Next-Generation Biomarkers for Cholangiocarcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
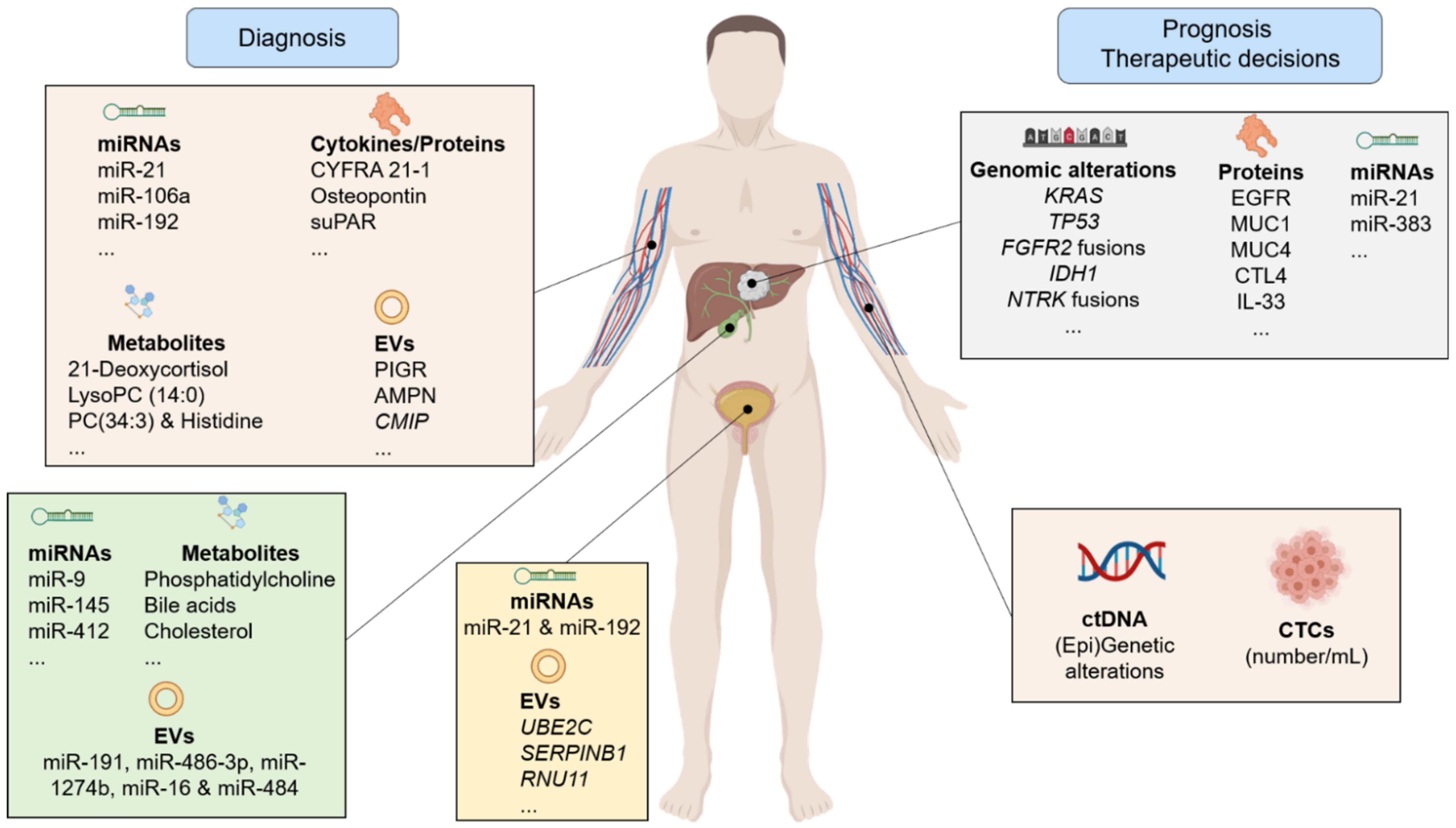
2. Biomarkers in the World of Clinical Needs
2.1. Diagnosis of CCA
2.2. CCA Treatment and Prognosis
3. Non-Invasive Biomarkers
3.1. Circulating Nucleic Acids
3.1.1. Circulating Tumor DNA
3.1.2. Cell-Free Non-Coding RNAs
| Source | miRNA | Levels | Comparison | AUC | Ref. |
|---|---|---|---|---|---|
| Serum/plasma | miR-21 | Up | iCCA (n = 74) vs. healthy controls (n = 74) | 0.908 | [49] |
| Up | iCCA (n = 25) vs. healthy controls (n = 7) | 0.940 | [50] | ||
| Up | Hepatolithiasis-CCA (n = 31) vs. hepatolithiasis (n = 40) | 0.610 | [51] | ||
| miR-221 | Up | Hepatolithiasis-CCA (n = 31) vs. hepatolithiasis (n = 40) | 0.767 | [51] | |
| miR-106a | Down | CCA (n = 103) vs. healthy controls (n = 20) | 0.890 | [62] | |
| miR-194 and miR-483-5p | Up | CCA (n = 30) vs. healthy controls (n = 30) | 0.810 | [63] | |
| miR-222 and miR-483-5p | Up | CCA (n = 30) vs. PSC (n = 30) | 0.770 | ||
| miR-122 | Up | CCA (n = 94) vs. healthy controls (n = 40) | 0.992 | [64] | |
| Down | CCA (n = 30) vs. PSC (n = 30) | 0.650 | [47] | ||
| miR-192 | Up | O. viverrini-related CCA (n = 51) vs. healthy controls (n = 32) | 0.803 | [70] | |
| miR-150 | Up | iCCA (n = 15) vs. healthy controls (n = 15) | 0.764 | [65] | |
| miR-26a | Up | CCA (n = 66) vs. healthy controls (n = 66) | 0.899 | [68] | |
| Down | CCA (n = 30) vs. PSC (n = 30) | 0.780 | [47] | ||
| miR-1281 | Down | CCA (n = 31) vs. PSC (n = 40) | 0.830 | [47] | |
| miR-126 | Down | CCA (n = 30) vs. PSC (n = 30) | 0.870 | ||
| miR-30b | Down | CCA (n = 30) vs. PSC (n = 30) | 0.780 | ||
| Bile | miR-9 | Up | Biliary tract cancer (n = 9) vs. choledocholithiasis (n = 9) | 0.975 | [46] |
| miR-145 | Up | Biliary tract cancer (n = 9) vs. choledocholithiasis (n = 9) | 0.975 | [46] | |
| miR-640 | Up | PSC-CCA (n = 12) vs. PSC (n = 52) | 0.810 | [47] | |
| miR-412 | Up | PSC-CCA (n = 12) vs. PSC (n = 52) | 0.810 | [47] | |
| miR-1537 | Up | PSC-CCA (n = 12) vs. PSC (n = 52) | 0.780 | [47] | |
| miR-3189 | Up | PSC-CCA (n = 12) vs. PSC (n = 52) | 0.800 | [47] | |
| miR-30d-5p | Up | CCA (n = 37) vs. obstructive benign biliary disease (n = 48) | 0.730 | [48] | |
| miR-92a-3p | Up | CCA (n = 37) vs. obstructive benign biliary disease (n = 48) | 0.652 | [48] | |
| Urine | miR-21 and miR-192 | Up | CCA (n = 22) vs. healthy controls (n = 21) | 0.849 | [52] |
3.2. Cytokines/Proteins
| Source | Protein/Ctyokine | Levels | Comparison | AUC | Ref. |
|---|---|---|---|---|---|
| Serum/plasma | CYFRA 21-1 | Up | Biliary tract cancer (n = 134) vs. benign biliary diseases (n = 52) | 0.851 | [75] |
| MMP7 | Up | CCA (n = 44) vs. benign biliary tract disease (n = 36) | 0.730 | [78] | |
| Up | CCA (n = 59) vs. benign biliary tract disease (n = 128) | 0.840 | [77] | ||
| Osteopontin | Up | CCA (n = 80) vs. healthy controls (n = 42) | 0.964 | [79] | |
| IL-6 | Up | Bile duct cancer (n = 26) vs. healthy controls (n = 23) | 0.875 | [87] | |
| S100A6 | Up | CCA (n = 29) vs. healthy controls (n = 22) | 0.909 | [82] | |
| DKK1 | Up | iCCA (n = 37) vs. healthy controls (n = 50) | 0.872 | [84] | |
| SSP411 | Up | CCA (n = 35) vs. “cholangitis (n = 13) and healthy controls (n = 23)” | 0.913 | [85] | |
| suPAR | Up | Biliary tract cancer (n = 95) vs. healthy controls (n = 66) | 0.969 | [81] | |
| TGF-β1 | Up | CCA (n = 45) vs. healthy controls (n = 45) | 0.668 | [88] |
3.3. Metabolites
3.4. Extracellular Vesicles
| Source | Metabolite | Levels | Comparison | AUC | Ref. |
|---|---|---|---|---|---|
| Serum | 21-Deoxycortisol | Down | CCA (n = 225) vs. healthy controls (n = 101) | 0.918 | [91] |
| Bilirubin | Up | CCA (n = 225) vs. healthy controls (n = 101) | 0.922 | [91] | |
| LysoPC (14:0) | Down | CCA (n = 225) vs. healthy controls (n = 101) | 0.954 | [91] | |
| LysoPC (15:0) | Up | CCA (n = 225) vs. healthy controls (n = 101) | 0.927 | [91] | |
| Glycocholic acid | Up | Biopsy-proven iCCA (n = 20) vs. healthy controls (n = 20) | DIS: 0.857 VAL: 0.991 | [92] | |
| Glycochenodeoxycholic acid | Up | Biopsy-proven iCCA (n = 20) vs. healthy controls (n = 20) | DIS: 0.823 VAL: 0.987 | [92] | |
| Androsterone sulfate II | Down | Biopsy-proven iCCA (n = 20) vs. healthy controls (n = 20) | DIS: 0.808 VAL: 0.800 | [92] | |
| Dehydroepiandrosterone | Up | Biopsy-proven iCCA (n = 20) vs. healthy controls (n = 20) | DIS: 0.790 VAL: 0.804 | [92] | |
| ChoE (22:6) | Down | Biopsy-proven iCCA (n = 20) vs. healthy controls (n = 20) | DIS: 0.763 VAL: 0.769 | [92] | |
| ChoE (20:4) | Down | Biopsy-proven iCCA (n = 20) vs. healthy controls (n = 20) | DIS: 0.760 VAL: 0.778 | [92] | |
| CMH (d18:1/16:0) | Up | Biopsy-proven iCCA (n = 20) vs. healthy controls (n = 20) | DIS: 0.798 VAL: 0.809 | [92] | |
| PC (16:0/16:0) | Up | Biopsy-proven iCCA (n = 20) vs. healthy controls (n = 20) | DIS: 0.773 VAL: 0.920 | [92] | |
