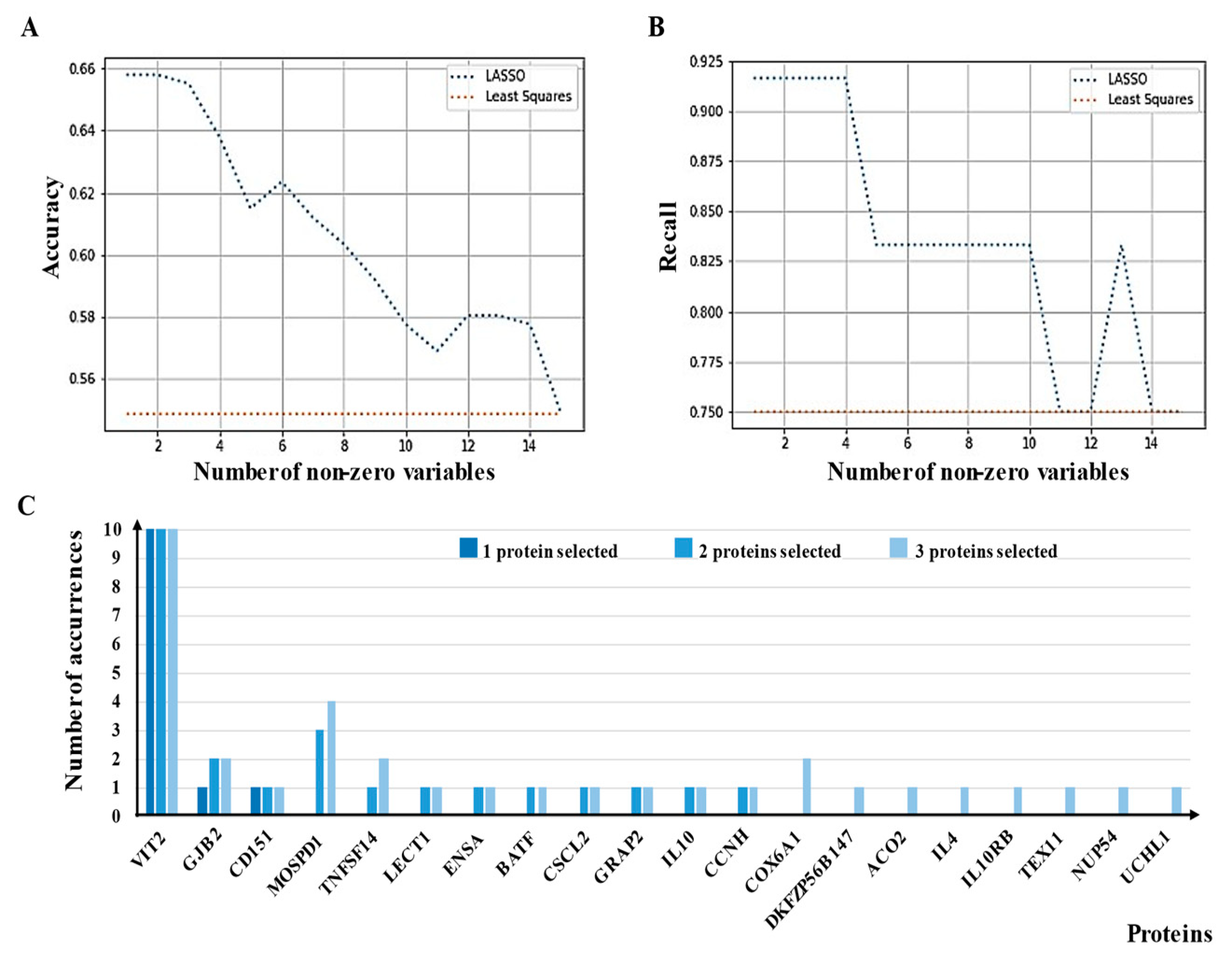
2.5. Feature Selection Algorithms and Linear Models
2.6. Data Visualization
2.7. Validation of a Featured Panel of TAAs
3. Results
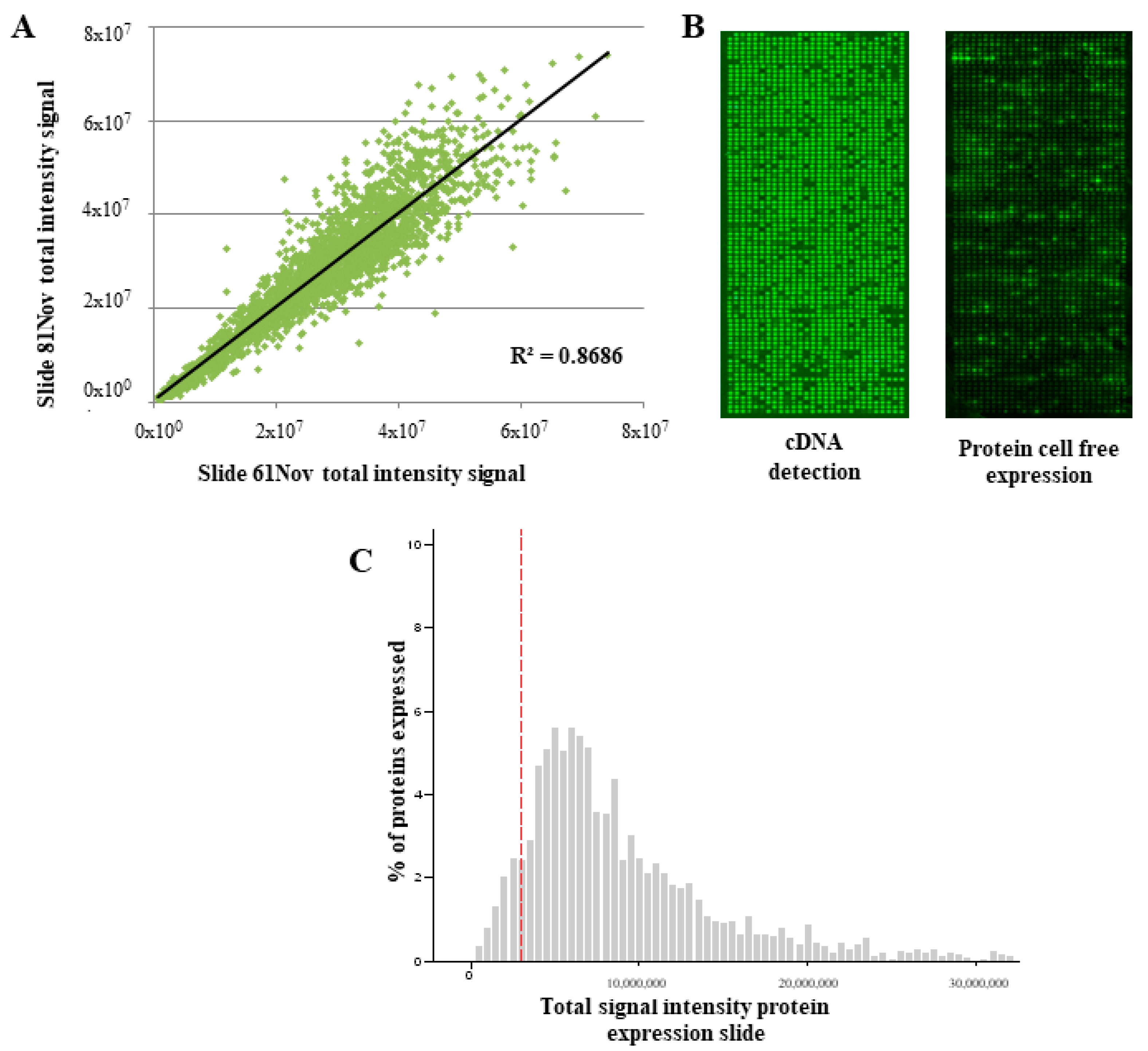
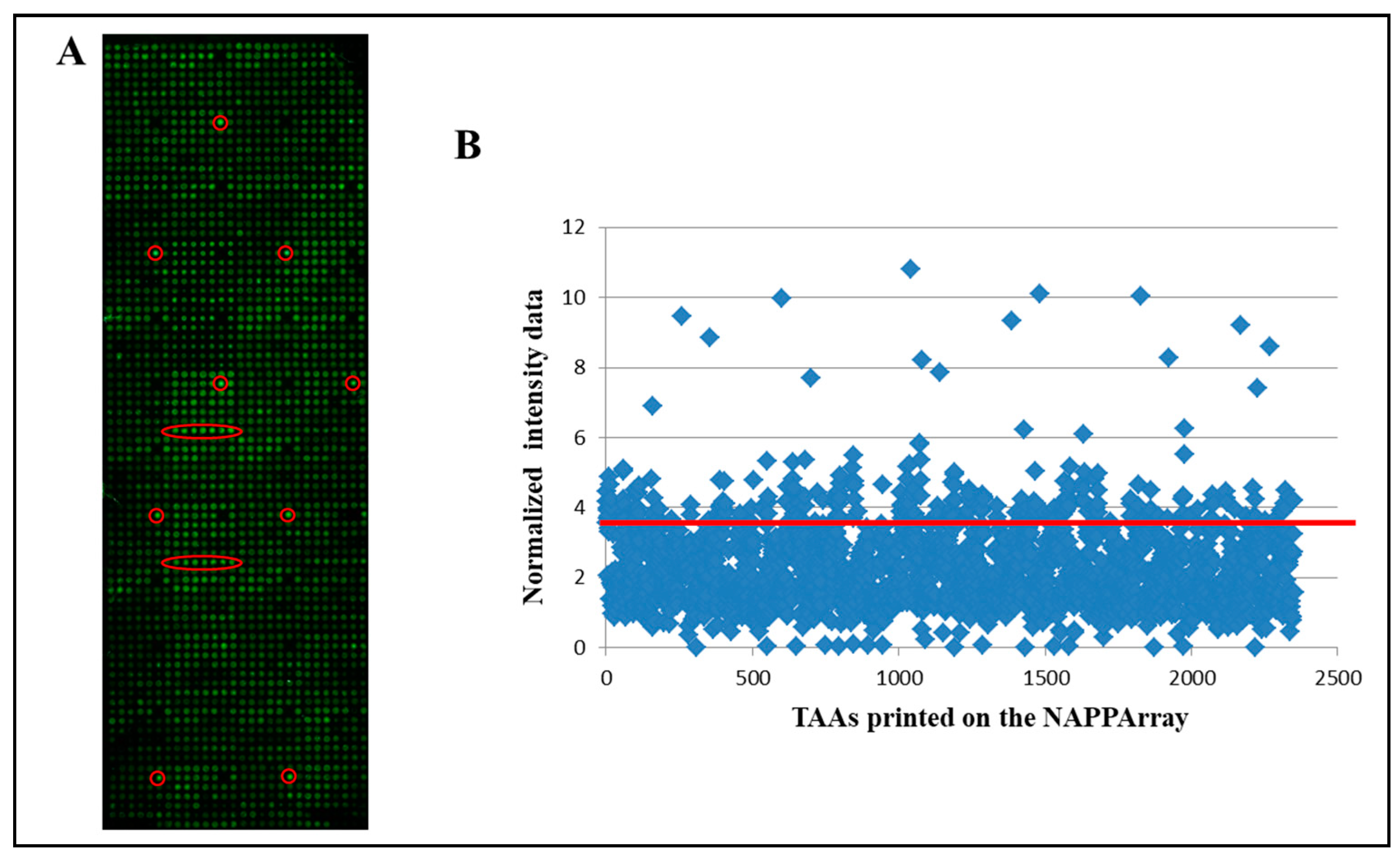
3.1. Performance of the NAPPArray for sCRC-Associated Plasma aAb Screening
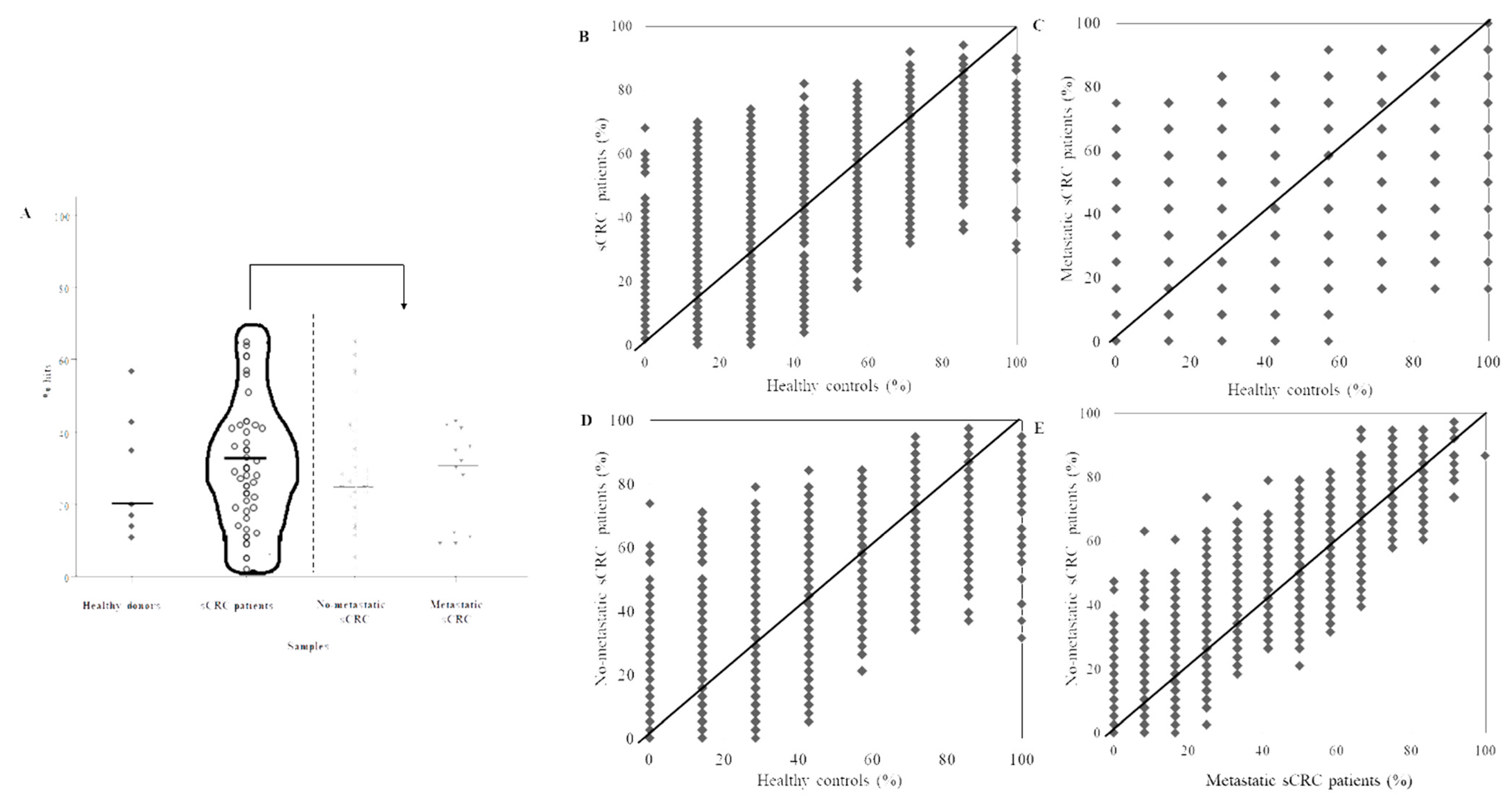
3.2. Identification of aAb Profiles in Healthy Donors vs. sCRC Plasma Samples
3.3. Determination of Differential aAb Profiles
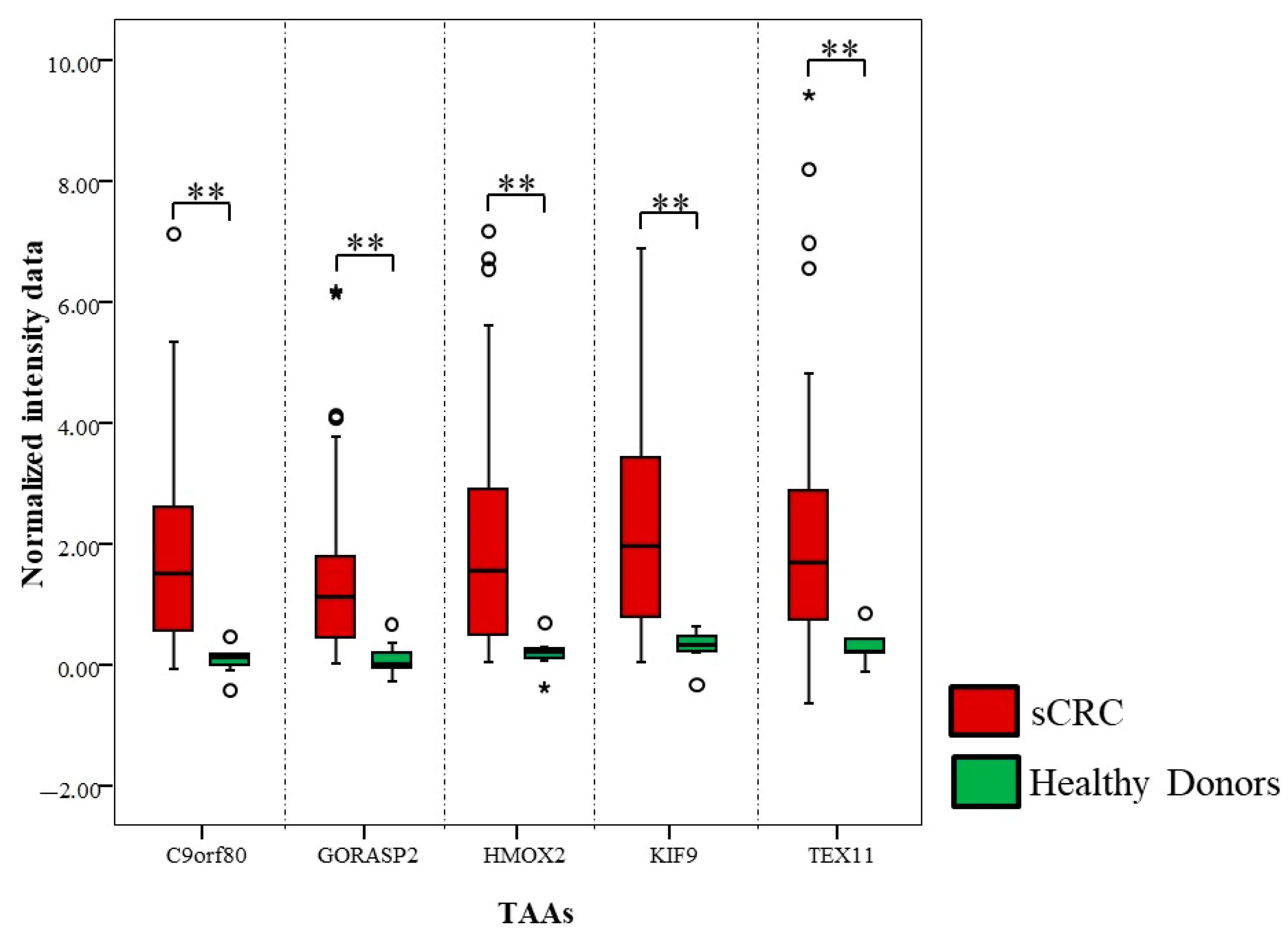
3.4. Functional In Silico Analysis of the Differential aAb Profiles Against TAAs in sCRC Patients
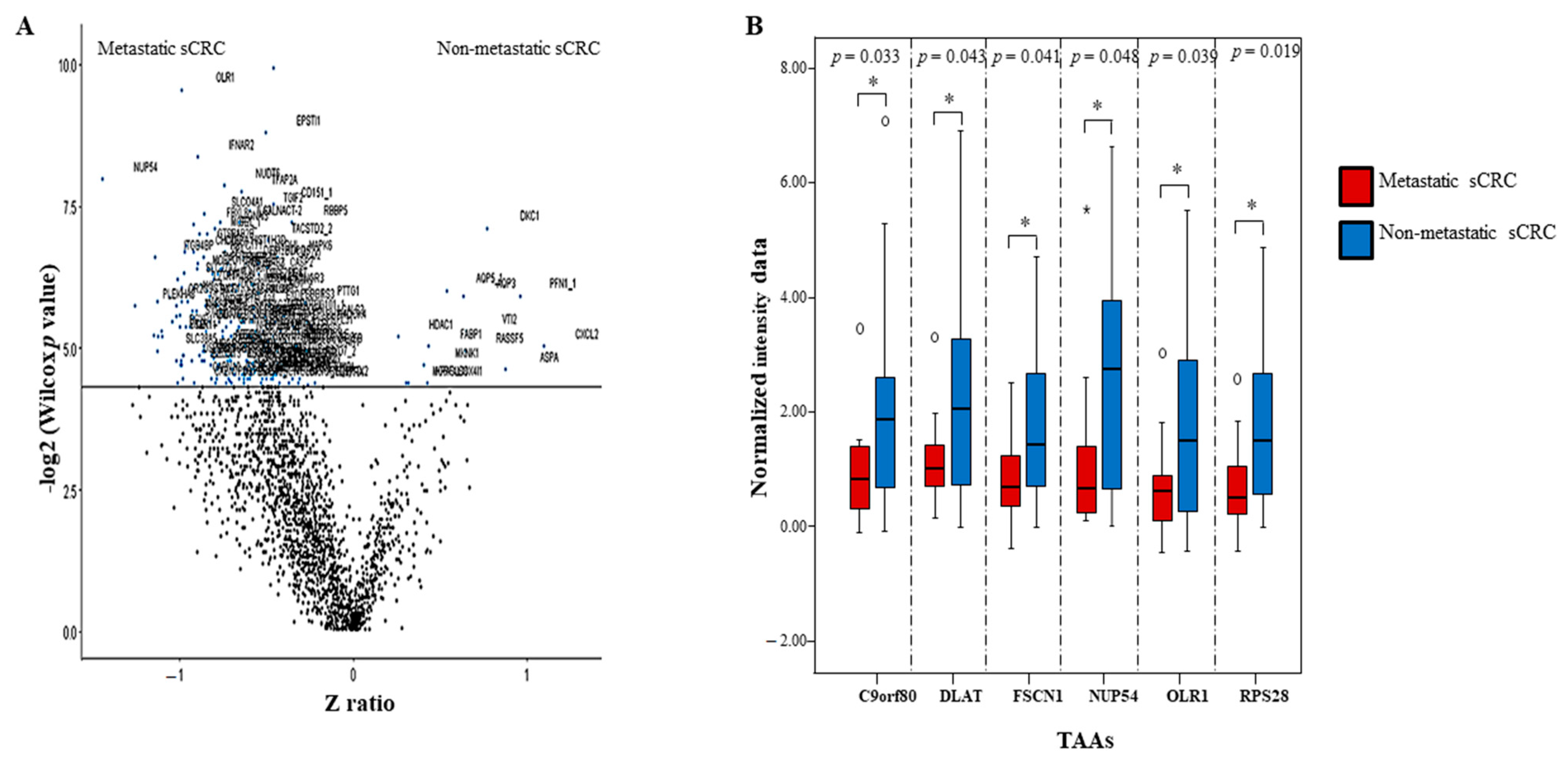
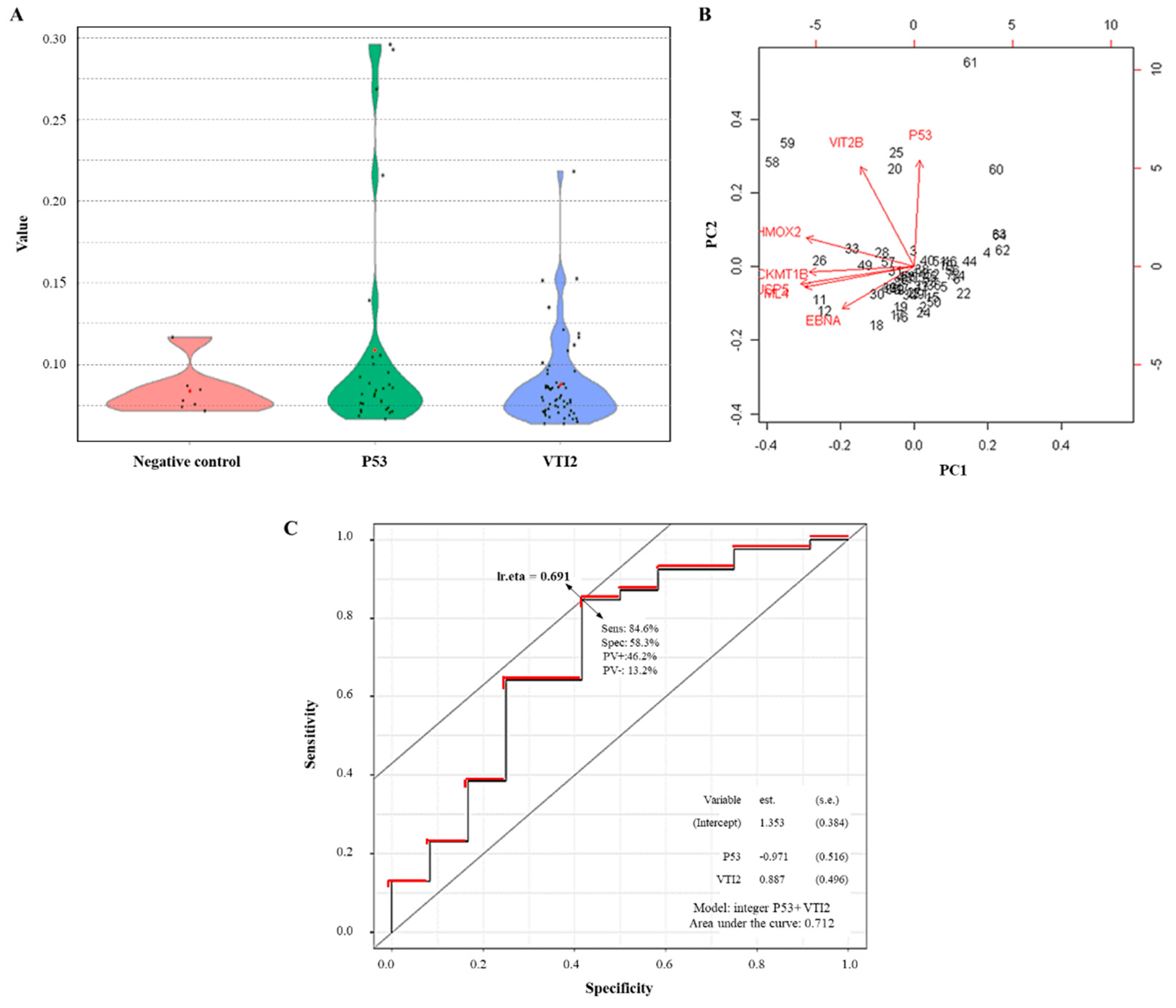
3.5. External Evaluation of a Potential Panel of aAbs as sCRC Useful Biomarkers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2015. CA A Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.; Spolverato, G.; Le, G.; Mavros, M.; Doyle, F.; Pawlik, T.; Winter, D. Synchronous colorectal liver metastasis: A network meta-analysis review comparing classical, combined, and liver-first surgical strategies. J. Surg. Oncol. 2015, 111, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Cervantes, A.; Nordlinger, B.; Arnold, D. Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2014, 25 (Suppl. 3), iii1–iii9. [Google Scholar] [CrossRef]
- Weitz, J.; Koch, M.; Debus, J.; Höhler, T.; Galle, P.R.; Büchler, M.W. Colorectal cancer. Lancet 2005, 365, 153–165. [Google Scholar] [CrossRef]
- De Wijkerslooth, T.R.; Bossuyt, P.M.; Dekker, E. Strategies in screening for colon carcinoma. Neth. J. Med. 2011, 69, 112–119. [Google Scholar]
- Tan, H.T.; Low, J.; Lim, S.G.; Chung, M.C.M. Serum autoantibodies as biomarkers for early cancer detection. FEBS J. 2009, 276, 6880–6904. [Google Scholar] [CrossRef]
- Anderson, K.S.; LaBaer, J. The sentinel within: Exploiting the immune system for cancer biomarkers. J. Proteome Res. 2005, 4, 1123–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, O.J. Immune response as a biomarker for cancer detection and a lot more. N. Engl. J. Med. 2005, 353, 1288–1290. [Google Scholar] [CrossRef]
- Qiu, J.; LaBaer, J. Nucleic Acid Programmable Protein Array. Methods Enzymol. 2011, 500, 151–163. [Google Scholar] [CrossRef] [Green Version]
- Miersch, S.; Bian, X.; Wallstrom, G.; Sibani, S.; Logvinenko, T.; Wasserfall, C.H.; Schatz, D.; Atkinson, M.; Qiu, J.; LaBaer, J. Serological autoantibody profiling of type 1 diabetes by protein arrays. J. Proteom. 2013, 94, 486–496. [Google Scholar] [CrossRef]
- Barderas, R.; Villar-Vázquez, R.; Fernández-Aceñero, M.J.; Babel, I.; Peláez-García, A.; Torres, S.; Casal, J.I. Sporadic colon cancer murine models demonstrate the value of autoantibody detection for preclinical cancer diagnosis. Sci. Rep. 2013, 3, srep02938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henjes, F.; Lourido, L.; Ruiz-Romero, C.; Fernández-Tajes, J.; Schwenk, J.M.; Gonzalez-Gonzalez, M.; Blanco, F.J.; Nilsson, P.; Fuentes, M. Analysis of Autoantibody Profiles in Osteoarthritis Using Comprehensive Protein Array Concepts. J. Proteome Res. 2014, 13, 5218–5229. [Google Scholar] [CrossRef] [PubMed]
