Global Alternative Splicing Defects in Human Breast Cancer Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture, RNA Extraction, and RT-PCR
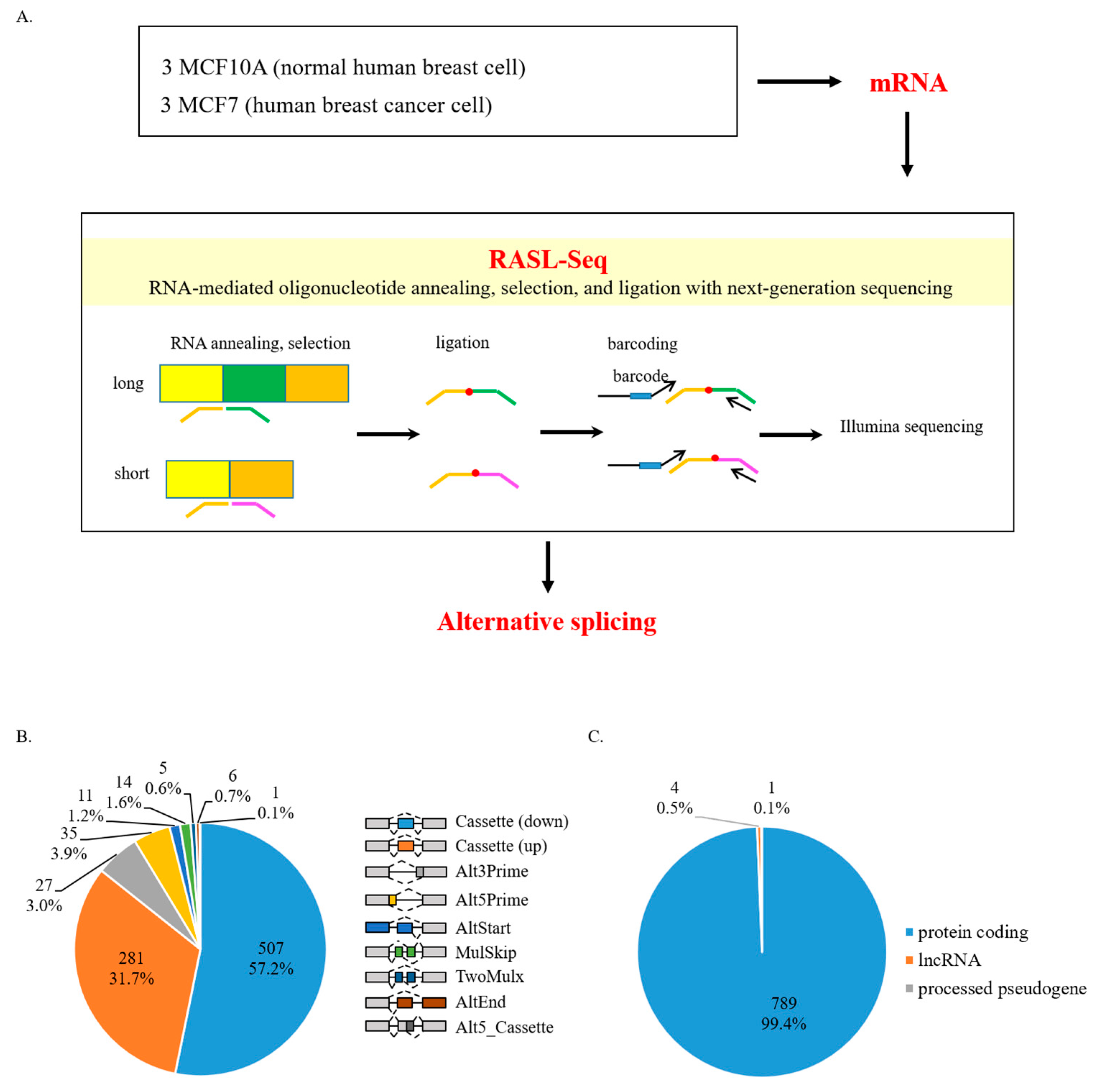
2.2. Alternative Splicing Analysis with RASL-Seq
2.3. Gene Ontology (GO) Analysis
2.4. Splicing Analysis of Transcriptomic Data
2.5. Prediction of Intrinsically Disordered Regions (IDRs)
2.6. Analysis of Protein-Protein Interaction Networks
2.7. Correlation of Drug Sensitivity and Transcript-Specific Expression of AS Isoforms
3. Results
3.1. AS Defects in Human Breast Cancer Cells
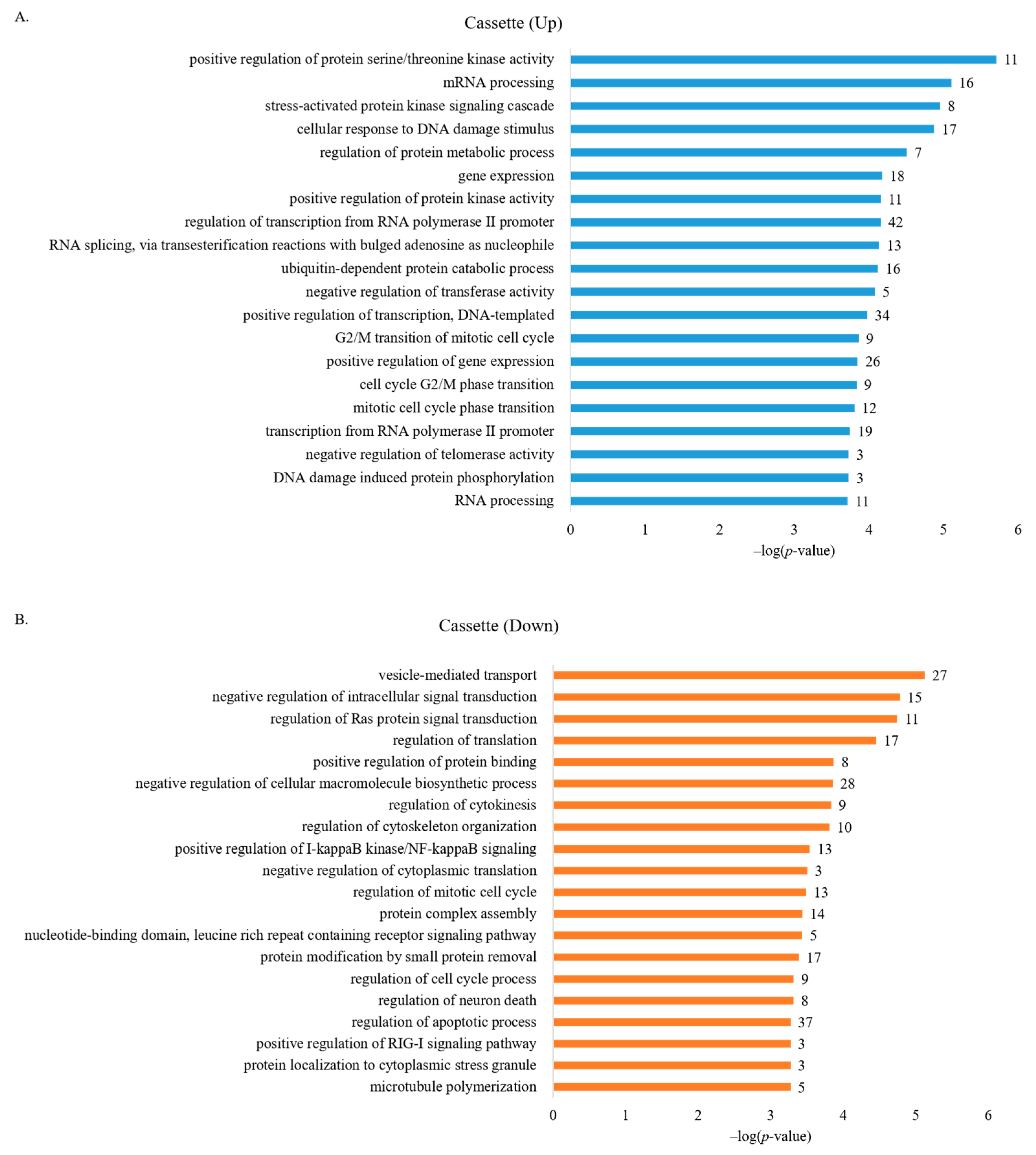
3.2. GO Analysis Identifies Functions Related to Oncogenesis in Genes with Altered AS Profiles in Breast Cancer Cells
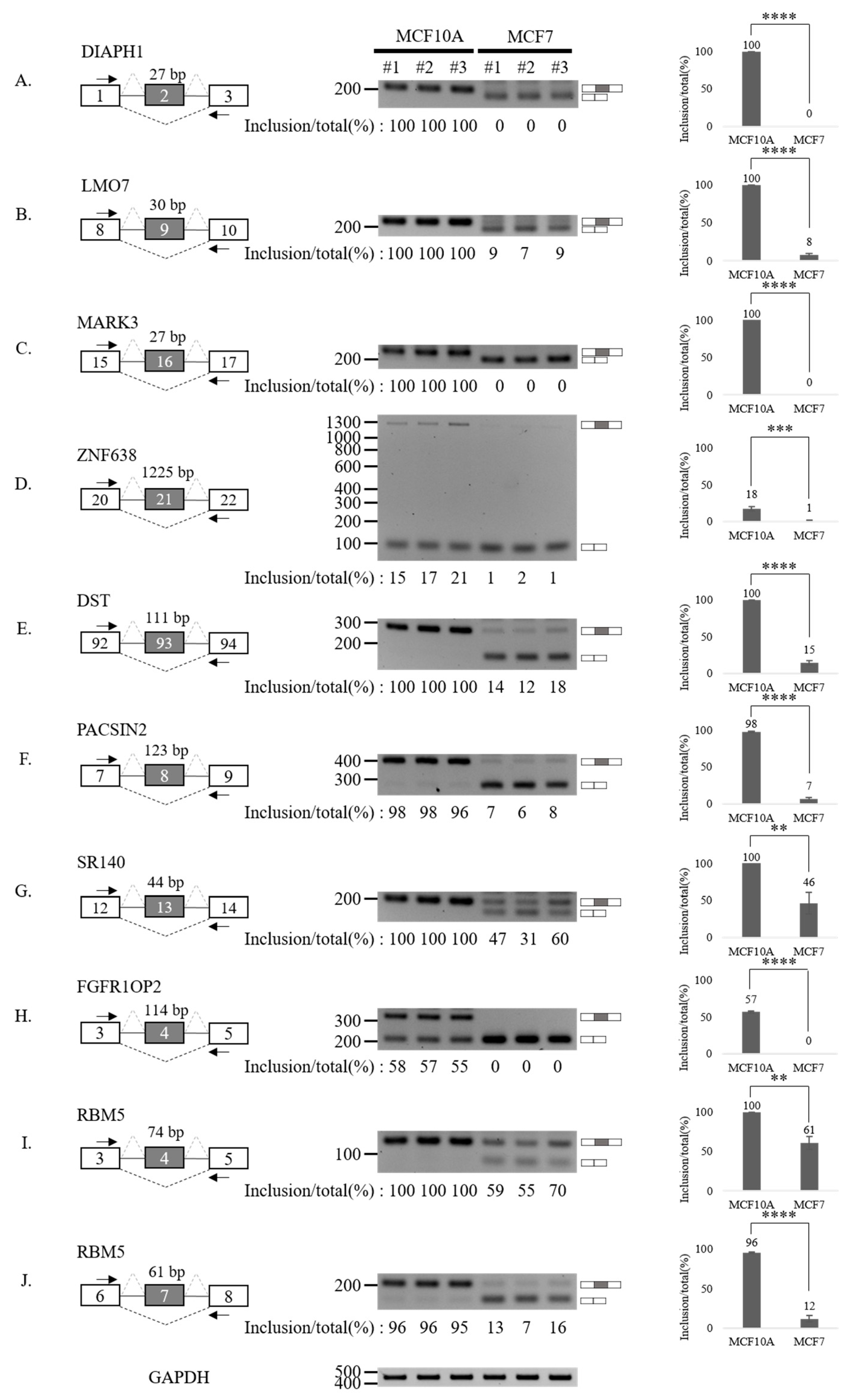
3.3. Validation of Aberrantly Regulated AS Events in Breast Tumor Cell Lines
3.4. Validation of Aberrantly Regulated AS in the TCGA-BRCA Dataset and in Breast Tumor Specimens
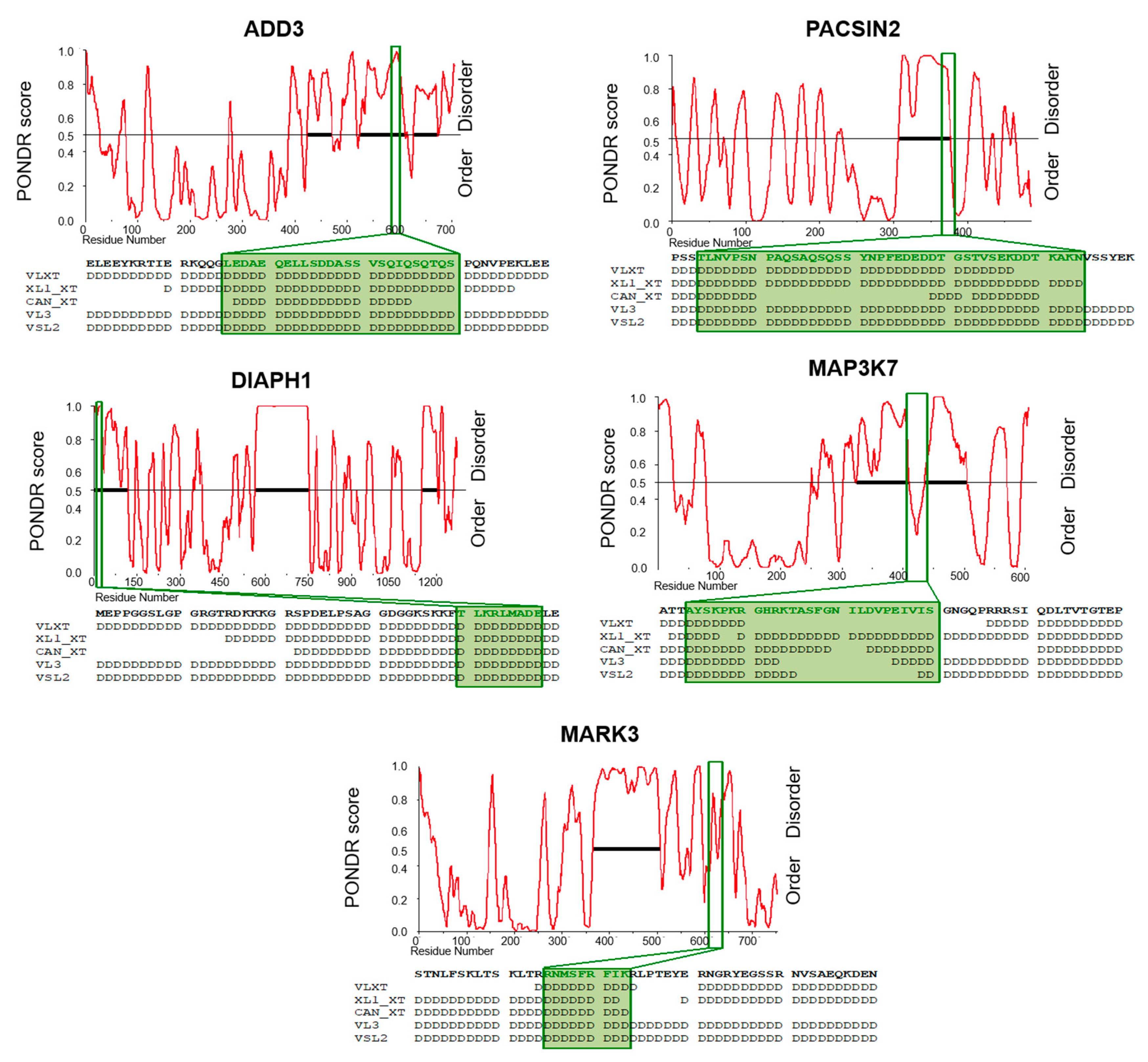
3.5. Aberrantly Expressed AS Exons Encode for Intrinsically Disordered Regions (IDRs)
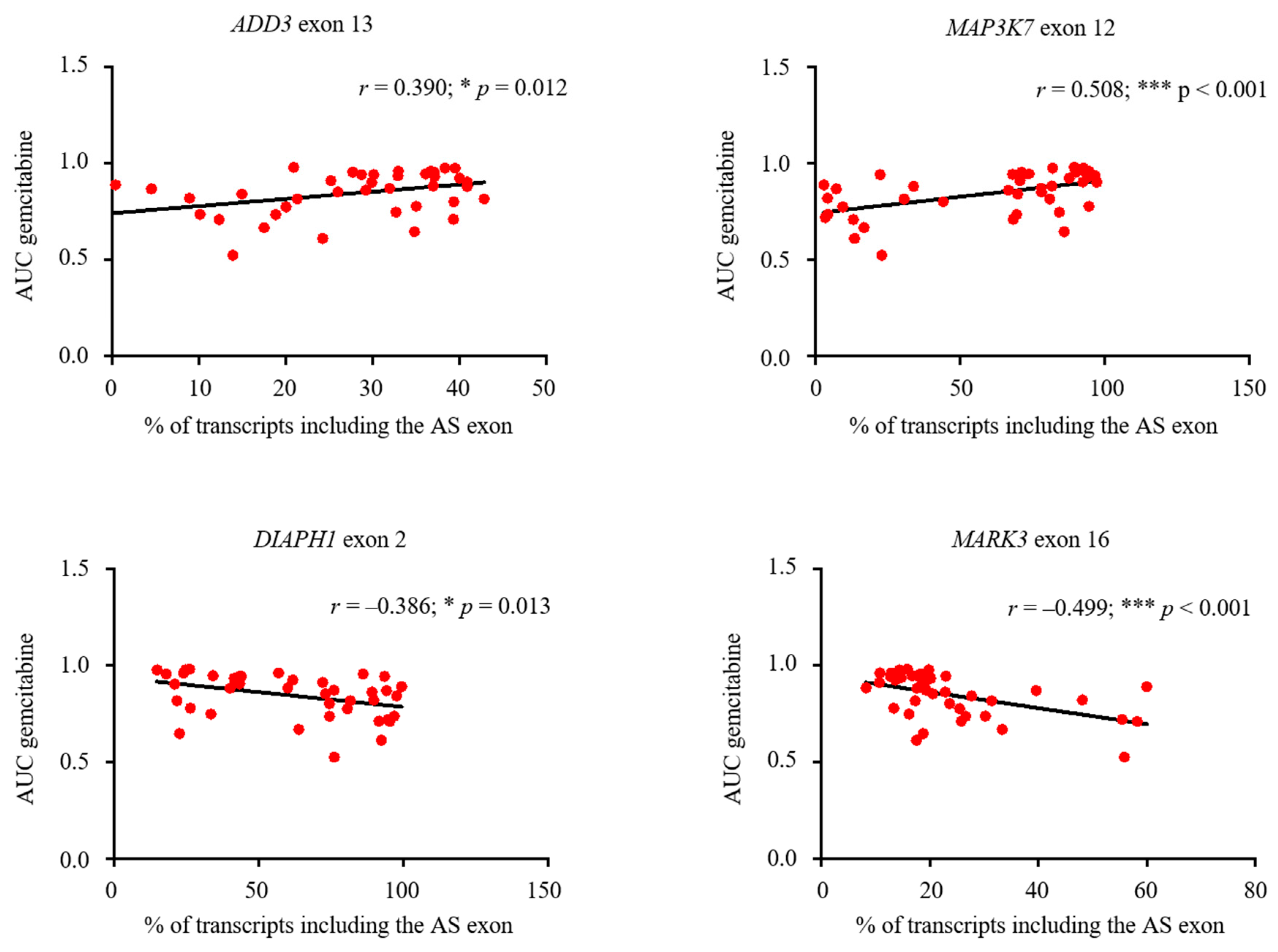
3.6. Expression of Aberrantly Expressed IDR-Encoding AS Exons Is Associated to Chemotherapeutic Sensitivity in Breast Cancer Cell Lines Databases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003, 72, 291–336. [Google Scholar] [CrossRef]
- Green, M.R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu. Rev. Cell Biol. 1991, 7, 559–599. [Google Scholar] [CrossRef]