| SM(43:2) PC(O-16:0/20:3) PC(O-18:0/18:2) SM(d18:2/16:0) Cer(d18:1/16:0) SM(42:3) | Up Down Down Up Up Up | iCCA (n = 20) vs. HCC (n = 20) (Biopsy-proven patients) | DIS:0.900 VAL: 0.981 | [92] | |
| PC(34:3) Histidine | Down | iCCA (n = 20) vs. PSC (n = 20) (Biopsy-proven patients) | DIS: 0.990 VAL: 0.995 | [92] | |
| Bile | Phosphatidylcholine Bile acids Cholesterol/lipid | Down | CCA (n = 16) vs. begin non-PSC biliary diseases (n = 27) | SEN: 88.9% SPE: 87.1% | [105] |
| Glycine-conjugated bile acids Phosphatidylcholines | Up Down | “Inoperable pCCA (n = 3) and dCCA (n = 2)” vs. non-malignant biliary diseases without cholestasis (n = 20) | SEN: 80% SPE: 95% | [106] |
| Source | EV Cargo | Biomarker Type | Levels | Comparison | AUC | Ref. |
|---|---|---|---|---|---|---|
| Serum | AnnexinV+ EpCAM+ ASGPR1+ | TAMP concentration | Up | CCA (n = 38) vs. cirrhosis (n = 49) | 0.630 | [104] |
| AMPN | Protein | Up | CCA (n = 43) vs. healthy controls (n = 32) | 0.878 | [107] | |
| VNN1 | Up | CCA (n = 43) vs. healthy controls (n = 32) | 0.876 | |||
| PIGR | Up | CCA (n = 43) vs. healthy controls (n = 32) | 0.844 | |||
| PIGR | Up | Early stage CCA (n = 13) vs. healthy controls (n = 22) | 0.905 | |||
| AMPN | Up | Early stage CCA (n = 13) vs. healthy controls (n = 22) | 0.833 | |||
| FIBG | Up | Early stage CCA (n = 13) vs. healthy controls (n = 22) | 0.833 | |||
| FIBG | Up | CCA (n = 43) vs. PSC (n = 30) | 0.796 | |||
| A1AG1 | Up | CCA (n = 43) vs. PSC (n = 30) | 0.794 | |||
| S100A8 | Up | CCA (n = 43) vs. PSC (n = 30) | 0.759 | |||
| FCN2 | Up | Early stage CCA (n = 13) vs. PSC (n = 30) | 0.956 | |||
| ITIH4 | Up | Early stage CCA (n = 13) vs. PSC (n = 30) | 0.881 | |||
| FIBG | Up | Early stage CCA (n = 13) vs. PSC (n = 30) | 0.881 | |||
| FIBG | Up | iCCA (n = 12) vs. HCC (n = 29) | 0.894 | |||
| A1AG1 | Up | iCCA (n = 12) vs. HCC (n = 29) | 0.845 | |||
| VTDB | Up | iCCA (n = 12) vs. HCC (n = 29) | 0.823 | |||
| CMIP | RNA | Up | CCA (n = 12) vs. (PSC + UC + healthy controls) (n = 23) | 0.957 | [108] | |
| GAD1 | Up | CCA (n = 12) vs. (PSC + UC + healthy controls) (n = 23) | 0.928 | |||
| NME1 | Up | CCA (n = 12) vs. (PSC + UC + healthy controls) (n = 23) | 0.899 | |||
| CDS1 | Up | CCA (n = 12) vs. (PSC + UC + healthy controls) (n = 23) | 0.893 | |||
| CKS1B | Up | CCA (n = 12) vs. (PSC + UC + healthy controls) (n = 23) | 0.891 | |||
| CMIP NME1 CKS1B | Up | CCA (n = 12) vs. (PSC + UC + healthy controls) (n = 23) | 1.000 | |||
| miR-551B | miRNA | Up | CCA (n = 12) vs. (PSC + UC + healthy controls) (n = 23) | 0.909 | [108] | |
| PMS2L4 | pseudogene | Up | CCA (n = 12) vs. (PSC + UC + healthy controls) (n = 23) | 0.880 | [108] | |
| LOC643955 | pseudogene | Up | CCA (n = 12) vs. (PSC + UC + healthy controls) (n = 23) | 0.873 | [108] | |
| LOC100134868 | lncRNA | Up | CCA (n = 12) vs. (PSC + UC + healthy controls) (n = 23) | 0.864 | [108] | |
| PTTG3P | pseudogene | Up | CCA (n = 12) vs. (PSC + UC + healthy controls) (n = 23) | 0.859 | [108] | |
| miR-200c-3p | miRNA | Up | CCA (n = 36) vs. healthy controls (n = 12) | 0.930 | [111] | |
| miR-96-5p | Up | CCA (n = 45) vs. healthy controls (n = 40) | 0.733 | [112] | ||
| miR-151a-5p | Up | CCA (n = 45) vs. healthy controls (n = 40) | 0.764 | |||
| miR-191-5p | Up | CCA (n = 45) vs. healthy controls (n = 40) | 0.542 | |||
| miR-4732-3p | Up | CCA (n = 45) vs. healthy controls (n = 40) | 0.654 | |||
| Urine | UBE2C | RNA | Up | CCA (n = 23) vs. (PSC + UC + healthy controls) (n = 22) | 0.779 | [108] |
| SERPINB1 | Up | CCA (n = 23) vs. (PSC + UC + healthy controls) (n = 22) | 0.654 | |||
| UBE2C SERPINB1 | Up | CCA (n = 23) vs. (PSC + UC + healthy controls) (n = 22) | 0.812 | |||
| RNU11 | snRNA | Up | CCA (n = 23) vs. (PSC + UC + healthy controls) (n = 22) | 0.830 | [108] | |
| LOC257358 | miscRNA | Up | CCA (n = 23) vs. (PSC + UC + healthy controls) (n = 22) | 0.812 | [108] | |
| VTRNA1-1 | vtRNA | Up | CCA (n = 23) vs. (PSC + UC + healthy controls) (n = 22) | 0.777 | [108] | |
| AURKAPS1 | Pseudogene | Down | CCA (n = 23) vs. (PSC + UC + healthy controls) (n = 22) | 0.771 | [108] | |
| miR-483 | miRNA | Down | CCA (n = 23) vs. (PSC + UC + healthy controls) (n = 22) | 0.763 | [108] | |
| Bile | miR-191 miR-486-3p miR-1274b miR-16 miR-484 | miRNA | Up | CCA (n = 46) vs. benign biliary diseases (n = 50) | SEN: 67% SPE: 96% | [110] |
3.5. Circulating Tumor Cells
4. Biomarkers in Tumor Tissue
5. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Banales, J.M.; Marin, J.J.G.; Lamarca, A.; Rodrigues, P.M.; Khan, S.A.; Roberts, L.R.; Cardinale, V.; Carpino, G.; Andersen, J.B.; Braconi, C.; et al. Cholangiocarcinoma 2020: The next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Banales, J.M.; Cardinale, V.; Carpino, G.; Marzioni, M.; Andersen, J.B.; Invernizzi, P.; Lind, G.E.; Folseraas, T.; Forbes, S.J.; Fouassier, L.; et al. Expert consensus document: Cholangiocarcinoma: Current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 261–280. [Google Scholar] [CrossRef] [PubMed]
- Valle, J.W.; Kelley, R.K.; Nervi, B.; Oh, D.-Y.; Zhu, A.X. Biliary tract cancer. Lancet 2021, 397, 428–444. [Google Scholar] [CrossRef]