- Katchman, B.A.; Barderas, R.; Alam, R.; Chowell, D.; Field, M.S.; Esserman, L.J.; Wallstrom, G.; LaBaer, J.; Cramer, D.W.; Hollingsworth, M.A.; et al. Proteomic mapping of p53 immunogenicity in pancreatic, ovarian, and breast cancers. Proteom. Clin. Appl. 2016, 10, 720–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, K.S.; Sibani, S.; Wallstrom, G.; Qiu, J.; Mendoza, E.A.; Raphael, J.; Hainsworth, E.; Montor, W.R.; Wong, J.; Park, J.G.; et al. Protein Microarray Signature of Autoantibody Biomarkers for the Early Detection of Breast Cancer. J. Proteome Res. 2010, 10, 85–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; O Petritis, B.; LaBaer, J. Advancing translational research with next-generation protein microarrays. Proteom. 2016, 16, 1238–1250. [Google Scholar] [CrossRef]
- Gonzalez-Gonzalez, M.; Jara-Acevedo, R.; Matarraz, S.; Jara-Acevedo, M.; Paradinas, S.; Sayagües, J.; Orfao, A.; Fuentes, M. Nanotechniques in proteomics: Protein microarrays and novel detection platforms. Eur. J. Pharm. Sci. 2012, 45, 499–506. [Google Scholar] [CrossRef]
- Ramachandran, N.; Raphael, J.V.; Hainsworth, E.; Demirkan, G.; Fuentes, M.G.; Rolfs, A.; Hu, Y.; LaBaer, J. Next-generation high-density self-assembling functional protein arrays. Nat. Methods 2008, 5, 535–538. [Google Scholar] [CrossRef] [Green Version]
- Manzano-Román, R.; Díaz-Martín, V.; González-González, M.; Matarraz, S.; Álvarez-Prado, A.F.; LaBaer, J.; Orfao, A.; Pérez-Sánchez, R.; Fuentes, M. Self-assembled Protein Arrays from an Ornithodoros moubata Salivary Gland Expression Library. J. Proteome Res. 2012, 11, 5972–5982. [Google Scholar] [CrossRef]
- Wang, D.; Yang, L.; Zhang, P.; LaBaer, J.; Hermjakob, H.; Li, D.; Yu, X. AAgAtlas 1.0: A human autoantigen database. Nucleic Acids Res. 2017, 45, D769–D776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tibshirani, R. Regression Shrinkage and Selection Via the Lasso. J. R. Stat. Soc. Ser. B Stat. Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Hansen, C.B.; Csuka, D.; Munthe-Fog, L.; Varga, L.; Farkas, H.; Hansen, K.M.; Koch, C.; Skjødt, K.; Garred, P.; Skjoedt, M.-O.; et al. The Levels of the Lectin Pathway Serine Protease MASP-1 and Its Complex Formation with C1 Inhibitor Are Linked to the Severity of Hereditary Angioedema. J. Immunol. 2015, 195, 3596–3604. [Google Scholar] [CrossRef] [Green Version]
- Ji, Q.; Wang, W.; Luo, Y.; Cai, F.; Lu, Y.; Deng, W.; Li, Q.; Su, S. Characteristic proteins in the plasma of postoperative colorectal and liver cancer patients with Yin deficiency of liver-kidney syndrome. Oncotarget 2017, 8, 103223–103235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Fan, J.; Chen, Q.; Lei, C.; Qiao, B.; Liu, Q. SPP1 and AGER as potential prognostic biomarkers for lung adenocarcinoma. Oncol. Lett. 2018, 15, 7028–7036. [Google Scholar] [CrossRef]
- Jiang, G.; Wang, P.; Wang, W.; Li, W.; Dai, L.; Chen, K. Annexin A13 promotes tumor cell invasion in vitro and is associated with metastasis in human colorectal cancer. Oncotarget 2017, 8, 21663–21673. [Google Scholar] [CrossRef] [Green Version]
- Miyoshi, N.; Yamamoto, H.; Mimori, K.; Yamashita, S.; Miyazaki, S.; Nakagawa, S.; Ishii, H.; Noura, S.; Ohue, M.; Yano, M.; et al. ANXA9 gene expression in colorectal cancer: A novel marker for prognosis. Oncol. Lett. 2014, 8, 2313–2317. [Google Scholar] [CrossRef]