- Sharp, P.A. Speculations on RNA splicing. Cell 1981, 23, 643–646. [Google Scholar] [CrossRef]
- Reed, R.; Maniatis, T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 1988, 2, 1268–1276. [Google Scholar] [CrossRef] [PubMed]
- Padgett, R.A.; Grabowski, P.J.; Konarska, M.M.; Seiler, S.; Sharp, P.A. Splicing of messenger RNA precursors. Annu. Rev. Biochem. 1986, 55, 1119–1150. [Google Scholar] [CrossRef]
- Maniatis, T.; Reed, R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature 1987, 325, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Will, C.L.; Luhrmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Rio, D.C. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef] [PubMed]
- Di Matteo, A.; Belloni, E.; Pradella, D.; Cappelletto, A.; Volf, N.; Zacchigna, S.; Ghigna, C. Alternative splicing in endothelial cells: Novel therapeutic opportunities in cancer angiogenesis. J. Exp. Clin. Cancer Res. 2020, 39, 275. [Google Scholar] [CrossRef]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Yabas, M.; Elliott, H.; Hoyne, G.F. The Role of Alternative Splicing in the Control of Immune Homeostasis and Cellular Differentiation. Int. J. Mol. Sci. 2015, 17, 3. [Google Scholar] [CrossRef] [PubMed]
- Parra, M.; Booth, B.W.; Weiszmann, R.; Yee, B.; Yeo, G.W.; Brown, J.B.; Celniker, S.E.; Conboy, J.G. An important class of intron retention events in human erythroblasts is regulated by cryptic exons proposed to function as splicing decoys. RNA 2018, 24, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- Shin, C.; Manley, J.L. Cell signalling and the control of pre-mRNA splicing. Nat. Rev. Mol. Cell Biol. 2004, 5, 727–738. [Google Scholar] [CrossRef]
- Lara-Pezzi, E.; Gomez-Salinero, J.; Gatto, A.; Garcia-Pavia, P. The alternative heart: Impact of alternative splicing in heart disease. J. Cardiovasc. Transl. Res. 2013, 6, 945–955. [Google Scholar] [CrossRef]
- Juan-Mateu, J.; Alvelos, M.I.; Turatsinze, J.V.; Villate, O.; Lizarraga-Mollinedo, E.; Grieco, F.A.; Marroqui, L.; Bugliani, M.; Marchetti, P.; Eizirik, D.L. SRp55 Regulates a Splicing Network That Controls Human Pancreatic beta-Cell Function and Survival. Diabetes 2018, 67, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Biamonti, G.; Amato, A.; Belloni, E.; Di Matteo, A.; Infantino, L.; Pradella, D.; Ghigna, C. Alternative splicing in Alzheimer’s disease. Aging Clin. Exp. Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Aifantis, I. RNA Splicing and Cancer. Trends Cancer 2020, 6, 631–644. [Google Scholar] [CrossRef]
- Ghigna, C.; Valacca, C.; Biamonti, G. Alternative splicing and tumor progression. Curr. Genom. 2008, 9, 556–570. [Google Scholar] [CrossRef]
- Ladomery, M. Aberrant alternative splicing is another hallmark of cancer. Int. J. Cell Biol. 2013, 2013, 463786. [Google Scholar] [CrossRef]
- Oltean, S.; Bates, D.O. Hallmarks of alternative splicing in cancer. Oncogene 2014, 33, 5311–5318. [Google Scholar] [CrossRef] [PubMed]
- Pradella, D.; Naro, C.; Sette, C.; Ghigna, C. EMT and stemness: Flexible processes tuned by alternative splicing in development and cancer progression. Mol. Cancer 2017, 16, 8. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef]
- Yang, Q.; Zhao, J.; Zhang, W.; Chen, D.; Wang, Y. Aberrant alternative splicing in breast cancer. J. Mol. Cell Biol. 2019, 11, 920–929. [Google Scholar] [CrossRef]
- Martinez-Montiel, N.; Anaya-Ruiz, M.; Perez-Santos, M.; Martinez-Contreras, R.D. Alternative Splicing in Breast Cancer and the Potential Development of Therapeutic Tools. Genes 2017, 8, 217. [Google Scholar] [CrossRef]
- Li, H.; Qiu, J.; Fu, X.D. RASL-seq for massively parallel and quantitative analysis of gene expression. Curr. Protoc. Mol. Biol. 2012, 13, 11–19. [Google Scholar] [CrossRef]
- Sun, S.; Ling, S.C.; Qiu, J.; Albuquerque, C.P.; Zhou, Y.; Tokunaga, S.; Li, H.; Qiu, H.; Bui, A.; Yeo, G.W.; et al. ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat. Commun. 2015, 6, 6171. [Google Scholar] [CrossRef]
- Qiu, J.; Zhou, B.; Thol, F.; Zhou, Y.; Chen, L.; Shao, C.; DeBoever, C.; Hou, J.; Li, H.; Chaturvedi, A.; et al. Distinct splicing signatures affect converged pathways in myelodysplastic syndrome patients carrying mutations in different splicing regulators. RNA 2016, 22, 1535–1549. [Google Scholar] [CrossRef]
- Soule, H.D.; Maloney, T.M.; Wolman, S.R.; Peterson, W.D., Jr.; Brenz, R.; McGrath, C.M.; Russo, J.; Pauley, R.J.; Jones, R.F.; Brooks, S.C. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990, 50, 6075–6086. [Google Scholar] [PubMed]
- Lee, A.V.; Oesterreich, S.; Davidson, N.E. MCF-7 cells—Changing the course of breast cancer research and care for 45 years. J. Natl. Cancer Inst. 2015, 107. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Ryan, M.; Wong, W.C.; Brown, R.; Akbani, R.; Su, X.; Broom, B.; Melott, J.; Weinstein, J. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res. 2016, 44, D1018–D1022. [Google Scholar] [CrossRef]
- Cancer Genome Atlas, Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Tapial, J.; Ha, K.C.H.; Sterne-Weiler, T.; Gohr, A.; Braunschweig, U.; Hermoso-Pulido, A.; Quesnel-Vallieres, M.; Permanyer, J.; Sodaei, R.; Marquez, Y.; et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 2017, 27, 1759–1768. [Google Scholar] [CrossRef]
- Garner, E.; Romero, P.; Dunker, A.K.; Brown, C.; Obradovic, Z. Predicting Binding Regions within Disordered Proteins. Genome Inform. Work. Genome Inform. 1999, 10, 41–50. [Google Scholar]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006, 7, 208. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013, 41, D955–D961. [Google Scholar] [CrossRef] [PubMed]
- Ghandi, M.; Huang, F.W.; Jane-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R., 3rd; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef]
- Van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef]
- Buljan, M.; Chalancon, G.; Dunker, A.K.; Bateman, A.; Balaji, S.; Fuxreiter, M.; Babu, M.M. Alternative splicing of intrinsically disordered regions and rewiring of protein interactions. Curr. Opin. Struct. Biol. 2013, 23, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Crown, J.; O’Leary, M.; Ooi, W.S. Docetaxel and paclitaxel in the treatment of breast cancer: A review of clinical experience. Oncologist 2004, 9 (Suppl. 2), 24–32. [Google Scholar] [CrossRef]
- Cameron, D.A.; Gabra, H.; Leonard, R.C. Continuous 5-fluorouracil in the treatment of breast cancer. Br. J. Cancer 1994, 70, 120–124. [Google Scholar] [CrossRef]
- Osborne, C.K. Tamoxifen in the treatment of breast cancer. N. Engl. J. Med. 1998, 339, 1609–1618. [Google Scholar] [CrossRef]
- Blackstein, M.; Vogel, C.L.; Ambinder, R.; Cowan, J.; Iglesias, J.; Melemed, A. Gemcitabine as first-line therapy in patients with metastatic breast cancer: A phase II trial. Oncology 2002, 62, 2–8. [Google Scholar] [CrossRef]
- Anczukow, O.; Akerman, M.; Clery, A.; Wu, J.; Shen, C.; Shirole, N.H.; Raimer, A.; Sun, S.; Jensen, M.A.; Hua, Y.; et al. SRSF1-Regulated Alternative Splicing in Breast Cancer. Mol. Cell 2015, 60, 105–117. [Google Scholar] [CrossRef]
- Zhao, W.; Hoadley, K.A.; Parker, J.S.; Perou, C.M. Identification of mRNA isoform switching in breast cancer. BMC Genom. 2016, 17, 181. [Google Scholar] [CrossRef] [PubMed]
- Sveen, A.; Kilpinen, S.; Ruusulehto, A.; Lothe, R.A.; Skotheim, R.I. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 2016, 35, 2413–2427. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Manley, J.L. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov. 2013, 3, 1228–1237. [Google Scholar] [CrossRef]
- Van Schie, E.H.; van Amerongen, R. Aberrant WNT/CTNNB1 Signaling as a Therapeutic Target in Human Breast Cancer: Weighing the Evidence. Front. Cell Dev. Biol. 2020, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Wang, W.; Zhang, Z.; Bai, Y.; Gao, J.; Zhao, C. Wnt signaling in human and mouse breast cancer: Focusing on Wnt ligands, receptors and antagonists. Cancer Sci. 2018, 109, 3368–3375. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, M.; Lacroix-Triki, M.; Driouch, K.; de la Grange, P.; Gratadou, L.; Beck, S.; Millevoi, S.; Tazi, J.; Lidereau, R.; Vagner, S.; et al. Exon-based clustering of murine breast tumor transcriptomes reveals alternative exons whose expression is associated with metastasis. Cancer Res. 2010, 70, 896–905. [Google Scholar] [CrossRef] [PubMed]
- Pang, B.; Cheng, S.; Sun, S.P.; An, C.; Liu, Z.Y.; Feng, X.; Liu, G.J. Prognostic role of PIK3CA mutations and their association with hormone receptor expression in breast cancer: A meta-analysis. Sci. Rep. 2014, 4, 6255. [Google Scholar] [CrossRef]
- Romero, P.R.; Zaidi, S.; Fang, Y.Y.; Uversky, V.N.; Radivojac, P.; Oldfield, C.J.; Cortese, M.S.; Sickmeier, M.; LeGall, T.; Obradovic, Z.; et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl. Acad. Sci. USA 2006, 103, 8390–8395. [Google Scholar] [CrossRef]
- Van der Lee, R.; Lang, B.; Kruse, K.; Gsponer, J.; Sanchez de Groot, N.; Huynen, M.A.; Matouschek, A.; Fuxreiter, M.; Babu, M.M. Intrinsically disordered segments affect protein half-life in the cell and during evolution. Cell Rep. 2014, 8, 1832–1844. [Google Scholar] [CrossRef]
- Wang, B.D.; Lee, N.H. Aberrant RNA Splicing in Cancer and Drug Resistance. Cancers 2018, 10, 458. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, J.; Pradella, D.; Kim, Y.; Shao, C.; Li, H.; Choi, N.; Ha, J.; Di Matteo, A.; Fu, X.-D.; Zheng, X.; et al. Global Alternative Splicing Defects in Human Breast Cancer Cells. Cancers 2021, 13, 3071. https://doi.org/10.3390/cancers13123071
Oh J, Pradella D, Kim Y, Shao C, Li H, Choi N, Ha J, Di Matteo A, Fu X-D, Zheng X, et al. Global Alternative Splicing Defects in Human Breast Cancer Cells. Cancers. 2021; 13(12):3071. https://doi.org/10.3390/cancers13123071
Chicago/Turabian StyleOh, Jagyeong, Davide Pradella, Yoonseong Kim, Changwei Shao, Hairi Li, Namjeong Choi, Jiyeon Ha, Anna Di Matteo, Xiang-Dong Fu, Xuexiu Zheng, and et al. 2021. "Global Alternative Splicing Defects in Human Breast Cancer Cells" Cancers 13, no. 12: 3071. https://doi.org/10.3390/cancers13123071
APA StyleOh, J., Pradella, D., Kim, Y., Shao, C., Li, H., Choi, N., Ha, J., Di Matteo, A., Fu, X.-D., Zheng, X., Ghigna, C., & Shen, H. (2021). Global Alternative Splicing Defects in Human Breast Cancer Cells. Cancers, 13(12), 3071. https://doi.org/10.3390/cancers13123071