- Clements, O.; Eliahoo, J.; Kim, J.U.; Taylor-Robinson, S.D.; Khan, S.A. Risk factors for intrahepatic and extrahepatic cholangiocarcinoma: A systematic review and meta-analysis. J. Hepatol. 2020, 72, 95–103. [Google Scholar] [CrossRef]
- De Oliveira, M.L.; Cunningham, S.C.; Cameron, J.L.; Kamangar, F.; Winter, J.M.; Lillemoe, K.D.; Choti, M.A.; Yeo, C.J.; Schulick, R.D. Cholangiocarcinoma: Thirty-one-year experience with 564 patients at a single institution. Ann. Surg. 2007, 245, 755–762. [Google Scholar] [CrossRef]
- Cillo, U.; Fondevila, C.; Donadon, M.; Gringeri, E.; Mocchegiani, F.; Schlitt, H.J.; Ijzermans, J.N.M.; Vivarelli, M.; Zieniewicz, K.; Damink, S.O.; et al. Surgery for cholangiocarcinoma. Liver Int. 2019, 39 (Suppl. 1), 143–155. [Google Scholar] [CrossRef] [Green Version]
- Lindnér, P.; Rizell, M.; Hafström, L. The Impact of Changed Strategies for Patients with Cholangiocarcinoma in This Millenium. HPB Surg. 2015, 2015, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Strijker, M.; Belkouz, A.; Van Der Geest, L.G.; Van Gulik, T.M.; Van Hooft, J.E.; De Meijer, V.E.; Mohammad, N.H.; De Reuver, P.R.; Verheij, J.; De Vos-Geelen, J.; et al. Treatment and survival of resected and unresected distal cholangiocarcinoma: A nationwide study. Acta Oncol. 2019, 58, 1048–1055. [Google Scholar] [CrossRef]
- Koerkamp, B.G.; Wiggers, J.K.; Allen, P.J.; Besselink, M.G.; Blumgart, L.H.; Busch, O.R.; Coelen, R.; D’Angelica, M.I.; DeMatteo, R.P.; Gouma, D.J.; et al. Recurrence Rate and Pattern of Perihilar Cholangiocarcinoma after Curative Intent Resection. J. Am. Coll. Surg. 2015, 221, 1041–1049. [Google Scholar] [CrossRef] [Green Version]
- Spolverato, G.; Kim, Y.; Alexandrescu, S.; Marques, H.P.; Lamelas, J.; Aldrighetti, L.; Gamblin, T.C.; Maithel, S.K.; Pulitano, C.; Bauer, T.W.; et al. Management and Outcomes of Patients with Recurrent Intrahepatic Cholangiocarcinoma Following Previous Curative-Intent Surgical Resection. Ann. Surg. Oncol. 2015, 23, 235–243. [Google Scholar] [CrossRef]
- Bertuccio, P.; Malvezzi, M.; Carioli, G.; Hashim, D.; Boffetta, P.; El-Serag, H.B.; La Vecchia, C.; Negri, E. Global trends in mortality from intrahepatic and extrahepatic cholangiocarcinoma. J. Hepatol. 2019, 71, 104–114. [Google Scholar] [CrossRef]
- Macias, R.I.; Banales, J.; Sangro, B.; Muntané, J.; Avila, M.; Lozano, E.; Perugorria, M.J.; Padillo, F.J.; Bujanda, L.; Marin, J.J. The search for novel diagnostic and prognostic biomarkers in cholangiocarcinoma. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1468–1477. [Google Scholar] [CrossRef]
- Macias, R.I.R.; Kornek, M.; Rodrigues, P.M.; Paiva, N.A.; Castro, R.E.; Urban, S.; Pereira, S.P.; Cadamuro, M.; Rupp, C.; Loosen, S.H.; et al. Diagnostic and prognostic biomarkers in cholangiocarcinoma. Liver Int. 2019, 39, 108–122. [Google Scholar] [CrossRef] [Green Version]
- Lazaridis, K.N.; Gores, G.J. Cholangiocarcinoma. Gastroenterology 2005, 128, 1655–1667. [Google Scholar] [CrossRef]
- Lamarca, A.; Barriuso, J.; Chander, A.; McNamara, M.G.; Hubner, R.; Óreilly, D.; Manoharan, P.; Valle, J.W. 18F-fluorodeoxyglucose positron emission tomography (18FDG-PET) for patients with biliary tract cancer: Systematic review and meta-analysis. J. Hepatol. 2019, 71, 115–129. [Google Scholar] [CrossRef] [Green Version]
- Rizvi, S.; Eaton, J.; Yang, J.D.; Chandrasekhara, V.; Gores, G.J. Emerging Technologies for the Diagnosis of Perihilar Cholangiocarcinoma. Semin. Liver Dis. 2018, 38, 160–169. [Google Scholar] [CrossRef]
- Brooks, C.; Gausman, V.; Kokoy-Mondragon, C.; Munot, K.; Amin, S.P.; Desai, A.; Kipp, C.; Poneros, J.; Sethi, A.; Gress, F.G.; et al. Role of Fluorescent In Situ Hybridization, Cholangioscopic Biopsies, and EUS-FNA in the Evaluation of Biliary Strictures. Dig. Dis. Sci. 2018, 63, 636–644. [Google Scholar] [CrossRef]
- Navaneethan, U.; Hasan, M.K.; Lourdusamy, V.; Njei, B.; Varadarajulu, S.; Hawes, R.H. Single-operator cholangioscopy and targeted biopsies in the diagnosis of indeterminate biliary strictures: A systematic review. Gastrointest. Endosc. 2015, 82, 608–614.e2. [Google Scholar] [CrossRef] [Green Version]
- Slivka, A.; Gan, I.; Jamidar, P.; Costamagna, G.; Cesaro, P.; Giovannini, M.; Caillol, F.; Kahaleh, M. Validation of the diagnostic accuracy of probe-based confocal laser endomicroscopy for the characterization of indeterminate biliary strictures: Results of a prospective multicenter international study. Gastrointest. Endosc. 2015, 81, 282–290. [Google Scholar] [CrossRef]
- Sethi, A.; Tyberg, A.; Slivka, A.; Adler, D.G.; Desai, A.P.; Sejpal, D.V.; Pleskow, D.K.; Bertani, H.; Gan, S.-I.; Shah, R.; et al. Digital Single-operator Cholangioscopy (DSOC) Improves Interobserver Agreement (IOA) and Accuracy for Evaluation of Indeterminate Biliary Strictures. The Monaco Classification. J. Clin. Gastroenterol. 2020. [Google Scholar] [CrossRef]