- Roessler, M.; Rollinger, W.; Mantovani-Endl, L.; Hagmann, M.-L.; Palme, S.; Berndt, P.; Engel, A.M.; Pfeffer, M.; Karl, J.; Bodenmüller, H.; et al. Identification of PSME3 as a Novel Serum Tumor Marker for Colorectal Cancer by Combining Two-dimensional Polyacrylamide Gel Electrophoresis with a Strictly Mass Spectrometry-based Approach for Data Analysis. Mol. Cell. Proteom. 2006, 5, 2092–2101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melero, I.; Gabari, I.; Corbí, A.L.; Relloso, M.; Mazzolini, G.; Schmitz, V.; Rodriguez-Calvillo, M.; Tirapu, I.; Camafeita, E.; Albar, J.P.; et al. An anti-ICAM-2 (CD102) monoclonal antibody induces immune-mediated regressions of transplanted ICAM-2-negative colon carcinomas. Cancer Res. 2002, 62, 3167–3174. [Google Scholar]
- Uhlén, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rank, K.M.; Shaukat, A. Stool Based Testing for Colorectal Cancer: An Overview of Available Evidence. Curr. Gastroenterol. Rep. 2017, 19, 39. [Google Scholar] [CrossRef]
- Song, L.-L.; Li, Y.-M. Current noninvasive tests for colorectal cancer screening: An overview of colorectal cancer screening tests. World J. Gastrointest. Oncol. 2016, 8, 793–800. [Google Scholar] [CrossRef]
- Brenner, H.; Niedermaier, T.; Chen, H. Strong subsite-specific variation in detecting advanced adenomas by fecal immunochemical testing for hemoglobin. Int. J. Cancer 2017, 140, 2015–2022. [Google Scholar] [CrossRef] [Green Version]
- Cohen, S.J.; Punt, C.J.A.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of Circulating Tumor Cells to Tumor Response, Progression-Free Survival, and Overall Survival in Patients With Metastatic Colorectal Cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef]
- Zhao, R.; Cai, Z.; Li, S.; Cheng, Y.; Gao, H.; Liu, F.; Wu, S.; Liu, S.; Dong, Y.; Zheng, L.; et al. Expression and clinical relevance of epithelial and mesenchymal markers in circulating tumor cells from colorectal cancer. Oncotarget 2016, 8, 9293–9302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thierry, A.R.; Mouliere, F.; El Messaoudi, S.; Mollevi, C.; Lopez-Crapez, E.; Rolet, F.; Gillet, B.; Gongora, C.; Dechelotte, P.; Robert, B.; et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat. Med. 2014, 20, 430–435. [Google Scholar] [CrossRef]
- LeComte, T.; Ceze, N.; Dorval, E.; Laurent-Puig, P. Circulating free tumor DNA and colorectal cancer. Gastroentérol. Clin. Biol. 2010, 34, 662–681. [Google Scholar] [CrossRef] [PubMed]
- Engvall, E.; Perlmann, P. Enzyme-linked immunosorbent assay (ELISA) quantitative assay of immunoglobulin G. Immunochemistry 1971, 8, 871–874. [Google Scholar] [CrossRef]
- Kobold, S.; Lütkens, T.; Cao, Y.; Bokemeyer, C.; Atanackovic, D. Autoantibodies against tumor-related antigens: Incidence and biologic significance. Hum. Immunol. 2010, 71, 643–651. [Google Scholar] [CrossRef]
- Zaenker, P.; Gray, E.; Ziman, M. Autoantibody Production in Cancer—The Humoral Immune Response toward Autologous Antigens in Cancer Patients. Autoimmun. Rev. 2016, 15, 477–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pancione, M.; Giordano, G.; Remo, A.; Febbraro, A.; Sabatino, L.; Manfrin, E.; Ceccarelli, M.; Colantuoni, V. Immune Escape Mechanisms in Colorectal Cancer Pathogenesis and Liver Metastasis. J. Immunol. Res. 2014, 2014, 1–11. [Google Scholar] [CrossRef]
- Paltridge, J.L.; Belle, L.; Khew-Goodall, Y. The secretome in cancer progression. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2013, 1834, 2233–2241. [Google Scholar] [CrossRef]
- Barderas, R.; Mendes, M.; Torres, S.; Bartolomé, R.A.; López-Lucendo, M.; Villar-Vázquez, R.; Peláez-García, A.; Fuente, E.; Bonilla, F.; Casal, J.I. In-depth Characterization of the Secretome of Colorectal Cancer Metastatic Cells Identifies Key Proteins in Cell Adhesion, Migration, and Invasion. Mol. Cell. Proteom. 2013, 12, 1602–1620. [Google Scholar] [CrossRef] [Green Version]
- Pimentel-Nunes, P.; Gonçalves, N.; Boal-Carvalho, I.; Afonso, L.; Lopes, P.; Jr, R.R.-A.; Soares, J.-B.; Cardoso, E.; Henrique, R.; Moreira-Dias, L.; et al. Decreased Toll-interacting protein and peroxisome proliferator-activated receptor γ are associated with increased expression of Toll-like receptors in colon carcinogenesis. J. Clin. Pathol. 2012, 65, 302–308. [Google Scholar] [CrossRef]
- Finlayson, A.E.; Freeman, K.W. A Cell Motility Screen Reveals Role for MARCKS-Related Protein in Adherens Junction Formation and Tumorigenesis. PLoS ONE 2009, 4, e7833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qualtrough, D.; Singh, K.; Banu, N.; Paraskeva, C.; Pignatelli, M. The actin-bundling protein fascin is overexpressed in colorectal adenomas and promotes motility in adenoma cells in vitro. Br. J. Cancer 2009, 101, 1124–1129. [Google Scholar] [CrossRef] [Green Version]
- Alajez, N.M. Significance of BMI1 and FSCN1 expression in colorectal cancer. Saudi J. Gastroenterol. 2016, 22, 288–293. [Google Scholar] [CrossRef]
- García-Solano, J.; García-Solano, M.E.; Torres-Moreno, D.; Carbonell, P.; Trujillo-Santos, J.; Pérez-Guillermo, M.; Conesa-Zamora, P. Biomarkers for the identification of precursor polyps of colorectal serrated adenocarcinomas. Cell. Oncol. 2016, 39, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.Y.; Kim, Y.B.; Suh, K.W.; Paek, O.J.; Moon, H.Y. Prognostic Impact of Fascin-1 Expression is More Significant in Advanced Colorectal Cancer. J. Surg. Res. 2012, 172, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Tan, V.Y.; Lewis, S.J.; Adams, J.C.; Martin, R.M. Association of fascin-1 with mortality, disease progression and metastasis in carcinomas: A systematic review and meta-analysis. BMC Med. 2013, 11, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baik, I.H.; Jo, G.-H.; Seo, D.; Ko, M.J.; Cho, C.H.; Lee, M.G.; Lee, Y.-H. Knockdown of RPL9 expression inhibits colorectal carcinoma growth via the inactivation of Id-1/NF-κB signaling axis. Int. J. Oncol. 2016, 49. [Google Scholar] [CrossRef] [Green Version]
- Warner, J.R.; McIntosh, K.B. How Common Are Extraribosomal Functions of Ribosomal Proteins? Mol. Cell 2009, 34, 3–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parsyan, A.; Hernández, G.; Meterissian, S. Translation initiation in colorectal cancer. Cancer Metastasis Rev. 2012, 31, 387–395. [Google Scholar] [CrossRef]
