Serum Exosomal microRNA-21, 222 and 124-3p as Noninvasive Predictive Biomarkers in Newly Diagnosed High-Grade Gliomas: A Prospective Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
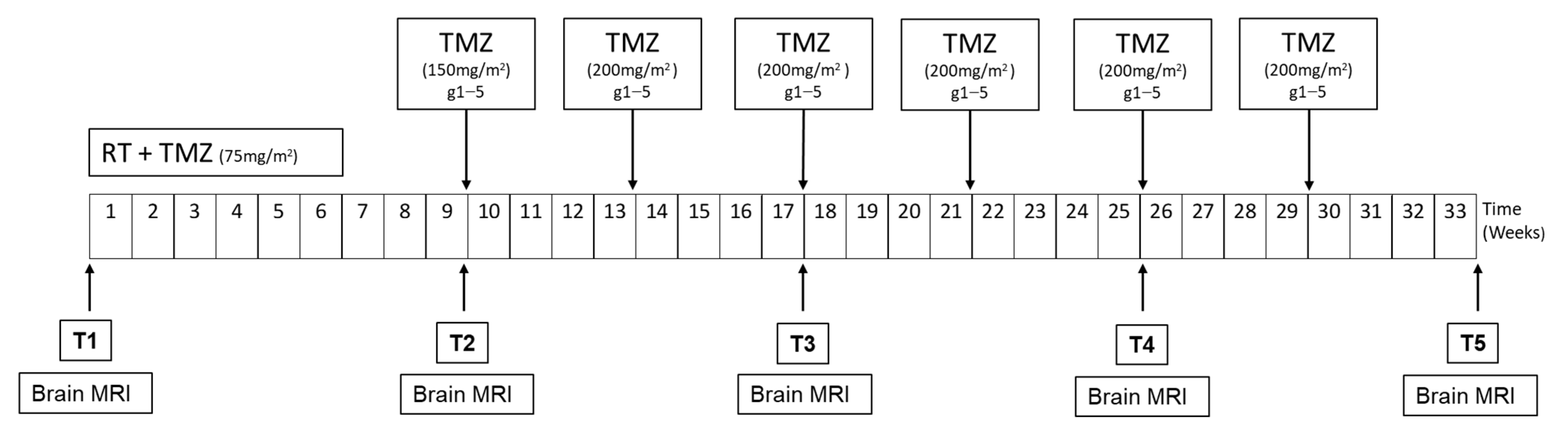
2.1. Study Design
2.2. Patients
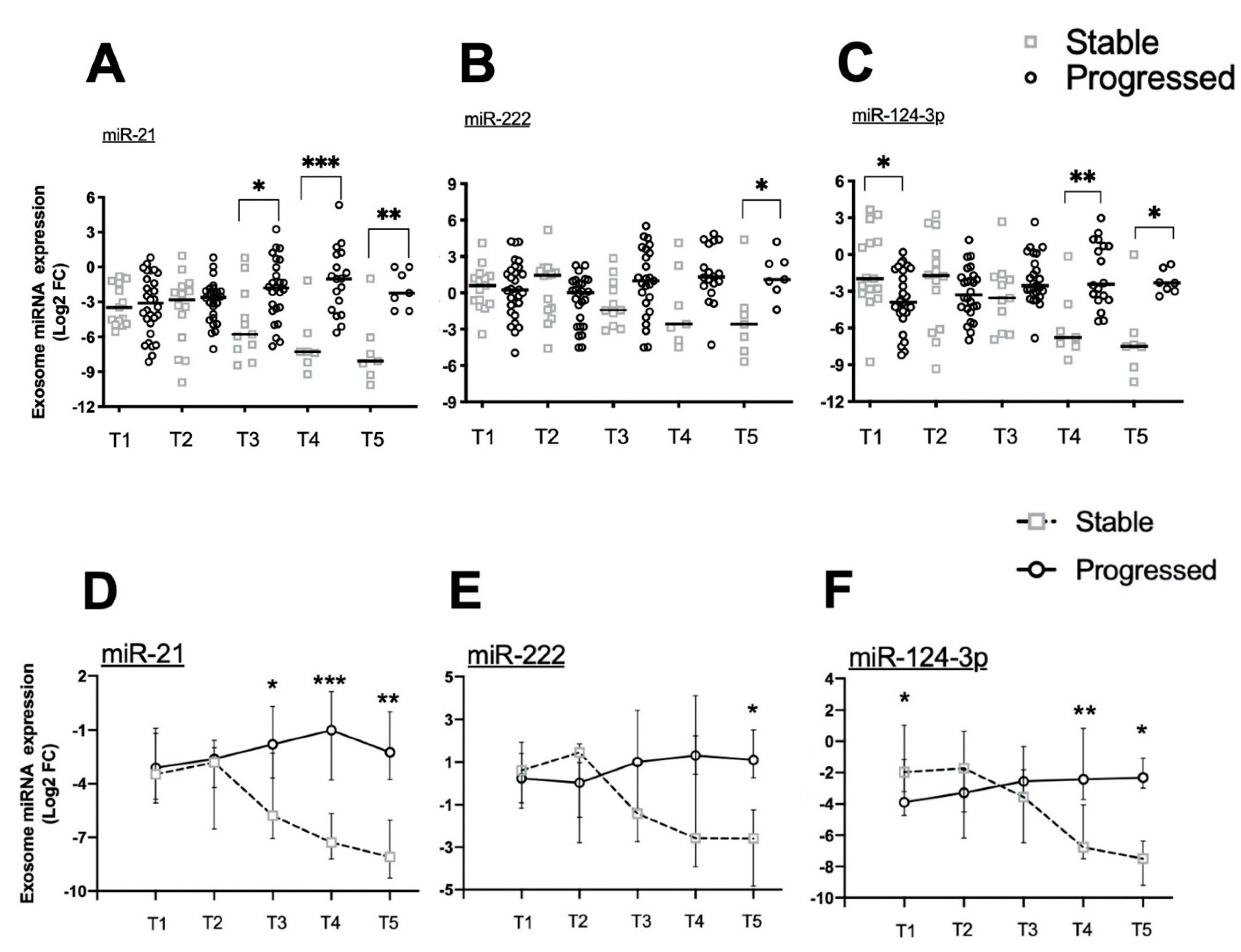
2.3. Association of Circulating miRNA Expression with PFS or OS
2.4. Exosomal miRNA Expression during Longitudinal Follow-Up
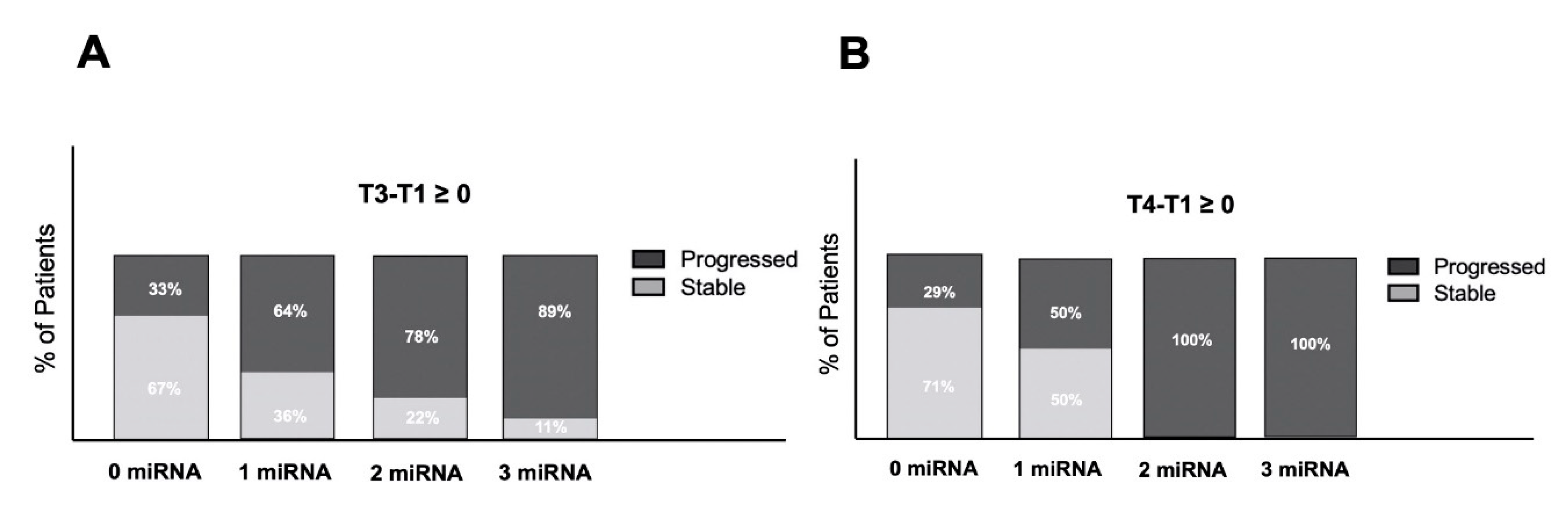
2.5. Exosomal miRNAs Predict Progression in HGG Patients
2.6. Exosomal miRNA Expression in Clinical Management of HGG Patients
3. Discussion
4. Materials and Methods
4.1. Patients and Sample Collection
4.2. Purification of Exosomes
4.3. RNA Isolation from Exosomes
4.4. miRNA Quantification by Real-Time qPCR
4.5. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schwartzbaum, J.A.; Fisher, J.L.; Aldape, K.D.; Wrensch, M. Epidemiology and molecular pathology of glioma. Nat. Clin. Pract. Neurol. 2006, 2, 494–503. [Google Scholar] [CrossRef]
- Stupp, R. Temozolomide: A milestone in neuro-oncology and beyond? Expert Rev. Anticancer Ther. 2006, 6, 1187–1204. [Google Scholar]
- De Rubis, G.; Rajeev Krishnan, S.; Bebawy, M. Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol. Sci. 2019, 40, 172–186. [Google Scholar] [CrossRef] [PubMed]
- Mattox, A.K.; Yan, H.; Bettegowda, C. The potential of cerebrospinal fluid–based liquid biopsy approaches in CNS tumors. Neuro Oncol. 2019, 21, 1509–1518. [Google Scholar] [CrossRef]
- André-Grégoire, G.; Gavard, J. Spitting out the demons: Extracellular vesicles in glioblastoma. Cell Adhes. Migr. 2016, 11, 164–172. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Morishita, M.; Takahashi, Y.; Nishikawa, M.; Takakura, Y. Pharmacokinetics of Exosomes—An Important Factor for Elucidating the Biological Roles of Exosomes and for the Development of Exosome-Based Therapeutics. J. Pharm. Sci. 2017, 106, 2265–2269. [Google Scholar] [CrossRef] [PubMed]