- Balderramo, D. Probe-based confocal laser endomicroscopy contribution in the evaluation of indeterminate biliary strictures. Gastrointest. Endosc. 2015, 82, 970. [Google Scholar] [CrossRef] [Green Version]
- Eloubeidi, M.A.; Chen, V.K.; Jhala, N.C.; Eltoum, I.E.; Jhala, D.; Chhieng, D.C.; Syed, S.A.; Vickers, S.M.; Wilcox, C.M. Endoscopic ultrasound-guided fine needle aspiration biopsy of suspected cholangiocarcinoma. Clin. Gastroenterol. Hepatol. 2004, 2, 209–213. [Google Scholar] [CrossRef]
- Rosen, C.B.; Murad, S.D.; Heimbach, J.K.; Nyberg, S.L.; Nagorney, D.M.; Gores, G.J. Neoadjuvant Therapy and Liver Transplantation for Hilar Cholangiocarcinoma: Is Pretreatment Pathological Confirmation of Diagnosis Necessary? J. Am. Coll. Surg. 2012, 215, 31–38. [Google Scholar] [CrossRef]
- Levy, C.; Lymp, J.; Angulo, P.; Gores, G.J.; LaRusso, N.; Lindor, K.D. The Value of Serum CA 19-9 in Predicting Cholangiocarcinomas in Patients with Primary Sclerosing Cholangitis. Dig. Dis. Sci. 2005, 50, 1734–1740. [Google Scholar] [CrossRef]
- Razumilava, N.; Gores, G.J. Cholangiocarcinoma. Lancet 2014, 383, 2168–2179. [Google Scholar] [CrossRef] [Green Version]
- Rahnemai-Azar, A.A.; Weisbrod, A.; Dillhoff, M.; Schmidt, C.; Pawlik, T.M. Intrahepatic cholangiocarcinoma: Molecular markers for diagnosis and prognosis. Surg. Oncol. 2017, 26, 125–137. [Google Scholar] [CrossRef]
- Rizvi, S.; Gores, G.J. Pathogenesis, Diagnosis, and Management of Cholangiocarcinoma. Gastroenterology 2013, 145, 1215–1229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakrabarti, S.; Kamgar, M.; Mahipal, A. Targeted Therapies in Advanced Biliary Tract Cancer: An Evolving Paradigm. Cancers 2020, 12, 2039. [Google Scholar] [CrossRef] [PubMed]
- Jusakul, A.; Cutcutache, I.; Yong, C.H.; Lim, J.Q.; Ni Huang, M.; Padmanabhan, N.; Nellore, V.; Kongpetch, S.; Ng, A.W.T.; Ng, L.M.; et al. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer Discov. 2017, 7, 1116–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowery, M.A.; Ptashkin, R.N.; Jordan, E.J.; Berger, M.F.; Zehir, A.; Capanu, M.; Kemeny, N.E.; O’Reilly, E.M.; El-Dika, I.; Jarnagin, W.R.; et al. Comprehensive Molecular Profiling of Intrahepatic and Extrahepatic Cholangiocarcinomas: Potential Targets for Intervention. Clin. Cancer Res. 2018, 24, 4154–4161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verlingue, L.; Malka, D.; Allorant, A.; Massard, C.; Ferté, C.; Lacroix, L.; Rouleau, E.; Auger, N.; Ngo, M.; Nicotra, C.; et al. Precision medicine for patients with advanced biliary tract cancers: An effective strategy within the prospective MOSCATO-01 trial. Eur. J. Cancer 2017, 87, 122–130. [Google Scholar] [CrossRef]
- Silverman, I.M.; Lihou, C.F.; Féliz, L.; Frampton, G.M.; Newton, R.C.; Murugesan, K.; Tada, H.; Lee, A.A.; Burn, T.C. Comprehensive genomic profiling in FIGHT-202 reveals the landscape of actionable alterations in advanced cholangiocarcinoma. J. Clin. Oncol. 2019, 37, 4080. [Google Scholar] [CrossRef]
- Javle, M.M.; Shroff, R.T.; Borad, M.J.; Abdel-Wahab, R.; Schrock, A.B.; Chung, J.; Goyal, L.; Frampton, G.M.; Kelley, R.K.; Miller, V.A.; et al. Profiling of 3634 cholangiocarcinomas (CCA) to identify genomic alterations (GA), tumor mutational burden (TMB), and genomic loss of heterozygosity (gLOH). J. Clin. Oncol. 2019, 37, 4087. [Google Scholar] [CrossRef]
- Abou-Alfa, G.K.; Javle, M.; Kelley, R.K.; Lubner, S.; Adeva, J.; Macarulla Mercade, T. LBA10_PRClarIDHy: A global, phase III, randomized, double-blind study of ivosidenib (IVO) vs. placebo in patients with advanced cholangiocarcinoma (CC) with an isocitrate dehydrogenase 1 (IDH1) mutation. Ann. Oncol. 2019, 30. [Google Scholar] [CrossRef]
- Zhu, A.X.; Macarulla, T.; Javle, M.M.; Kelly, R.K.; Lubner, S.J.; Adeva, J.; Cleary, J.M.; Catenacci, D.V.T.; Borad, M.J.; Wu, B.; et al. Final results from ClarIDHy, a global, phase III, randomized, double-blind study of ivosidenib (IVO) versus placebo (PBO) in patients (pts) with previously treated cholangiocarcinoma (CCA) and an isocitrate dehydrogenase 1 (IDH1) mutation. J. Clin. Oncol. 2021, 39, 266. [Google Scholar]
- Abou-Alfa, G.K.; Sahai, V.; Hollebecque, A.; Vaccaro, G.; Melisi, D.; Al-Rajabi, R.; Paulson, A.S.; Borad, M.J.; Gallinson, D.; Murphy, A.G.; et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: A multicentre, open-label, phase 2 study. Lancet Oncol. 2020, 21, 671–684. [Google Scholar] [CrossRef]
- Kinugasa, H.; Nouso, K.; Ako, S.; Dohi, C.; Matsushita, H.; Matsumoto, K.; Kato, H.; Okada, H. Liquid biopsy of bile for the molecular diagnosis of gallbladder cancer. Cancer Biol. Ther. 2018, 19, 934–938. [Google Scholar] [CrossRef]
- Yarlagadda, B.; Kamatham, V.; Ritter, A.; Shahjehan, F.; Kasi, P.M. Trastuzumab and pertuzumab in circulating tumor DNA ERBB2-amplified HER2-positive refractory cholangiocarcinoma. NPJ Precis. Oncol. 2019, 3, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Zill, O.A.; Greene, C.; Sebisanovic, D.; Siew, L.M.; Leng, J.; Vu, M.; Hendifar, A.E.; Wang, Z.; Atreya, C.E.; Kelley, R.K.; et al. Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas. Cancer Discov. 2015, 5, 1040–1048. [Google Scholar] [CrossRef] [Green Version]