- Chen, J.; Wei, Y.; Feng, Q.; Ren, L.; He, G.; Chang, W.; Zhu, D.; Yi, T.; Lin, Q.; Tang, W.; et al. Ribosomal protein S15A promotes malignant transformation and predicts poor outcome in colorectal cancer through misregulation of p53 signaling pathway. Int. J. Oncol. 2016, 48, 1628–1638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coronell, J.A.L.; Sergelen, K.; Höfer, P.; Gyurján, I.; Brezina, S.; Hettegger, P.; Leeb, G.; Mach, K.; Gsur, A.; Weinhäusel, A. The Immunome of Colon Cancer: Functional In Silico Analysis of Antigenic Proteins Deduced from IgG Microarray Profiling. Genom. Proteom. Bioinform. 2018, 16, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Garranzo-Asensio, M.; Segundo-Acosta, P.S.; Povés, C.; Fernández-Aceñero, M.J.; Martínez-Useros, J.; Montero-Calle, A.; Fernandez, G.S.; Sanchez-Martinez, M.; Rodríguez, N.; Cerón, M.Á.; et al. Identification of tumor-associated antigens with diagnostic ability of colorectal cancer by in-depth immunomic and seroproteomic analysis. J. Proteom. 2020, 214, 103635. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Kim, D.J.; Adams, J.C. The roles of fascins in health and disease. J. Pathol. 2011, 224, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Mukohyama, J.; Shimono, Y.; Minami, H.; Kakeji, Y.; Suzuki, A. Roles of microRNAs and RNA-Binding Proteins in the Regulation of Colorectal Cancer Stem Cells. Cancers 2017, 9, 143. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, Y.; Skacel, M.; Lavery, I.C.; Mukherjee, A.L.; Casey, G.; Adams, J.C. Prognostic significance of fascin expression in advanced colorectal cancer: An immunohistochemical study of colorectal adenomas and adenocarcinomas. BMC Cancer 2006, 6, 241. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.S.; Jung, E.-J.; Lee, J.-H.; Min, B.-W.; Kim, Y.-S. Clinicopathologic significance of fascin, extracellular matrix metalloproteinase inducer, and ezrin expressions in colorectal adenocarcinoma. Indian J. Pathol. Microbiol. 2011, 54. [Google Scholar] [CrossRef]
- Machesky, L.M.; Li, A. Fascin. Commun. Integr. Biol. 2010, 3, 263–270. [Google Scholar] [CrossRef]
- Gerke, V.; Creutz, C.E.; Moss, S.E. Annexins: Linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 2005, 6, 449–461. [Google Scholar] [CrossRef]
- Yu, S.; Bian, H.; Gao, X.; Gui, L. Annexin A9 promotes invasion and metastasis of colorectal cancer and predicts poor prognosis. Int. J. Mol. Med. 2018, 41, 2185–2192. [Google Scholar] [CrossRef] [Green Version]
- Cicardi, M.; Bisiani, G.; Cugno, M.; Spath, P.; Agostoni, A. Autoimmune C1 inhibitor deficiency: Report of eight patients. Am. J. Med. 1993, 95, 169–175. [Google Scholar] [CrossRef]
- Lubin, R.; Schlichtholz, B.; Bengoufa, D.; Zalcman, G.; Trédaniel, J.; Hirsch, A.; De Fromentel, C.C.; Preudhomme, C.; Fenaux, P.; Fournier, G.; et al. Analysis of p53 antibodies in patients with various cancers define B-cell epitopes of human p53: Distribution on primary structure and exposure on protein surface. Cancer Res. 1993, 53, 5872–5876. [Google Scholar]
- Cha, S.W.; Bonissone, S.; Na, S.; Pevzner, P.A.; Bafna, V. The Antibody Repertoire of Colorectal Cancer. Mol. Cell. Proteom. 2017, 16, 2111–2124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poynter, J.N.; Gruber, S.B.; Higgins, P.D.; Almog, R.; Bonner, J.D.; Rennert, H.S.; Low, M.; Greenson, J.K.; Rennert, G. Statins and the Risk of Colorectal Cancer. N. Engl. J. Med. 2005, 352, 2184–2192. [Google Scholar] [CrossRef] [Green Version]
- Cominacini, L.; Rigoni, A.; Pasini, A.F.; Garbin, U.; Davoli, A.; Campagnola, M.; Pastorino, A.M.; Cascio, V.L.; Sawamura, T. The Binding of Oxidized Low Density Lipoprotein (ox-LDL) to ox-LDL Receptor-1 Reduces the Intracellular Concentration of Nitric Oxide in Endothelial Cells through an Increased Production of Superoxide. J. Biol. Chem. 2001, 276, 13750–13755. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, K.; Ito, Y.; Wakai, K.; Kawado, M.; Hashimoto, S.; Toyoshima, H.; Kojima, M.; Tokudome, S.; Hayakawa, N.; Watanabe, Y.; et al. Serum oxidized low-density lipoprotein levels and risk of colorectal cancer: A case-control study nested in the Japan Collaborative Cohort Study. Cancer Epidemiol. Biomark. Prev. 2004, 13, 1781–1787. [Google Scholar]
| Clinical Characteristics | Metastatic sCRC (n = 12) | Non-Metastatic sCRC (n = 38) | p-Value | Total Cases (n = 50) |
|---|---|---|---|---|
| Age (years) | 63 (28–76) | 71 (27–85) | NS | 72 (28–85) |
| Gender | ||||
| Female | 6 (50%) | 27 (72%) | NS | 33 (67%) |
| Male | 6 (50%) | 11 (28%) | 17 (33%) | |
| Tumor Localization | ||||
| Rectum | 6 (50%) | 23 (61%) | NS | 29 (59%) |
| Left colon | 2 (33%) | 1 (3%) | 3 (6%) | |
| Right colon | 4 (17%) | 14 (36%) | 18 (35%) | |
| Histological grade | ||||
| Well-differentiated | 3 (53%) | 1 (3%) | p = 0.03 | 4 (8%) |
| Moderate- differentiated | 2 (28%) | 10 (26%) | 12 (40%) | |
| Poorly- differentiated | 2 (29%) | 12 (31%) | 14 (47%) | |
| Lymph node status (TNM) | ||||
| pN0 | 4 (33%) | 18 (46%) | NS | 22 (43%) |
| pN1 | 6 (50%) | 16 (44%) | 22 (45%) | |
| pN2 | 2 (17%) | 4 (10%) | 6 (12%) | |
| Tumor Size (cm) | 5 (2–11) | 3,5 (1,7–7) | NS | 4.7 (1.7–11) |
| Serum CEA (ng/mL) * | 11.8 (1.2–344) | 4.9 (0.9–90) | NS | 5.5 (0.9–344) |
| N. of Deaths | 9 (75%) | 9 (23%) | p = 0.001 | 18 (35%) |
| Median OS (months) * | 14 (0–64) | 22 (0–40) | p = 0.001 | Not reached |
| Sample ID | Diagnosis | N° of aAb | % of aAb |
|---|---|---|---|
| 1 | sCRC | 862 | 43 |
| 2 | sCRC | 846 | 42 |
| 3 | sCRC | 822 | 41 |
| 4 | sCRC | 191 | 9 |
| 5 | sCRC | 110 | 5 |
| 6 | sCRC | 466 | 23 |
| 7 | sCRC | 609 | 30 |
| 8 | sCRC | 832 | 41 |
| 9 | sCRC | 704 | 35 |
| 10 | sCRC | 285 | 14 |
| 11 | sCRC | 497 | 25 |
| 12 | sCRC | 851 | 42 |
| 13 | sCRC | 249 | 12 |
| 14 | sCRC | 717 | 35 |
| 15 | sCRC | 227 | 11 |
| 16 | sCRC | 646 | 32 |
| 17 | sCRC | 1040 | 51 |
| 18 | sCRC | 447 | 22 |
| 19 | sCRC | 593 | 29 |
| 20 | sCRC | 174 | 9 |
| 21 | sCRC | 1228 | 61 |
| 22 | sCRC | 1288 | 64 |
| 23 | sCRC | 471 | 23 |
| 24 | sCRC | 1225 | 61 |
| 25 | sCRC | 615 | 30 |
| 26 | sCRC | 458 | 23 |
| 27 | sCRC | 521 | 26 |
| 28 | sCRC | 864 | 43 |
| 29 | sCRC | 266 | 13 |
| 30 | sCRC | 1150 | 57 |
| 31 | sCRC | 813 | 40 |
| 32 | sCRC | 608 | 30 |
| 33 | sCRC | 671 | 33 |
| 34 | sCRC | 1141 | 56 |
| 35 | sCRC | 700 | 35 |
| 36 | sCRC | 381 | 19 |
| 37 | sCRC | 1310 | 65 |
| 38 | sCRC | 217 | 11 |
| 39 | sCRC | 327 | 16 |
| 40 | sCRC | 562 | 28 |
| 41 | sCRC | 541 | 27 |
| 42 | sCRC | 755 | 37 |
| 43 | sCRC | 557 | 28 |
| 44 | sCRC | 415 | 21 |
| 45 | sCRC | 720 | 36 |
| 46 | sCRC | 93 | 5 |
| 47 | sCRC | 363 | 18 |
| 48 | sCRC | 505 | 25 |
| 49 | sCRC | 48 | 2 |
| 50 | sCRC | 381 | 19 |
| 51 | Healthy donor | 715 | 35 |
| 52 | Healthy donor | 398 | 20 |
| 53 | Healthy donor | 880 | 43 |
| 54 | Healthy donor | 1149 | 57 |
| 55 | Healthy donor | 229 | 11 |
| 56 | Healthy donor | 278 | 14 |
| 57 | Healthy donor | 345 | 17 |
| TAA ID | p-Value (U-Mann-Whitney) | N° sCRC (n = 50) with aAb Positive | Fold Change (FC) | |
|---|---|---|---|---|
| Median FC sCRC | % sCRC > FC1 | |||
| TEX11 | 0.0005 | 8/50 | 2.6 | 90 |
| GORASP2 | 0.0007 | 2/50 | 3.1 | 82 |
| C9orf80 | 0.0007 | 3/50 | 3.9 | 92 |
| HMOX2 | 0.0008 | 13/50 | 3.0 | 92 |
| KIF9 | 0.0008 | 14/50 | 2.7 | 92 |
| ICAM2 | 0.0012 | 16/50 | 2.4 | 94 |
| MARCKSL1 | 0.0012 | 10/50 | 2.4 | 88 |
| RB1 | 0.0012 | 2/50 | 2.6 | 92 |
| SPP1 | 0.0012 | 3/50 | 2.8 | 86 |
| STC2 | 0.0013 | 10/50 | 2.0 | 84 |
| BHMT2 | 0.0014 | 10/50 | 2.2 | 86 |
| D21S2056E | 0.0016 | 10/50 | 2.2 | 84 |
| DLAT | 0.0016 | 2/50 | 2.3 | 86 |
| CKMT1B | 0.0017 | 27/50 | 2.6 | 86 |
| GTF2H1 | 0.0017 | 8/50 | 2.3 | 82 |
| ALDOA | 0.0019 | 8/50 | 2.0 | 88 |
| COX11 | 0.0020 | 14/50 | 2.4 | 88 |
| RPL11 | 0.0020 | 2/50 | 2.4 | 86 |
| ASB3 | 0.0020 | 8/50 | 2.3 | 84 |
| PRCP | 0.0024 | 21/50 | 1.9 | 86 |
| PDEF | 0.0024 | 10/50 | 2.2 | 86 |
| ANP32A | 0.0024 | 2/50 | 2.3 | 82 |
| GNAI3 | 0.0026 | 5/50 | 2.5 | 86 |
| HBG1 | 0.0026 | 2/50 | 3.5 | 84 |
| ARHI | 0.0028 | 11/50 | 1.9 | 86 |
| RCV1 | 0.0028 | 3/50 | 2.9 | 86 |
| RAB8A | 0.0028 | 5/50 | 2.4 | 84 |
| GDEP | 0.0030 | 11/50 | 2.2 | 82 |
| PLAC1 | 0.0030 | 7/50 | 2.0 | 84 |
| HSD17B3 | 0.0030 | 2/50 | 2.3 | 80 |
| SH3BP1 | 0.0033 | 3/50 | 2.0 | 84 |
| USP5 | 0.0033 | 2/50 | 2.1 | 82 |
| TCEAL1 | 0.0033 | 5/50 | 2.5 | 84 |
| KPNA6 | 0.0035 | 18/50 | 2.3 | 84 |
| Progranulin | 0.0038 | 4/50 | 1.8 | 86 |
| CHODL | 0.0038 | 4/50 | 2.2 | 86 |
| KCNE2 | 0.0041 | 13/50 | 2.0 | 82 |
| SERPINA5 | 0.0041 | 5/50 | 2.2 | 80 |
| SLCO4A1 | 0.0041 | 4/50 | 2.5 | 80 |
| SDPR | 0.0041 | 10/50 | 1.8 | 86 |
| JUP | 0.0041 | 3/50 | 2.5 | 82 |
| FRK | 0.0041 | 6/50 | 2.1 | 84 |
| DDX39 | 0.0044 | 2/50 | 2.1 | 80 |
| PSAP | 0.0048 | 2/50 | 2.1 | 84 |
| SCARB1 | 0.0048 | 2/50 | 2.6 | 84 |
| AMY2A | 0.0052 | 9/50 | 2.1 | 84 |
| BECN1 | 0.0056 | 16/50 | 2.0 | 84 |
| ST14 | 0.0056 | 16/50 | 1.7 | 80 |
| LDHB | 0.0056 | 8/50 | 2.3 | 84 |
| SNX10 | 0.0056 | 2/50 | 2.1 | 82 |
| PSTPIP1 | 0.0060 | 17/50 | 2.1 | 84 |
| SLC6A1 | 0.0065 | 6/50 | 2.0 | 78 |
| HM13 | 0.0070 | 10/50 | 1.7 | 84 |
| RHOH | 0.0070 | 7/50 | 1.9 | 82 |
| KIF22 | 0.0075 | 5/50 | 2.4 | 78 |
| SYTL1 | 0.0075 | 4/50 | 2.1 | 80 |
| Mage3 | 0.0075 | 10/50 | 1.9 | 82 |
| HIST1H3D | 0.0075 | 4/50 | 2.3 | 82 |
| RWDD1 | 0.0080 | 9/50 | 2.1 | 80 |
| RILP | 0.0080 | 6/50 | 2.0 | 76 |
| RPL35 | 0.0080 | 4/50 | 1.9 | 82 |
| PLAGL1 | 0.0086 | 21/50 | 1.6 | 82 |
| DDX56 | 0.0086 | 9/50 | 1.7 | 84 |
| CTNNA1 | 0.0086 | 9/50 | 2.2 | 82 |
| BET1 | 0.0086 | 4/50 | 2.0 | 78 |
| HLA-DOB | 0.0093 | 28/50 | 1.6 | 80 |
| HSPC047 | 0.0093 | 4/50 | 1.9 | 80 |
| Healthy Donors vs. Pathological sCCR | |||||
|---|---|---|---|---|---|
| GO Term | Function | p-Value | TAAs Number | % TAA | TAAs ID |
| GO:0006974 | Response to DNA damage stimulus | 0.0011 | 7 | 14.6 | SUMO1, UBE2A, CCNH, GTF2H4, MLH1, GTSE1, RAD17 |
| GO:0006281 | DNA repair | 0.0020 | 6 | 12.5 | SUMO1, UBE2A, CCNH, GTF2H4, MLH1, RAD17 |
| GO:0033554 | Cellular response to stress | 0.0087 | 7 | 14.6 | SUMO1, UBE2A, CCNH, GTF2H4, MLH1, GTSE1, RAD17 |
| GO:0006259 | DNA metabolic process | 0.0051 | 7 | 14.6 | SUMO1, UBE2A, CCNH, GTF2H4, MLH1, IGF1, RAD17 |
| GO:0006950 | Response to stress | 0.0053 | 13 | 27.1 | UBE2A, CCNH, GTF2H4, LYZ, MLH1, IGF1, SMAD1, GTSE1, SUMO1, DARC, CA2, ENTPD2, RAD17 |
| GO:0051716 | Cellular response to stimulus | 0.0143 | 8 | 16.7 | SUMO1, UBE2A, CCNH, GTF2H4, MLH1, SMAD1, GTSE1, RAD17 |
| GO:0009056 | Catabolic process | 0.0158 | 10 | 20.8 | ALDOA, SUMO1, UBE2A, CCNH, LYZ, GTF2H4, MLH1, SAE1, PSME3, ENTPD2 |
| GO:0006367 | Transcription initiation from RNA polymerase II promoter | 0.098 | 3 | 6.3 | MED4, CCNH, GTF2H4 |
| GO:0044265 | Cellular macromolecule catabolic process | 0.0266 | 7 | 14.6 | SUMO1, UBE2A, CCNH, GTF2H4, MLH1, SAE1, PSME3 |
| GO:0042770 | DNA damage response, signal transduction | 0.0268 | 3 | 6.3 | MLH1, GTSE1, RAD17 |
| GO:0016070 | RNA metabolic process | 0.0278 | 8 | 16.7 | MED4, CCNH, GTF2H4, MLH1, WBP11, SMAD1, SCGB1A1, RPS7 |
| GO:0006352 | Transcription initiation | 0.0287 | 3 | 6.3 | MED4, CCNH, GTF2H4 |
| GO:0009892 | Negative regulation of metabolic process | 0.0362 | 7 | 14.6 | RPS26, SUMO1, MLH1, PSME3, SMAD1, SCGB1A1, RAD17 |
| GO:0010605 | Negative regulation of macromolecule metabolic process | 0.0280 | 7 | 14.6 | RPS26, SUMO1, MLH1, PSME3, SMAD1, SCGB1A1, RAD17 |
| GO:0009057 | Macromolecule catabolic process | 0.0364 | 7 | 14.6 | SUMO1, UBE2A, CCNH, GTF2H4, MLH1, SAE1, PSME3 |
| GO:0006414 | Translational elongation | 0.0411 | 3 | 6.3 | RPS26, RPL35, RPS7 |
| GO:0044248 | Cellular catabolic process | 0.0418 | 8 | 16.7 | SUMO1, UBE2A, CCNH, GTF2H4, MLH1, SAE1, PSME3, ENTPD2 |
| GO:0014902 | Myotube differentiation | 0.0498 | 2 | 4.2 | CAST, IGF1 |
| GO:0030901 | Midbrain development | 0.0498 | 2 | 4.2 | SMAD1, PITX3 |
| Non-metastatic sCRC vs. Synchronic Metastasis sCRC | |||||
| GO term | Function | p-value | Aab number | % Aab | Aab ID |
| GO:0007548 | Sex differentiation | 0.0209 | 3 | 13.0 | HSD17B3, EIF2B2, TEX11 |
| GO:0045137 | Development of primary sexual characteristics | 0.0151 | 3 | 13.0 | HSD17B3, EIF2B2, TEX11 |
| GO:0006833 | Water transport | 0.0236 | 2 | 8.7 | AQP5, AQP3 |
| GO:0042044 | Fluid transport | 0.0250 | 2 | 8.7 | AQP5, AQP3 |
| GO:0048513 | Organ development | 0.0366 | 7 | 30.4 | PFN1, CRIP2, FHL3, HSD17B3, EIF2B2, TEX11, TIMP1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-González, M.; Sayagués, J.M.; Muñoz-Bellvís, L.; Pedreira, C.E.; de Campos, M.L.R.; García, J.; Alcázar, J.A.; Braz, P.F.; Galves, B.L.; González, L.M.; et al. Tracking the Antibody Immunome in Sporadic Colorectal Cancer by Using Antigen Self-Assembled Protein Arrays. Cancers 2021, 13, 2718. https://doi.org/10.3390/cancers13112718
González-González M, Sayagués JM, Muñoz-Bellvís L, Pedreira CE, de Campos MLR, García J, Alcázar JA, Braz PF, Galves BL, González LM, et al. Tracking the Antibody Immunome in Sporadic Colorectal Cancer by Using Antigen Self-Assembled Protein Arrays. Cancers. 2021; 13(11):2718. https://doi.org/10.3390/cancers13112718
Chicago/Turabian StyleGonzález-González, María, José María Sayagués, Luis Muñoz-Bellvís, Carlos Eduardo Pedreira, Marcello L. R. de Campos, Jacinto García, José Antonio Alcázar, Patrick F. Braz, Breno L. Galves, Luis Miguel González, and et al. 2021. "Tracking the Antibody Immunome in Sporadic Colorectal Cancer by Using Antigen Self-Assembled Protein Arrays" Cancers 13, no. 11: 2718. https://doi.org/10.3390/cancers13112718