- Petrescu, G.E.D.; Sabo, A.A.; Torsin, L.I.; Calin, G.A.; Dragomir, M.P. MicroRNA based theranostics for brain cancer: Basic principles. J. Exp. Clin. Cancer Res. 2019, 38, 1–21. [Google Scholar] [CrossRef]
- Godlewski, J.; Krichevsky, A.M.; Johnson, M.D.; Chiocca, E.A.; Bronisz, A. Belonging to a network--microRNAs, extracellular vesicles, and the glioblas-toma microenvironment. Neuro Oncol. 2015, 17, 652–662. [Google Scholar] [CrossRef]
- Goodall, E.F.; Leach, V.; Wang, C.; Cooper-Knock, J.; Heath, P.R.; Baker, D.; Drew, D.R.; Saffrey, M.J.; Simpson, J.E.; Romero, I.A.; et al. Age-Associated mRNA and miRNA Expression Changes in the Blood-Brain Barrier. Int. J. Mol. Sci. 2019, 20, 3097. [Google Scholar] [CrossRef]
- Aili, Y.; Maimaitiming, N.; Mahemuti, Y.; Qin, H.; Wang, Y.; Wang, Z. Liquid biopsy in central nervous system tumors: The potential roles of circulating miRNA and exosomes. Am. J. Cancer Res. 2020, 10, 4134–4150. [Google Scholar]
- Swellam, M.; El Arab, L.E.; Al-Posttany, A.S.; Said, S.B. Clinical impact of circulating oncogenic MiRNA-221 and MiRNA-222 in glioblastoma multiform. J. Neuro Oncol. 2019, 144, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Khalil, S.; Fabbri, E.; Santangelo, A.; Bezzerri, V.; Cantù, C.; Di Gennaro, G.; Finotti, A.; Ghimenton, C.; Eccher, A.; Dechecchi, M.; et al. miRNA array screening reveals cooperative MGMT-regulation between miR-181d-5p and miR-409-3p in glioblastoma. Oncotarget 2016, 7, 28195–28206. [Google Scholar] [CrossRef]
- Santangelo, A.; Imbrucè, P.; Gardenghi, B.; Belli, L.; Agushi, R.; Tamanini, A.; Munari, S.; Bossi, A.M.; Scambi, I.; Benati, D.; et al. A microRNA signature from serum exosomes of patients with glioma as com-plementary diagnostic biomarker. J. Neurooncol. 2018, 136, 51–62. [Google Scholar] [CrossRef]