- Mody, K.; Surapaneni, P.K.; Bekaii-Saab, T.S.; Ramanathan, R.K.; Ahn, D.H.; Mahipal, A.; Starr, J.S.; Ritter, A.; McMillan, J.; Wylie, N. Landscape of circulating tumor DNA and tissue-based profiling in advanced cholangiocarcinoma. J. Clin. Oncol. 2019, 37, 291. [Google Scholar] [CrossRef]
- Goyal, L.; Saha, S.K.; Liu, L.Y.; Siravegna, G.; Leshchiner, I.; Ahronian, L.G.; Lennerz, J.K.; Vu, P.; Deshpande, V.; Kambadakone, A.; et al. Polyclonal Secondary FGFR2 Mutations Drive Acquired Resistance to FGFR Inhibition in Patients with FGFR2 Fusion–Positive Cholangiocarcinoma. Cancer Discov. 2017, 7, 252–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, N.; Zhang, D.; Yin, L.; Qiu, Y.; Liu, J.; Yu, W.; Fu, X.; Zhu, B.; Xu, X.; Duan, A.; et al. Bile cell-free DNA as a novel and powerful liquid biopsy for detecting somatic variants in biliary tract cancer. Oncol. Rep. 2019, 42, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Afonso, M.B.; Rodrigues, P.M.; Simão, A.L.; Castro, R.E. Circulating microRNAs as Potential Biomarkers in Non-Alcoholic Fatty Liver Disease and Hepatocellular Carcinoma. J. Clin. Med. 2016, 5, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Z.; Liu, X.; Zhang, Q.; Wang, C.; Zhao, Y. Diagnostic value of microRNAs as biomarkers for cholangiocarcinoma. Dig. Liver Dis. 2016, 48, 1227–1232. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, Z.; Yang, S.; Li, X. Identification of microRNAs as biomarkers for cholangiocarcinoma detection: A diagnostic meta-analysis. Clin. Res. Hepatol. Gastroenterol. 2017, 41, 156–162. [Google Scholar] [CrossRef]
- Shigehara, K.; Yokomuro, S.; Ishibashi, O.; Mizuguchi, Y.; Arima, Y.; Kawahigashi, Y.; Kanda, T.; Akagi, I.; Tajiri, T.; Yoshida, H.; et al. Real-Time PCR-Based Analysis of the Human Bile MicroRNAome Identifies miR-9 as a Potential Diagnostic Biomarker for Biliary Tract Cancer. PLoS ONE 2011, 6, e23584. [Google Scholar] [CrossRef]
- Voigtländer, T.; Gupta, S.K.; Thum, S.; Fendrich, J.; Manns, M.P.; Lankisch, T.O.; Thum, T. MicroRNAs in Serum and Bile of Patients with Primary Sclerosing Cholangitis and/or Cholangiocarcinoma. PLoS ONE 2015, 10, e0139305. [Google Scholar] [CrossRef] [Green Version]
- Han, H.S.; Kim, M.J.; Han, J.-H.; Yun, J.; Kim, H.K.; Yang, Y.; Kim, K.B.; Park, S.M. Bile-derived circulating extracellular miR-30d-5p and miR-92a-3p as potential biomarkers for cholangiocarcinoma. Hepatobiliary Pancreat. Dis. Int. 2020, 19, 41–50. [Google Scholar] [CrossRef]
- Wang, L.-J.; He, C.-C.; Sui, X.; Cai, M.-J.; Zhou, C.-Y.; Ma, J.-L.; Wu, L.; Wang, H.; Han, S.-X.; Zhu, Q. MiR-21 promotes intrahepatic cholangiocarcinoma proliferation and growth in vitro and in vivo by targeting PTPN14 and PTEN. Oncotarget 2015, 6, 5932–5946. [Google Scholar] [CrossRef] [Green Version]
- Correa, J.C.; Maddalo, D.; Doussot, A.; Kemeny, N.; Kingham, T.P.; Allen, P.J.; D’Angelica, M.I.; DeMatteo, R.P.; Betel, D.; Klimstra, D.; et al. Circulating Plasma Levels of MicroRNA-21 and MicroRNA-221 Are Potential Diagnostic Markers for Primary Intrahepatic Cholangiocarcinoma. PLoS ONE 2016, 11, e0163699. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; Deng, X.; Zhu, T.; Wei, Y.; Lei, Z.; Guo, M.; Yang, J. Identification of Cholangiocarcinoma Associated with Hepatolithiasis via the Combination of miRNA and Ultrasound. Cancer Manag. Res. 2020, 12, 1845–1853. [Google Scholar] [CrossRef] [Green Version]
- Silakit, R.; Loilome, W.; Yongvanit, P.; Thongchot, S.; Sithithaworn, P.; Boonmars, T.; Koonmee, S.; Titapun, A.; Khuntikeo, N.; Chamadol, N.; et al. Urinary microRNA-192 and microRNA-21 as potential indicators for liver fluke-associated cholangiocarcinoma risk group. Parasitol. Int. 2017, 66, 479–485. [Google Scholar] [CrossRef]
- Huang, Q.; Liu, C.-H.; Jin, Z.-Y.; Xie, F.; Zhu, C.-L.; Liu, Z.; Wang, C. Circulating microRNA-21 as a prognostic, biological marker in cholangiocarcinoma. J. Cancer Res. Ther. 2018, 14, 220. [Google Scholar] [CrossRef]
- Guo, X.; Lv, X.; Lv, X.; Ma, Y.; Chen, L.; Chen, Y. Circulating miR-21 serves as a serum biomarker for hepatocellular carcinoma and correlated with distant metastasis. Oncotarget 2017, 8, 44050–44058. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, C.; Jiang, W.; Huang, D.; Xu, L.; Yang, Q.; Zheng, L.; Wang, X.; Hu, L. Serum miR-21, miR-26a and miR-101 as potential biomarkers of hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 2016, 40, 386–396. [Google Scholar] [CrossRef]
- Bihrer, V.; Waidmann, O.; Friedrich-Rust, M.; Forestier, N.; Susser, S.; Haupenthal, J.; Welker, M.; Shi, Y.; Peveling-Oberhag, J.; Polta, A.; et al. Serum MicroRNA-21 as Marker for Necroinflammation in Hepatitis C Patients with and without Hepatocellular Carcinoma. PLoS ONE 2011, 6, e26971. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Wu, C.; Che, X.; Wang, L.; Yu, D.; Zhang, T.; Huang, L.; Li, H.; Tan, W.; Wang, C.; et al. Circulating MicroRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol. Carcinog. 2011, 50, 136–142. [Google Scholar] [CrossRef]
- Goto, T.; Fujiya, M.; Konishi, H.; Sasajima, J.; Fujibayashi, S.; Hayashi, A.; Utsumi, T.; Sato, H.; Iwama, T.; Ijiri, M.; et al. An elevated expression of serum exosomal microRNA-191, 21, 451 a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer 2018, 18, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Sierzega, M.; Kaczor, M.; Kolodziejczyk, P.; Kulig, J.; Sanak, M.; Richter, P. Evaluation of serum microRNA biomarkers for gastric cancer based on blood and tissue pools profiling: The importance of miR-21 and miR-331. Br. J. Cancer 2017, 117, 266–273. [Google Scholar] [CrossRef] [Green Version]
- Han, J.-G.; Jiang, Y.-D.; Zhang, C.-H.; Yang, Y.-M.; Pang, D.; Song, Y.-N.; Zhang, G.-Q. A novel panel of serum miR-21/miR-155/miR-365 as a potential diagnostic biomarker for breast cancer. Ann. Surg. Treat. Res. 2017, 92, 55–66. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.; Huang, Z.; Zhu, D.; Zhou, X.; Shan, X.; Qi, L.-W.; Wu, L.; Cheng, W.; Zhu, J.; Zhang, L.; et al. A panel of microRNA signature in serum for colorectal cancer diagnosis. Oncotarget 2017, 8, 17081–17091. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Q.; Feng, F.; Zhu, L.; Zheng, Y.; Luo, X.; Liu, C.; Yi, B.; Jiang, X. Circulating miR-106a is a Novel Prognostic and Lymph Node Metastasis Indicator for Cholangiocarcinoma. Sci. Rep. 2015, 5, 16103. [Google Scholar] [CrossRef]
- Bernuzzi, F.; Marabita, F.; Lleo, A.; Carbone, M.; Mirolo, M.; Marzioni, M.; Alpini, G.; Alvaro, D.; Boberg, K.M.; Locati, M.; et al. Serum microRNAs as novel biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Clin. Exp. Immunol. 2016, 185, 61–71. [Google Scholar] [CrossRef] [Green Version]
- Loosen, S.H.; Lurje, G.; Wiltberger, G.; Vucur, M.; Koch, A.; Kather, J.N.; Paffenholz, P.; Tacke, F.; Ulmer, F.T.; Trautwein, C.; et al. Serum levels of miR-29, miR-122, miR-155 and miR-192 are elevated in patients with cholangiocarcinoma. PLoS ONE 2019, 14, e0210944. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Yin, J.; Li, T.; Yuan, L.; Wang, N.; He, J.; Du, X.; Lu, J. Upregulated circulating miR-150 is associated with the risk of intrahepatic cholangiocarcinoma. Oncol. Rep. 2014, 33, 819–825. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Guangxia, C.; Chen, D.; Wu, F.; Lv, Z.; Zhan, Q.; Jiao, Y.; Wang, W.; Chen, G.; Dayang, C. Profiling of downregulated blood-circulating miR-150-5p as a novel tumor marker for cholangiocarcinoma. Tumor Biol. 2016, 37, 15019–15029. [Google Scholar] [CrossRef]
- Salem, P.E.S.; Ghazala, R.A.; El Gendi, A.M.; Emara, D.M.; Ahmed, N.M. The association between circulating MicroRNA-150 level and cholangiocarcinoma. J. Clin. Lab. Anal. 2020, 34, e23397. [Google Scholar] [CrossRef]
- Wang, L.-J.; Zhang, K.-L.; Zhang, N.; Ma, X.-W.; Yan, S.-W.; Cao, D.-H.; Shi, S.-J. Serum miR-26a as a diagnostic and prognostic biomarker in cholangiocarcinoma. Oncotarget 2015, 6, 18631–18640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plieskatt, J.; Rinaldi, G.; Feng, Y.; Peng, J.; Easley, S.; Jia, X.; Potriquet, J.; Pairojkul, C.; Bhudhisawasdi, V.; Sripa, B.; et al. A microRNA profile associated with Opisthorchis viverrini-induced cholangiocarcinoma in tissue and plasma. BMC Cancer 2015, 15, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silakit, R.; Loilome, W.; Yongvanit, P.; Chusorn, P.; Techasen, A.; Boonmars, T.; Khuntikeo, N.; Chamadol, N.; Pairojkul, C.; Namwat, N. Circulating miR-192 in liver fluke-associated cholangiocarcinoma patients: A prospective prognostic indicator. J. Hepato Biliary Pancreat. Sci. 2014, 21, 864–872. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Zhong, L.; He, Q.; Wang, S.; Pan, Z.; Wang, T.; Zhao, Y. Diagnostic Accuracy of Serum CA19-9 in Patients with Cholangiocarcinoma: A Systematic Review and Meta-Analysis. Med. Sci. Monit. 2015, 21, 3555–3563. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Li, D.-J.; Chen, J.; Liu, W.; Li, J.-W.; Jiang, P.; Zhao, X.; Guo, F.; Li, X.-W.; Wang, S.-G. Application of Joint Detection of AFP, CA19-9, CA125 and CEA in Identification and Diagnosis of Cholangiocarcinoma. Asian Pac. J. Cancer Prev. 2015, 16, 3451–3455. [Google Scholar] [CrossRef] [Green Version]
- Ince, A.T.; Yildiz, K.; Baysal, B.; Danalioglu, A.; Kocaman, O.; Tozlu, M.; Gangarapu, V.; Kemik, A.S.; Uysal, O.; Senturk, H. Roles of serum and biliary CEA, CA19-9, VEGFR3, and TAC in differentiating between malignant and benign biliary obstructions. Turk. J. Gastroenterol. 2014, 25, 162–169. [Google Scholar] [CrossRef] [Green Version]
- Loosen, S.H.; Roderburg, C.; Kauertz, K.L.; Koch, A.; Vucur, M.; Schneider, A.T.; Binnebösel, M.; Ulmer, T.F.; Lurje, G.; Schoening, W.; et al. CEA but not CA19-9 is an independent prognostic factor in patients undergoing resection of cholangiocarcinoma. Sci. Rep. 2017, 7, 16975. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Chen, W.; Liang, P.; Hu, W.; Zhang, K.; Shen, S.; Chen, J.; Zhang, Z.; Chen, B.; Han, Y.; et al. Serum CYFRA 21-1 in Biliary Tract Cancers: A Reliable Biomarker for Gallbladder Carcinoma and Intrahepatic Cholangiocarcinoma. Dig. Dis. Sci. 2014, 60, 1273–1283. [Google Scholar] [CrossRef] [PubMed]
- Uenishi, T.; Yamazaki, O.; Tanaka, H.; Takemura, S.; Yamamoto, T.; Tanaka, S.; Nishiguchi, S.; Kubo, S. Serum Cytokeratin 19 Fragment (CYFRA21-1) as a Prognostic Factor in Intrahepatic Cholangiocarcinoma. Ann. Surg. Oncol. 2007, 15, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Leelawat, K.; Narong, S.; Wannaprasert, J.; Ratanashu-Ek, T. Prospective study of MMP7 serum levels in the diagnosis of cholangiocarcinoma. World J. Gastroenterol. 2010, 16, 4697–4703. [Google Scholar] [CrossRef]
- Leelawat, K.; Sakchinabut, S.; Narong, S.; Wannaprasert, J. Detection of serum MMP-7 and MMP-9 in cholangiocarcinoma patients: Evaluation of diagnostic accuracy. BMC Gastroenterol. 2009, 9, 30. [Google Scholar] [CrossRef] [Green Version]
- Loosen, S.H.; Roderburg, C.; Kauertz, K.L.; Pombeiro, I.; Leyh, C.; Benz, F.; Vucur, M.; Longerich, T.; Koch, A.; Braunschweig, T.; et al. Elevated levels of circulating osteopontin are associated with a poor survival after resection of cholangiocarcinoma. J. Hepatol. 2017, 67, 749–757. [Google Scholar] [CrossRef]
- Chang, T.; Cheng, J.; Tsai, H.; Young, K.; Hsieh, S.; Ho, C. Plasma proteome plus site-specific N-glycoprofiling for hepatobiliary carcinomas. J. Pathol. Clin. Res. 2019, 5, 199–212. [Google Scholar] [CrossRef] [Green Version]
- Loosen, S.H.; Breuer, A.; Tacke, F.; Kather, J.N.; Gorgulho, J.; Alizai, P.H.; Bednarsch, J.; Roeth, A.A.; Lurje, G.; Schmitz, S.M.; et al. Circulating levels of soluble urokinase plasminogen activator receptor predict outcome after resection of biliary tract cancer. JHEP Rep. 2020, 2, 100080. [Google Scholar] [CrossRef] [Green Version]
- Onsurathum, S.; Haonon, O.; Pinlaor, P.; Pairojkul, C.; Khuntikeo, N.; Thanan, R.; Roytrakul, S.; Pinlaor, S. Proteomics detection of S100A6 in tumor tissue interstitial fluid and evaluation of its potential as a biomarker of cholangiocarcinoma. Tumor. Biol. 2018, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loosen, S.H.; Benz, F.; Niedeggen, J.; Schmeding, M.; Schüller, F.; Koch, A.; Vucur, M.; Tacke, F.; Trautwein, C.; Roderburg, C.; et al. Serum levels of S100A6 are unaltered in patients with resectable cholangiocarcinoma. Clin. Transl. Med. 2016, 5, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, R.-Y.; Yang, X.-R.; Shen, Q.-J.; Yang, L.-X.; Xu, Y.; Qiu, S.-J.; Sun, Y.-F.; Zhang, X.; Wang, Z.; Zhu, K.; et al. High expression of Dickkopf-related protein 1 is related to lymphatic metastasis and indicates poor prognosis in intrahepatic cholangiocarcinoma patients after surgery. Cancer 2012, 119, 993–1003. [Google Scholar] [CrossRef]
- Shen, J.; Wang, W.; Wu, J.; Feng, B.; Chen, W.; Wang, M.; Tang, J.; Wang, F.; Cheng, F.; Pu, L.; et al. Comparative Proteomic Profiling of Human Bile Reveals SSP411 as a Novel Biomarker of Cholangiocarcinoma. PLoS ONE 2012, 7, e47476. [Google Scholar] [CrossRef]
- Kobayashi, S.; Werneburg, N.W.; Bronk, S.F.; Kaufmann, S.H.; Gores, G.J. Interleukin-6 Contributes to Mcl-1 Up-regulation and TRAIL Resistance via an Akt-Signaling Pathway in Cholangiocarcinoma Cells. Gastroenterology 2005, 128, 2054–2065. [Google Scholar] [CrossRef]
- Cheon, Y.K.; Cho, Y.D.; Moon, J.H.; Jang, J.Y.; Kim, Y.S.; Kim, Y.S.; Lee, M.S.; Lee, J.S.; Shim, C.S. Diagnostic Utility of Interleukin-6 (IL-6) for Primary Bile Duct Cancer and Changes in Serum IL-6 Levels Following Photodynamic Therapy. Am. J. Gastroenterol. 2007, 102, 2164–2170. [Google Scholar] [CrossRef]
- Kimawaha, P.; Jusakul, A.; Junsawang, P.; Loilome, W.; Khuntikeo, N.; Techasen, A. Circulating TGF-β1 as the potential epithelial mesenchymal transition-biomarker for diagnosis of cholangiocarcinoma. J. Gastrointest. Oncol. 2020, 11, 304–318. [Google Scholar] [CrossRef]
- Intuyod, K.; Armartmuntree, N.; Jusakul, A.; Sakonsinsiri, C.; Thanan, R.; Pinlaor, S. Current omics-based biomarkers for cholangiocarcinoma. Expert Rev. Mol. Diagn. 2019, 19, 997–1005. [Google Scholar] [CrossRef]
- Chang, Y.-C.; Chen, M.-H.; Yeh, C.-N.; Hsiao, M. Omics-Based Platforms: Current Status and Potential Use for Cholangiocarcinoma. Biomolecues 2020, 10, 1377. [Google Scholar] [CrossRef]
- Liang, Q.; Liu, H.; Zhang, T.; Jiang, Y.; Xing, H.; Zhang, H. Serum metabolomics uncovering specific metabolite signatures of intra- and extrahepatic cholangiocarcinoma. Mol. BioSyst. 2015, 12, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Banales, J.S.; Iñarrairaegui, M.; Arbelaiz, A.; Milkiewicz, P.; Muntane, J.; Muñoz-Bellvis, L.; La Casta, A.; Gonzalez, L.M.; Arretxe, E.; Alonso, C.; et al. Serum Metabolites as Diagnostic Biomarkers for Cholangiocarcinoma, Hepatocellular Carcinoma, and Primary Sclerosing Cholangitis. Hepatolongy 2019, 70, 547–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urman, J.M.; Herranz, J.M.; Uriarte, I.; Rullán, M.; Oyón, D.; González, B.; Fernandez-Urién, I.; Carrascosa, J.; Bolado, F.; Zabalza, L.; et al. Pilot Multi-Omic Analysis of Human Bile from Benign and Malignant Biliary Strictures: A Machine-Learning Approach. Cancers 2020, 12, 1644. [Google Scholar] [CrossRef] [PubMed]
- Alsaleh, M.; Leftley, Z.; Barbera, T.A.; Koomson, L.K.; Zabron, A.; Crossey, M.M.; Reeves, H.L.; Cramp, M.; Ryder, S.; Greer, S.; et al. Characterisation of the Serum Metabolic Signature of Cholangiocarcinoma in a United Kingdom Cohort. J. Clin. Exp. Hepatol. 2020, 10, 17–29. [Google Scholar] [CrossRef]
- Haznadar, M.; Diehl, C.M.; Parker, A.L.; Krausz, K.W.; Bowman, E.D.; Rabibhadana, S.; Forgues, M.; Bhudhisawasdi, V.; Gonzalez, F.J.; Mahidol, C.; et al. Urinary Metabolites Diagnostic and Prognostic of Intrahepatic Cholangiocarcinoma. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1704–1711. [Google Scholar] [CrossRef] [Green Version]
- Lapitz, A.; Arbelaiz, A.; Olaizola, P.; Aranburu, A.; Bujanda, L.; Perugorria, M.J.; Banales, J.M. Extracellular Vesicles in Hepatobiliary Malignancies. Front. Immunol. 2018, 9, 2270. [Google Scholar] [CrossRef] [Green Version]
- Caby, M.-P.; Lankar, D.; Vincendeau-Scherrer, C.; Raposo, G.; Bonnerot, C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005, 17, 879–887. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, Y.; Miura, Y.; Harazono, A.; Kanai-Azuma, M.; Akimoto, Y.; Kawakami, H.; Yamaguchi, T.; Toda, T.; Endo, T.; Tsubuki, M.; et al. Proteomic Analysis of Two Types of Exosomes in Human Whole Saliva. Biol. Pharm. Bull. 2011, 34, 13–23. [Google Scholar] [CrossRef] [Green Version]
- Pisitkun, T.; Shen, R.-F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar] [CrossRef] [Green Version]
- Masyuk, A.I.; Huang, B.Q.; Ward, C.J.; Gradilone, S.A.; Banales, J.; Masyuk, T.V.; Radtke, B.; Splinter, P.L.; LaRusso, N.F. Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia. Am. J. Physiol. Liver Physiol. 2010, 299, G990–G999. [Google Scholar] [CrossRef] [Green Version]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Severino, V.; Dumonceau, J.-M.; Delhaye, M.; Moll, S.; Annessi-Ramseyer, I.; Robin, X.; Frossard, J.-L.; Farina, A. Extracellular Vesicles in Bile as Markers of Malignant Biliary Stenoses. Gastroenterology 2017, 153, 495–504.e8. [Google Scholar] [CrossRef] [Green Version]
- Julich-Haertel, H.; Urban, S.K.; Krawczyk, M.; Willms, A.; Jankowski, K.; Patkowski, W.; Kruk, B.; Krasnodębski, M.; Ligocka, J.; Schwab, R.; et al. Cancer-associated circulating large extracellular vesicles in cholangiocarcinoma and hepatocellular carcinoma. J. Hepatol. 2017, 67, 282–292. [Google Scholar] [CrossRef]
- Albiin, N.; Smith, I.C.P.; Arnelo, U.; Lindberg, B.; Bergquist, A.; Dolenko, B.; Bryksina, N.; Bezabeh, T. Detection of cholangiocarcinoma with magnetic resonance spectroscopy of bile in patients with and without primary sclerosing cholangitis. Acta Radiol. 2008, 49, 855–862. [Google Scholar] [CrossRef]
- Sharif, A.W.; Williams, H.R.; Lampejo, T.; Khan, S.A.; Bansi, D.S.; Westaby, D.; Thillainayagam, A.V.; Thomas, H.C.; Cox, I.J.; Taylor-Robinson, S.D. Metabolic profiling of bile in cholangiocarcinoma using in vitro magnetic resonance spectroscopy. HPB 2010, 12, 396–402. [Google Scholar] [CrossRef] [Green Version]
- Arbelaiz, A.; Azkargorta, M.; Krawczyk, M.; Santos-Laso, A.; Lapitz, A.; Perugorria, M.J.; Erice, O.; Gonzalez, E.; Jimenez-Agüero, R.; La Casta, A.; et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology 2017, 66, 1125–1143. [Google Scholar] [CrossRef]
- Lapitz, A.; Arbelaiz, A.; O’Rourke, C.J.; Lavin, J.L.; La Casta, A.; Ibarra, C.; Jimeno, J.P.; Santos-Laso, A.; Izquierdo-Sanchez, L.; Krawczyk, M.; et al. Patients with Cholangiocarcinoma Present Specific RNA Profiles in Serum and Urine Extracellular Vesicles Mirroring the Tumor Expression: Novel Liquid Biopsy Biomarkers for Disease Diagnosis. Cells 2020, 9, 721. [Google Scholar] [CrossRef] [Green Version]
- Ge, X.; Wang, Y.; Nie, J.; Li, Q.; Tang, L.; Deng, X.; Wang, F.; Xu, B.; Wu, X.; Zhang, X.; et al. The diagnostic/prognostic potential and molecular functions of long non-coding RNAs in the exosomes derived from the bile of human cholangiocarcinoma. Oncotarget 2017, 8, 69995–70005. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Masica, D.; Ishida, M.; Tomuleasa, C.; Umegaki, S.; Kalloo, A.N.; Georgiades, C.; Singh, V.K.; Khashab, M.; Amateau, S.; et al. Human bile contains MicroRNA-laden extracellular vesicles that can be used for cholangiocarcinoma diagnosis. Hepatology 2014, 60, 896–907. [Google Scholar] [CrossRef]
- Shen, L.; Chen, G.; Xia, Q.; Shao, S.; Fang, H. Exosomal miR-200 family as serum biomarkers for early detection and prognostic prediction of cholangiocarcinoma. Int. J. Clin. Exp. Pathol. 2019, 12, 3870–3876. [Google Scholar] [PubMed]
- Xue, X.-Y.; Liu, Y.-X.; Wang, C.; Gu, X.-J.; Xue, Z.-Q.; Zang, X.-L.; Ma, X.-D.; Deng, H.; