5′ Region Large Genomic Rearrangements in the BRCA1 Gene in French Families: Identification of a Tandem Triplication and Nine Distinct Deletions with Five Recurrent Breakpoints
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Family Selection and Screening Methods
2.2. Dedicated Zoom-In CGH Array
2.3. qPCR and Breakpoint Sequencing
2.4. Molecular Combing
2.5. Haplotype Study
2.6. RNA Studies
2.7. Tumoral Studies
2.8. Screening of the 5′ Region and Exons 1a and 1b
2.9. Identification of Sensitive Regions
2.10. Nomenclature
2.11. Variant Classification
3. Results
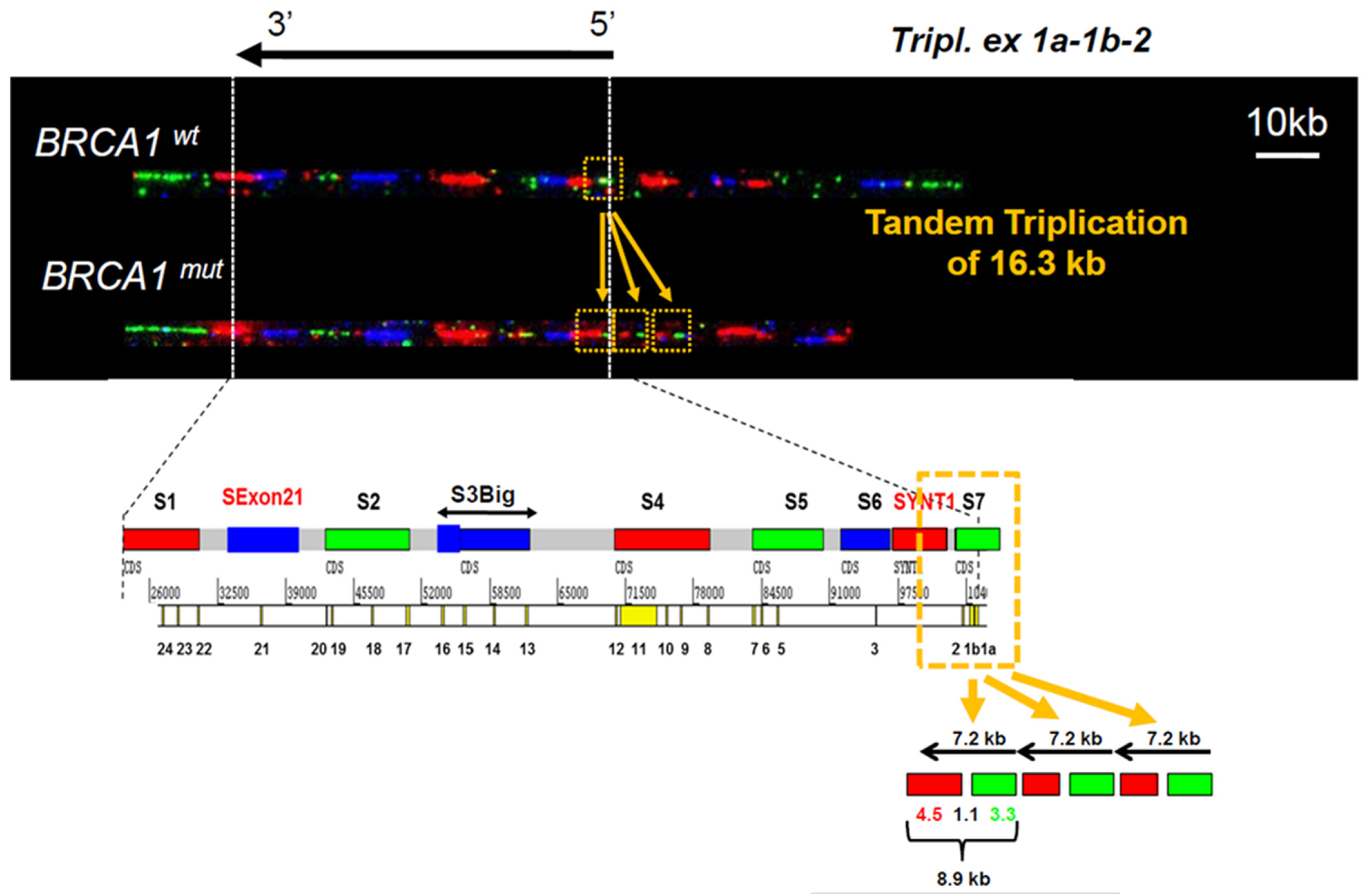
3.1. Identification of a Triplication of BRCA1 5′ Region
3.2. Characterization of Different Deletions in the BRCA1 5′ Region
3.3. Identification of Five Regions Highly Sensitive to LGR in BRCA1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caputo:, S.; Benboudjema, L.; Sinilnikova, O.; Rouleau, E.; Béroud, C.; Lidereau, R. Description and analysis of genetic variants in French hereditary breast and ovarian cancer families recorded in the UMD-BRCA1/BRCA2 databases. Nucleic Acids Res. 2012, 40, D992–D1002. [Google Scholar] [CrossRef]
- Futreal, P.A.; Liu, Q.; Shattuck-Eidens, D.; Cochran, C.; Harshman, K.; Tavtigian, S.; Bennett, L.M.; Haugen-Strano, A.; Swensen, J.; Miki, Y. BRCA1 mutations in primary breast and ovarian carcinomas. Science 1994, 266, 120–122. [Google Scholar] [CrossRef] [PubMed]
- Miki, Y.; Swensen, J.; Shattuck-Eidens, D.; Futreal, P.A.; Harshman, K.; Tavtigian, S.; Liu, Q.; Cochran, C.; Bennett, L.M.; Ding, W. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994, 266, 66–71. [Google Scholar] [CrossRef] [Green Version]
- Smith, T.M.; Lee, M.K.; Szabo, C.I.; Jerome, N.; McEuen, M.; Taylor, M.; Hood, L.; King, M.C. Complete genomic sequence and analysis of 117 kb of human DNA containing the gene BRCA1. Genome Res. 1996, 6, 1029–1049. [Google Scholar] [CrossRef] [Green Version]
- Puget, N.; Gad, S.; Perrin-Vidoz, L.; Sinilnikova, O.M.; Stoppa-Lyonnet, D.; Lenoir, G.M.; Mazoyer, S. Distinct BRCA1 rearrangements involving the BRCA1 pseudogene suggest the existence of a recombination hot spot. Am. J. Hum. Genet. 2002, 70, 858–865. [Google Scholar] [CrossRef] [Green Version]
- Sobczak, K.; Krzyzosiak, W.J. Structural determinants of BRCA1 translational regulation. J. Biol. Chem. 2002, 277, 17349–17358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puget, N.; Torchard, D.; Serova-Sinilnikova, O.M.; Lynch, H.T.; Feunteun, J.; Lenoir, G.M.; Mazoyer, S. A 1-kb Alu-mediated germ-line deletion removing BRCA1 exon 17. Cancer Res. 1997, 57, 828–831. [Google Scholar]
- Swensen, J.; Hoffman, M.; Skolnick, M.H.; Neuhausen, S.L. Identification of a 14 kb deletion involving the promoter region of BRCA1 in a breast cancer family. Hum. Mol. Genet. 1997, 6, 1513–1517. [Google Scholar] [CrossRef] [Green Version]
- Ewald, I.P.; Ribeiro, P.L.I.; Palmero, E.I.; Cossio, S.L.; Giugliani, R.; Ashton-Prolla, P. Genomic rearrangements in BRCA1 and BRCA2: A literature review. Genet. Mol. Biol. 2009, 32, 437–446. [Google Scholar] [CrossRef] [Green Version]
- Hogervorst, F.B.L.; Nederlof, P.M.; Gille, J.J.P.; McElgunn, C.J.; Grippeling, M.; Pruntel, R.; Regnerus, R.; van Welsem, T.; van Spaendonk, R.; Menko, F.H.; et al. Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res. 2003, 63, 1449–1453. [Google Scholar]
- Moisan, A.-M.; Fortin, J.; Dumont, M.; Samson, C.; Bessette, P.; Chiquette, J.; Laframboise, R.; Lépine, J.; Lespérance, B.; Pichette, R.; et al. No Evidence of BRCA1/2 genomic rearrangements in high-risk French-Canadian breast/ovarian cancer families. Genet. Test. 2006, 10, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Engert, S.; Wappenschmidt, B.; Betz, B.; Kast, K.; Kutsche, M.; Hellebrand, H.; Goecke, T.O.; Kiechle, M.; Niederacher, D.; Schmutzler, R.K.; et al. MLPA screening in the BRCA1 gene from 1,506 German hereditary breast cancer cases: Novel deletions, frequent involvement of exon 17, and occurrence in single early-onset cases. Hum. Mutat. 2008, 29, 948–958. [Google Scholar] [CrossRef]
- Sluiter, M.D.; van Rensburg, E.J. Large genomic rearrangements of the BRCA1 and BRCA2 genes: Review of the literature and report of a novel BRCA1 mutation. Breast Cancer Res. Treat. 2011, 125, 325–349. [Google Scholar] [CrossRef]
- Gad, S.; Bièche, I.; Barrois, M.; Casilli, F.; Pages-Berhouet, S.; Dehainault, C.; Gauthier-Villars, M.; Bensimon, A.; Aurias, A.; Lidereau, R.; et al. Characterisation of a 161 kb deletion extending from the NBR1 to the BRCA1 genes in a French breast-ovarian cancer family. Hum. Mutat. 2003, 21, 654. [Google Scholar] [CrossRef]
- Preisler-Adams, S.; Schönbuchner, I.; Fiebig, B.; Welling, B.; Dworniczak, B.; Weber, B.H.F. Gross rearrangements in BRCA1 but not BRCA2 play a notable role in predisposition to breast and ovarian cancer in high-risk families of German origin. Cancer Genet. Cytogenet. 2006, 168, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Villy, M.-C.; Mouret-Fourme, E.; Golmard, L.; Becette, V.; Callet, N.; Marx, G.; Colas, C.; Lamarque, D.; Rouleau, E.; Stoppa-Lyonnet, D. Co-occurrence of germline BRCA1 and CDH1 pathogenic variants. J. Med. Genet. 2020, 58, 357–361. [Google Scholar] [CrossRef] [PubMed]
- Mazoyer, S. Genomic rearrangements in the BRCA1 and BRCA2 genes. Hum. Mutat. 2005, 25, 415–422. [Google Scholar] [CrossRef]
- French Unicancer Genetics Group (UGG) of Cancer Genetics Clinic and Laboratory Network. Available online: http://www.unicancer.fr/la-recherche-unicancer/les-groupes-transversaux/groupe-genetique-cancer-ggc (accessed on 22 June 2021).
- Béroud, C.; Letovsky, S.I.; Braastad, C.D.; Caputo, S.M.; Beaudoux, O.; Bignon, Y.J.; Bressac-De Paillerets, B.; Bronner, M.; Buell, C.M.; Collod-Béroud, G.; et al. BRCA Share: A Collection of Clinical BRCA Gene Variants. Hum. Mutat. 2016, 37, 1318–1328. [Google Scholar] [CrossRef]
- Rouleau, E.; Jesson, B.; Briaux, A.; Nogues, C.; Chabaud, V.; Demange, L.; Sokolowska, J.; Coulet, F.; Barouk-Simonet, E.; Bignon, Y.J.; et al. Rare germline large rearrangements in the BRCA1/2 genes and eight candidate genes in 472 patients with breast cancer predisposition. Breast Cancer Res. Treat. 2012, 133, 1179–1190. [Google Scholar] [CrossRef]
- The BRCA1 Exon 13 Duplication Screening Group. The exon 13 duplication in the BRCA1 gene is a founder mutation present in geographically diverse populations. Am. J. Hum. Genet. 2000, 67, 207–212. [Google Scholar] [CrossRef] [Green Version]
- Puget, N.; Stoppa-Lyonnet, D.; Sinilnikova, O.M.; Pagès, S.; Lynch, H.T.; Lenoir, G.M.; Mazoyer, S. Screening for germ-line rearrangements and regulatory mutations in BRCA1 led to the identification of four new deletions. Cancer Res. 1999, 59, 455–461. [Google Scholar]
- Montagna, M.; Dalla Palma, M.; Menin, C.; Agata, S.; De Nicolo, A.; Chieco-Bianchi, L.; D’Andrea, E. Genomic rearrangements account for more than one-third of the BRCA1 mutations in northern Italian breast/ovarian cancer families. Hum. Mol. Genet. 2003, 12, 1055–1061. [Google Scholar] [CrossRef] [Green Version]
- van den Ouweland, A.M.W.; Dinjens, W.N.M.; Dorssers, L.C.J.; van Veghel-Plandsoen, M.M.; Brüggenwirth, H.T.; Withagen-Hermans, C.J.; Collée, J.M.; Joosse, S.A.; Terlouw-Kromosoeto, J.N.R.; Nederlof, P.M. Deletion of exons 1a-2 of BRCA1: A rather frequent pathogenic abnormality. Genet. Test. Mol. Biomark. 2009, 13, 399–406. [Google Scholar] [CrossRef]
- Casilli, F.; Di Rocco, Z.C.; Gad, S.; Tournier, I.; Stoppa-Lyonnet, D.; Frebourg, T.; Tosi, M. Rapid detection of novel BRCA1 rearrangements in high-risk breast-ovarian cancer families using multiplex PCR of short fluorescent fragments. Hum. Mutat. 2002, 20, 218–226. [Google Scholar] [CrossRef]
- Rouleau, E.; Lefol, C.; Tozlu, S.; Andrieu, C.; Guy, C.; Copigny, F.; Nogues, C.; Bieche, I.; Lidereau, R. High-resolution oligonucleotide array-CGH applied to the detection and characterization of large rearrangements in the hereditary breast cancer gene BRCA1. Clin. Genet. 2007, 72, 199–207. [Google Scholar] [CrossRef]
- Staaf, J.; Törngren, T.; Rambech, E.; Johansson, U.; Persson, C.; Sellberg, G.; Tellhed, L.; Nilbert, M.; Borg, A. Detection and precise mapping of germline rearrangements in BRCA1, BRCA2, MSH2, and MLH1 using zoom-in array comparative genomic hybridization (aCGH). Hum. Mutat. 2008, 29, 555–564. [Google Scholar] [CrossRef]
- Rouleau, E.; Lefol, C.; Bourdon, V.; Coulet, F.; Noguchi, T.; Soubrier, F.; Bièche, I.; Olschwang, S.; Sobol, H.; Lidereau, R. Quantitative PCR high-resolution melting (qPCR-HRM) curve analysis, a new approach to simultaneously screen point mutations and large rearrangements: Application to MLH1 germline mutations in Lynch syndrome. Hum. Mutat. 2009, 30, 867–875. [Google Scholar] [CrossRef] [Green Version]
- Barrois, M.; Bièche, I.; Mazoyer, S.; Champème, M.-H.; Bressac-de Paillerets, B.; Lidereau, R. Real-time PCR-based gene dosage assay for detecting BRCA1 rearrangements in breast-ovarian cancer families. Clin. Genet. 2004, 65, 131–136. [Google Scholar] [CrossRef]
- Tilkin, A.F.; Bagot, M.; Picard, F.; Dreyfus, F.; Vernant, J.P.; Lévy, J.P. B cells from leukemic patients are relatively resistant to in-vitro EBV transformation. Leuk. Res. 1986, 10, 1059–1060. [Google Scholar] [CrossRef]
- Lebofsky, R.; Heilig, R.; Sonnleitner, M.; Weissenbach, J.; Bensimon, A. DNA replication origin interference increases the spacing between initiation events in human cells. Mol. Biol. Cell 2006, 17, 5337–5345. [Google Scholar] [CrossRef] [Green Version]
- Cheeseman, K.; Rouleau, E.; Vannier, A.; Thomas, A.; Briaux, A.; Lefol, C.; Walrafen, P.; Bensimon, A.; Lidereau, R.; Conseiller, E.; et al. A diagnostic genetic test for the physical mapping of germline rearrangements in the susceptibility breast cancer genes BRCA1 and BRCA2. Hum. Mutat. 2012, 33, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Popova, T.; Manié, E.; Rieunier, G.; Caux-Moncoutier, V.; Tirapo, C.; Dubois, T.; Delattre, O.; Sigal-Zafrani, B.; Bollet, M.; Longy, M.; et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012, 72, 5454–5462. [Google Scholar] [CrossRef] [Green Version]
- Caputo, S.M.; Léone, M.; Damiola, F.; Ehlen, A.; Carreira, A.; Gaidrat, P.; Martins, A.; Brandão, R.D.; Peixoto, A.; Vega, A.; et al. Full in-frame exon 3 skipping of BRCA2 confers high risk of breast and/or ovarian cancer. Oncotarget 2018, 9, 17334–17348. [Google Scholar] [CrossRef]
- Petitalot, A.; Dardillac, E.; Jacquet, E.; Nhiri, N.; Guirouilh-Barbat, J.; Julien, P.; Bouazzaoui, I.; Bonte, D.; Feunteun, J.; Schnell, J.A.; et al. Combining Homologous Recombination and Phosphopeptide-binding Data to Predict the Impact of BRCA1 BRCT Variants on Cancer Risk. Mol. Cancer Res. 2019, 17, 54–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tubeuf, H.; Caputo, S.M.; Sullivan, T.; Rondeaux, J.; Krieger, S.; Caux-Moncoutier, V.; Hauchard, J.; Castelain, G.; Fiévet, A.; Meulemans, L.; et al. Calibration of Pathogenicity Due to Variant-Induced Leaky Splicing Defects by Using BRCA2 Exon 3 as a Model System. Cancer Res. 2020, 80, 3593–3605. [Google Scholar] [CrossRef] [PubMed]
- Meulemans, L.; Mesman, R.L.S.; Caputo, S.M.; Krieger, S.; Guillaud-Bataille, M.; Caux-Moncoutier, V.; Léone, M.; Boutry-Kryza, N.; Sokolowska, J.; Révillion, F.; et al. Skipping nonsense to maintain function: The paradigm of BRCA2 exon 12. Cancer Res. 2020, 80, 1374–1386. [Google Scholar] [CrossRef]
- Thompson, D.; Easton, D.F.; Goldgar, D.E. A full-likelihood method for the evaluation of causality of sequence variants from family data. Am. J. Hum. Genet. 2003, 73, 652–655. [Google Scholar] [CrossRef] [Green Version]
- Belman, S.; Parsons, M.T.; Spurdle, A.B.; Goldgar, D.E.; Feng, B.-J. Considerations in assessing germline variant pathogenicity using cosegregation analysis. Genet. Med. 2020, 22, 2052–2059. [Google Scholar] [CrossRef]
- Goldgar, D.E.; Easton, D.F.; Deffenbaugh, A.M.; Monteiro, A.N.A.; Tavtigian, S.V.; Couch, F.J. Breast Cancer Information Core (BIC) Steering Committee Integrated evaluation of DNA sequence variants of unknown clinical significance: Application to BRCA1 and BRCA2. Am. J. Hum. Genet. 2004, 75, 535–544. [Google Scholar] [CrossRef] [Green Version]
- Easton, D.F.; Deffenbaugh, A.M.; Pruss, D.; Frye, C.; Wenstrup, R.J.; Allen-Brady, K.; Tavtigian, S.V.; Monteiro, A.N.A.; Iversen, E.S.; Couch, F.J.; et al. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am. J. Hum. Genet. 2007, 81, 873–883. [Google Scholar] [CrossRef] [Green Version]
- Spurdle, A.B.; Whiley, P.J.; Thompson, B.; Feng, B.; Healey, S.; Brown, M.A.; Pettigrew, C.; kConFab; Van Asperen, C. J.; Ausems, M.G.E.M.; et al. BRCA1 R1699Q variant displaying ambiguous functional abrogation confers intermediate breast and ovarian cancer risk. J. Med. Genet. 2012, 49, 525–532. [Google Scholar] [CrossRef]
- Spurdle, A.B.; Couch, F.J.; Parsons, M.T.; McGuffog, L.; Barrowdale, D.; Bolla, M.K.; Wang, Q.; Healey, S.; Schmutzler, R.; Wappenschmidt, B.; et al. Refined histopathological predictors of BRCA1 and BRCA2 mutation status: A large-scale analysis of breast cancer characteristics from the BCAC, CIMBA, and ENIGMA consortia. Breast Cancer Res. 2014, 16, 3419. [Google Scholar] [CrossRef] [Green Version]
- Parsons, M.T.; Tudini, E.; Li, H.; Hahnen, E.; Wappenschmidt, B.; Feliubadaló, L.; Aalfs, C.M.; Agata, S.; Aittomäki, K.; Alducci, E.; et al. Large scale multifactorial likelihood quantitative analysis of BRCA1 and BRCA2 variants: An ENIGMA resource to support clinical variant classification. Hum. Mutat. 2019, 40, 1557–1578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindor, N.M.; Guidugli, L.; Wang, X.; Vallée, M.P.; Monteiro, A.N.A.; Tavtigian, S.; Goldgar, D.E.; Couch, F.J. A review of a multifactorial probability-based model for classification of BRCA1 and BRCA2 variants of uncertain significance (VUS). Hum. Mutat. 2012, 33, 8–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tavtigian, S.V.; Deffenbaugh, A.M.; Yin, L.; Judkins, T.; Scholl, T.; Samollow, P.B.; de Silva, D.; Zharkikh, A.; Thomas, A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J. Med. Genet. 2006, 43, 295–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallée, M.P.; Sera, T.L.D.; Nix, D.A.; Paquette, A.M.; Parsons, M.T.; Bell, R.; Hoffman, A.; Hogervorst, F.B.L.; Goldgar, D.E.; Spurdle, A.B.; et al. Adding In Silico Assessment of Potential Splice Aberration to the Integrated Evaluation of BRCA Gene Unclassified Variants. Hum. Mutat. 2016, 37, 627–639. [Google Scholar] [CrossRef] [Green Version]
- Plon, S.E.; Eccles, D.M.; Easton, D.; Foulkes, W.D.; Genuardi, M.; Greenblatt, M.S.; Hogervorst, F.B.L.; Hoogerbrugge, N.; Spurdle, A.B.; Tavtigian, S.V.; et al. Sequence variant classification and reporting: Recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum. Mutat. 2008, 29, 1282–1291. [Google Scholar] [CrossRef] [Green Version]
- Ramus, S.J.; Harrington, P.A.; Pye, C.; DiCioccio, R.A.; Cox, M.J.; Garlinghouse-Jones, K.; Oakley-Girvan, I.; Jacobs, I.J.; Hardy, R.M.; Whittemore, A.S.; et al. Contribution of BRCA1 and BRCA2 mutations to inherited ovarian cancer. Hum. Mutat. 2007, 28, 1207–1215. [Google Scholar] [CrossRef] [PubMed]
- Marino, M.; Rabacchi, C.; Simone, M.L.; Medici, V.; Cortesi, L.; Calandra, S. A novel deletion of BRCA1 gene that eliminates the ATG initiation codon without affecting the promoter region. Clin. Chim. Acta 2009, 403, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Palanca Suela, S.; Esteban Cardeñosa, E.; Barragán González, E.; Oltra Soler, S.; de Juan Jiménez, I.; Chirivella González, I.; Segura Huerta, A.; Guillén Ponce, C.; Martínez de Dueñas, E.; Bolufer Gilabert, P.; et al. Identification of a novel BRCA1 large genomic rearrangement in a Spanish breast/ovarian cancer family. Breast Cancer Res. Treat. 2008, 112, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Fachal, L.; Blanco, A.; Santamariña, M.; Carracedo, A.; Vega, A. Large genomic rearrangements of BRCA1 and BRCA2 among patients referred for genetic analysis in Galicia (NW Spain): Delimitation and mechanism of three novel BRCA1 rearrangements. PLoS ONE 2014, 9, e93306. [Google Scholar] [CrossRef]
- del Valle, J.; Feliubadaló, L.; Nadal, M.; Teulé, A.; Miró, R.; Cuesta, R.; Tornero, E.; Menéndez, M.; Darder, E.; Brunet, J.; et al. Identification and comprehensive characterization of large genomic rearrangements in the BRCA1 and BRCA2 genes. Breast Cancer Res. Treat. 2010, 122, 733–743. [Google Scholar] [CrossRef]
- Richardson, M.E.; Chong, H.; Mu, W.; Conner, B.R.; Hsuan, V.; Willett, S.; Lam, S.; Tsai, P.; Pesaran, T.; Chamberlin, A.C.; et al. DNA breakpoint assay reveals a majority of gross duplications occur in tandem reducing VUS classifications in breast cancer predisposition genes. Genet. Med. 2019, 21, 683–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tessereau, C.; Léoné, M.; Buisson, M.; Duret, L.; Sinilnikova, O.M.; Mazoyer, S. Occurrence of a non deleterious gene conversion event in the BRCA1 gene. Genes. Chromosomes Cancer 2015, 54, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Judkins, T.; Rosenthal, E.; Arnell, C.; Burbidge, L.A.; Geary, W.; Barrus, T.; Schoenberger, J.; Trost, J.; Wenstrup, R.J.; Roa, B.B. Clinical significance of large rearrangements in BRCA1 and BRCA2. Cancer 2012, 118, 5210–5216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, W.; Li, B.; Wu, S.; Chen, J.; Sain, D.; Xu, D.; Black, M.H.; Karam, R.; Gillespie, K.; Farwell Hagman, K.D.; et al. Detection of structural variation using target captured next-generation sequencing data for genetic diagnostic testing. Genet. Med. 2019, 21, 1603–1610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolussi, A.; Belardinilli, F.; Silvestri, V.; Mahdavian, Y.; Valentini, V.; D’Inzeo, S.; Petroni, M.; Zani, M.; Ferraro, S.; Di Giulio, S.; et al. Identification of novel BRCA1 large genomic rearrangements by a computational algorithm of amplicon-based Next-Generation Sequencing data. PeerJ 2019, 7, e7972. [Google Scholar] [CrossRef] [PubMed]
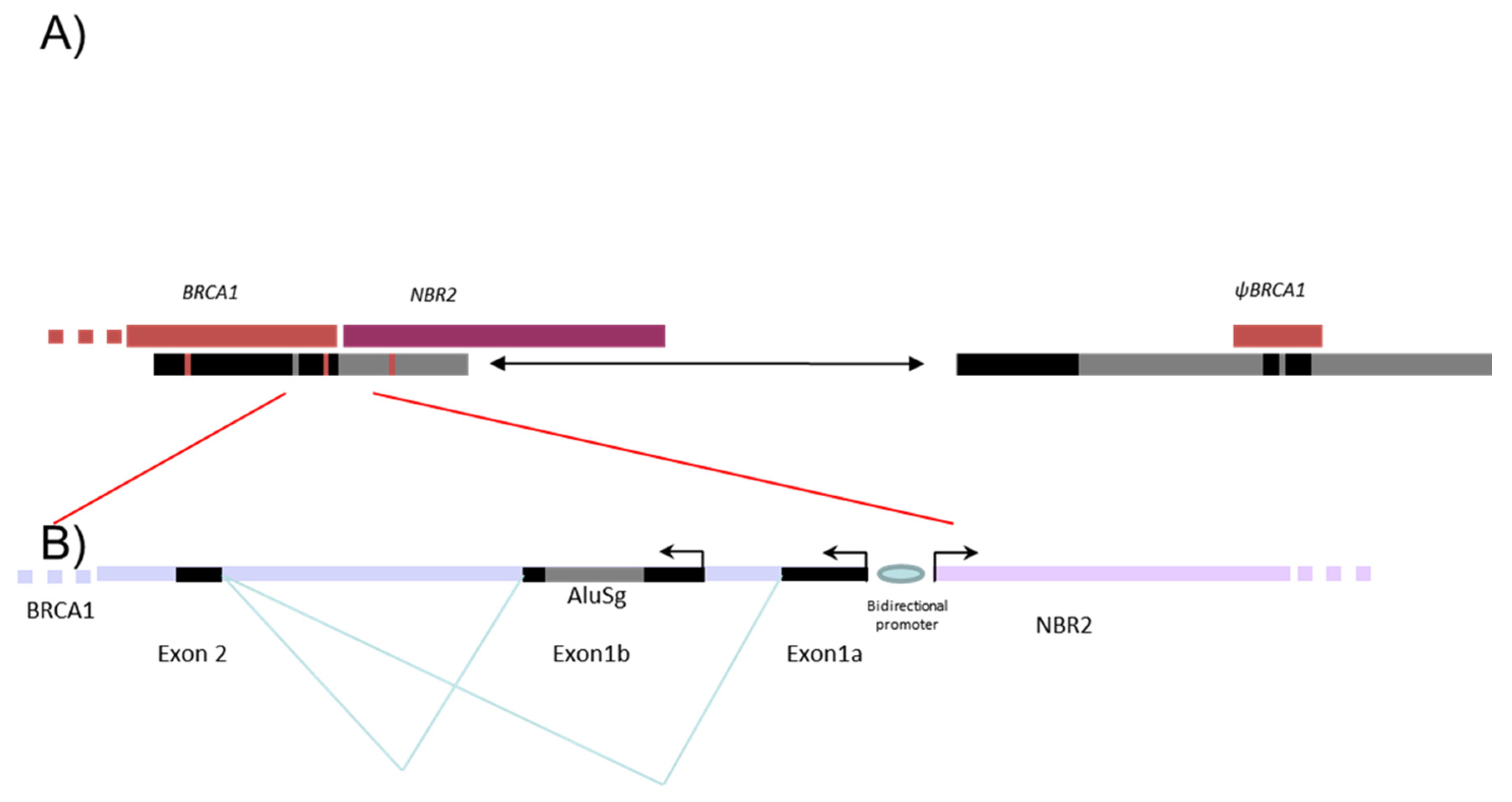



| Family | F (5′-3′): | R (5′-3′): |
|---|---|---|
| F8, F9, F10, F11, F12 | TCCCTAACCAAGATCTCTTTAAC | CTAAAACAGGCTGAAAACCTTAC |
| F1, F5 | ACCTAAAATTCCTTCTGCTGGAC | TTGGATAAAGTCCAGGCAAGAT |
| F2, F3, F4 * | CTTCAGAAAATACATCACCCA | ATAAGGTTACTGTCCCCGA |
| F14 | TCAAAGGATGAGTTGGGCATA | AAGGTCAATTGTGTTCATTTGCAT |
| Lit6 | CCACTGAGGACCTAAAGCATAA | GATATTGTAGGGAAAGACTATCAG |
| F13 | ATGAGTATGGGGCTAAGACA | CATTTGACCTGTGGAGTTTC |
| F6, F7 | AGACTTCCTGGACGGGGGAC | TAAGGAACACTGTGGCGAAGA |
| Exon 1 screening | CCTAACCTTTCCCAGTGACCTG | TAAGGAACACTGTGGCGAAGA |
| F15, F16, F17, F18 | GTTCAAGTTCAAGCGCTTCTC | GTGTCTAGCTTGGGGTTTGG |
| Family | Deleted Exons (MLPA, QMPSF) | Frequency | Estimated Size in CGH Array (kb) | Size of the Deletion (bp) | Deleted Locus in (From 3′ to 5′ of the BRCA1 Gene) chr17.hg38 | Sensitive Regions in Which the 5′ and 3′ Breakpoints Are Localized | Class | Reference |
|---|---|---|---|---|---|---|---|---|
| A/ | ||||||||
| F1 | Exons 1a-2 ~38 kb | 1 | 33–38 | 36,934 | g.43120187_43157120del | A1–A5 | 5 * | This report |
| F2 F3 F4 | 3 | 33–38 | 36,770 | g.43118776_43155546del | A0–A5 | 5 | This report | |
| F5 | 1 | 33–38 | 36,934 | g.43119898_43156832del | A1–A5 | 5 | This report and in [5,12,23,24] | |
| Lit1 | - | - | 36,934 | g.43119950_43156884del | A1–A5 | 5 | [5] | |
| Lit2 | - | - | 36,934 | g.43119629_43156562del | A1–A5 | 5 | [17] | |
| F6 F7 | Exon 2 <1 kb | 2 | <2 | 1407 | g.43123453_43124859del | A2–A2 | 5 | This report |
| Lit3 | Exon 2 | - | - | 2535 | g.43122409_43124943del | S–A2 | 5 | [49] |
| Lit4 | Exons 2–3 | - | - | 9113 | g.43115107_43124219del | A2–S | 5 | [50] |
| F8 F9 F10 | Exons 1a–2 ~5–10 kb | 3 | 4–8 | 6757 | g.43120817_43127573del | A1–A3 | 5 | This report |
| F11 | 1 | 4–8 | 6650 | g.43121312_43127961del | A1–A3 | 5 | This report | |
| F12 | 1 | 4–8 | 7248 | g.43120725-43127972del | A1–A3 | 5 | This report | |
| Lit5 | Exons 1–2 ~5–10 kb | - | 8168 | g.43123549_43131716delinsAAAAAAAAA | A2–A4 | 5 | [24] | |
| F13 | Exons 1a–2 13–14 kb | 1 | 13–14 | 13,408 | g.43118518_43131925del | A0–A4 | 5 | This report |
| Lit6 | - | - | 13,815 | g.43118829_43132643del | A0–A4 | 5 | [8,51] | |
| F14 | Exon 1a–1b | 1 | 4–6 | 5568 | g.43124936_43130504del | A2–A4 | 5 | This report |
| B/ | ||||||||
| Family | Triplicated Exons (MLPA) | Frequency | Estimated Size in CGH Array (kb) | Size of the Fragment Triplicated (bp) | Triplicated locus in (From 3′ to 5′ of the BRCA1 Gene) | Sensitive Regions in Which the 5′ and 3′ Breakpoints Are Localized | Reference | |
| F15, F16, F17, F18, F19, F20 Lit7 | Exon 1–2 | 7 | 6–8 kb | 7259 | g.:43,120,714–43,127,972 | A1–A3 | This report and [52,53] | |
| Lit8 | 5′UTR | 1 | - | 44,430 | g.43,124,439–43,168,868 | A2–S | [54] | |
| Lit9 | Exon 1 | 3 | - | 36,915 | g.43,119,759–43,156,673 | A1–A5 | [54] | |
| Tumor 1 (F16) | Tumor 2 (F17) | |
|---|---|---|
| Family | F16 | F17 |
| Cellularity | 60% | 50% |
| LST score | 18 | 5 |
| Tumor ploidy | 4 | 2 |
| Tumoral cells | 46% | 45% |
| Conclusion | LST low, no evidence of BRCAness | LST low, no evidence of BRCAness |
| Variant | Prior Probability | LR Cosegregation | LR Pathology | LR Family History | LR Combined | Posterior Probability of Pathogenicity | Classification |
|---|---|---|---|---|---|---|---|
| Exons 1a–2 ~38 kb | 0.5 | 119.105 | 15.4049 | 2.4 | 4403.52147 | 0.99977296058 | 5 (Pathogenic) |
| Exons 1a–2 ~5–10 kb | 0.5 | 118.28357 | 3.377408 | 4.170588 | 1666.11616 | 0.99940016 | 5 (Pathogenic) |
| Triplication | 0.5 | 0.060189299 | 0.006830208 | 0.786824 | 0.000323467 | 0.00032336283 | 1 (Benign) |
| Locus | Position | % Alu Sequence | Size Pb | Segmental Duplication and Repeated Sequence | Number of Breakpoints |
|---|---|---|---|---|---|
| A0 | 43118500–43118900 | 94% | 400 | AluJr4-AluSz | 3 |
| A1 | 43120150–43121400 | 68% | 1250 | AluYk3-AluSg-AluYm1 | 9 |
| A2 | 43123400–43125000 | 31% | 1600 | AluSc5-[Exon 2]-AluSx | 7 |
| A3 | 43127500–43128000 | 100% | 500 | LTR12C-AluY | 4 |
| A4 | 43130500–43132700 | 59% | 2200 | AluSz-MIRc-LIME3C2-[NBR2 exon]-AluY-AluSz6-MIRc-AlSc | 4 |
| A5 | 43155500–43157200 | 50% | 1700 | AluJr4-AluSz-AluSx-MIRc | 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caputo, S.M.; Telly, D.; Briaux, A.; Sesen, J.; Ceppi, M.; Bonnet, F.; Bourdon, V.; Coulet, F.; Castera, L.; Delnatte, C.; et al. 5′ Region Large Genomic Rearrangements in the BRCA1 Gene in French Families: Identification of a Tandem Triplication and Nine Distinct Deletions with Five Recurrent Breakpoints. Cancers 2021, 13, 3171. https://doi.org/10.3390/cancers13133171
Caputo SM, Telly D, Briaux A, Sesen J, Ceppi M, Bonnet F, Bourdon V, Coulet F, Castera L, Delnatte C, et al. 5′ Region Large Genomic Rearrangements in the BRCA1 Gene in French Families: Identification of a Tandem Triplication and Nine Distinct Deletions with Five Recurrent Breakpoints. Cancers. 2021; 13(13):3171. https://doi.org/10.3390/cancers13133171
Chicago/Turabian StyleCaputo, Sandrine M., Dominique Telly, Adrien Briaux, Julie Sesen, Maurizio Ceppi, Françoise Bonnet, Violaine Bourdon, Florence Coulet, Laurent Castera, Capucine Delnatte, and et al. 2021. "5′ Region Large Genomic Rearrangements in the BRCA1 Gene in French Families: Identification of a Tandem Triplication and Nine Distinct Deletions with Five Recurrent Breakpoints" Cancers 13, no. 13: 3171. https://doi.org/10.3390/cancers13133171
APA StyleCaputo, S. M., Telly, D., Briaux, A., Sesen, J., Ceppi, M., Bonnet, F., Bourdon, V., Coulet, F., Castera, L., Delnatte, C., Hardouin, A., Mazoyer, S., Schultz, I., Sevenet, N., Uhrhammer, N., Bonnet, C., Tilkin-Mariamé, A.-F., Houdayer, C., Moncoutier, V., ... Rouleau, E. (2021). 5′ Region Large Genomic Rearrangements in the BRCA1 Gene in French Families: Identification of a Tandem Triplication and Nine Distinct Deletions with Five Recurrent Breakpoints. Cancers, 13(13), 3171. https://doi.org/10.3390/cancers13133171