- Santangelo, A.; Rossato, M.; Lombardi, G.; Benfatto, S.; Lavezzari, D.; De Salvo, G.L.; Indraccolo, S.; Dechecchi, M.C.; Prandini, P.; Gambari, R.; et al. A molecular signature associated with prolonged survival in glioblastoma patients treated with regorafenib. Neuro Oncol. 2021, 23, 264–276. [Google Scholar] [CrossRef]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The Promising Role of miR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. Nucleic Acids 2020, 20, 409–420. [Google Scholar] [CrossRef]
- Qu, K.; Lin, T.; Pang, Q.; Liu, T.; Wang, Z.; Tai, M.; Meng, F.; Zhang, J.; Wan, Y.; Mao, P.; et al. Extracellular miRNA-21 as a novel biomarker in glioma: Evidence from meta-analysis, clinical validation and experimental investigations. Oncotarget 2016, 7, 33994–34010. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Lu, X.; Liu, L.; Xu, J.; Feng, D.; Shu, Y. miRNA-21: A biomarker predictive for platinum-based adjuvant chemotherapy response in patients with non-small cell lung cancer. Cancer Biol. Ther. 2012, 13, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhuang, L.; Zhang, J.; Fan, J.; Luo, J.; Chen, H.; Wang, K.; Liu, L.; Chen, Z.; Meng, Z. The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancre-atic cancer and miR-21 expression confers chemoresistance by targeting FasI. Mol. Oncol. 2013, 7, 334–345. [Google Scholar] [CrossRef]
- Liu, B.; Su, F.; Lv, X.; Zhang, W.; Shang, X.; Zhang, Y.; Zhang, J. Serum microRNA-21 predicted treatment outcome and survival in HER2-positive breast cancer pa-tients receiving neoadjuvant chemotherapy combined with trastuzumab. Cancer Chemother. Pharmacol. 2019, 84, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Krichevsky, A.M.; Gabriely, G. miR-21: A small multi-faceted RNA. J. Cell. Mol. Med. 2008, 13, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Chao, T.-F.; Xiong, H.-H.; Liu, W.; Chen, Y.; Zhang, J.-X. MiR-21 mediates the radiation resistance of glioblastoma cells by regulating PDCD4 and hMSH2. Acta Acad. Med. Wuhan 2013, 33, 525–529. [Google Scholar] [CrossRef] [PubMed]

| Patient Characteristics | N (%) |
|---|---|
| Total | 57 |
| GENDER | |
| Male Female | 41 (72) 16 (28) |
| Median Age (range) HISTOLOGY Glioblastoma Anaplastic Astrocytoma | 63 (23–88) 52 (91) 5 (9) |
| EXTENT OF RESECTION | |
| Radical Non-Radical Unknown | 18 (32) 39 (68) 1 (2) |
| ECOG PS | |
| 0–1 2 | 45 (79) 12 (21) |
| MGMT | |
| Methylated Unmethylated Unknown | 26 (46) 16 (28) 15 (26) |
| IDH 1/2 | |
| Wildtype Mutated Unknown | 49 (86) 3 (5) 5 (9) |
| PFS Ratio | OS Ratio | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| miRNA | Expression | Median (Months) | High/Low | 95% CI | Log-Rank p | Median (Months) | High/Low | 95% CI | Log-Rank p |
| T1 | |||||||||
| 21 | high low | 9.1 7.9 | 1.1 | (0.24–1.49) | 0.64 | 17.8 13.1 | 1.4 | (0.75–2.46) | 0.51 |
| 222 | high low | 9.1 6.1 | 1.5 | (0.13–1.43) | 0.16 | 14.6 16.2 | 0.9 | (0.46–1.74) | 0.24 |
| 124-3p | high low | 8.6 8.3 | 1.0 | (0.59–1.81) | 0.31 | 17.3 15.6 | 1.1 | (0.62–2.0) | 0.77 |
| T2 | |||||||||
| 21 | high low | 9.1 7.9 | 1.1 | (0.61–2.13) | 0.76 | 17.8 12.9 | 1.4 | (0.71–2.68) | 0.96 |
| 222 | high low | 8.6 8.7 | 1.0 | (0.53–1.83) | 0.71 | 14.6 16.2 | 0.9 | (0.46–1.74) | 0.24 |
| 124-3p | high low | 8.8 8.3 | 1.1 | (0.57–1.97) | 0.68 | 17.3 13.1 | 1.4 | (0.70–2.63) | 0.68 |
| T3 | |||||||||
| 21 | high low | 7.3 10.2 | 0.7 | (0.34–1.41) | 0.006 | 16.9 23.5 | 0.7 | (0.33–1.51) | 0.048 |
| 222 | high low | 8.4 9.4 | 0.9 | (0.44–1.78) | 0.18 | 16.9 22.1 | 0.8 | (0.36–1.61) | 0.13 |
| 124-3p | high low | 8.1 8.8 | 0.9 | (0.43–1.85) | 0.87 | 17.8 18.8 | 0.9 | (0.45–1.99) | 0.59 |
| T4 | |||||||||
| 21 | high low | 7.9 11.0 | 0.7 | (0.31–1.68) | 0.032 | 14.6 26.2 | 0.6 | (0.21–1.40) | 0.043 |
| 222 | high low | 9.5 9.7 | 1.0 | (0.42–2.27) | 0.29 | 16.9 26.2 | 0.6 | (0.25–1.83) | 0.047 |
| 124-3p | high low | 8.4 10.4 | 0.8 | (0.35–1.82) | 0.048 | 14.6 23.9 | 0.7 | (0.24–1.49) | 0.031 |
| T5 | |||||||||
| 21 | high low | 10.2 23.1 | 0.4 | (0.13–1.43) | 0.017 | 21.3 Und * | Und * | Und * | 0.032 |
| 222 | high low | 10.2 23.1 | 0.4 | (0.13–1.38) | 0.003 | 17.3 35.3 | 0.5 | (0.13–1.74) | 0.047 |
| 124-3p | high low | 10.2 23.2 | 0.4 | (0.13–1.38) | 0.022 | 21.0 Und * | Und * | Und * | 0.022 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olioso, D.; Caccese, M.; Santangelo, A.; Lippi, G.; Zagonel, V.; Cabrini, G.; Lombardi, G.; Dechecchi, M.C. Serum Exosomal microRNA-21, 222 and 124-3p as Noninvasive Predictive Biomarkers in Newly Diagnosed High-Grade Gliomas: A Prospective Study. Cancers 2021, 13, 3006. https://doi.org/10.3390/cancers13123006
Olioso D, Caccese M, Santangelo A, Lippi G, Zagonel V, Cabrini G, Lombardi G, Dechecchi MC. Serum Exosomal microRNA-21, 222 and 124-3p as Noninvasive Predictive Biomarkers in Newly Diagnosed High-Grade Gliomas: A Prospective Study. Cancers. 2021; 13(12):3006. https://doi.org/10.3390/cancers13123006
Chicago/Turabian StyleOlioso, Debora, Mario Caccese, Alessandra Santangelo, Giuseppe Lippi, Vittorina Zagonel, Giulio Cabrini, Giuseppe Lombardi, and Maria Cristina Dechecchi. 2021. "Serum Exosomal microRNA-21, 222 and 124-3p as Noninvasive Predictive Biomarkers in Newly Diagnosed High-Grade Gliomas: A Prospective Study" Cancers 13, no. 12: 3006. https://doi.org/10.3390/cancers13123006
APA StyleOlioso, D., Caccese, M., Santangelo, A., Lippi, G., Zagonel, V., Cabrini, G., Lombardi, G., & Dechecchi, M. C. (2021). Serum Exosomal microRNA-21, 222 and 124-3p as Noninvasive Predictive Biomarkers in Newly Diagnosed High-Grade Gliomas: A Prospective Study. Cancers, 13(12), 3006. https://doi.org/10.3390/cancers13123006








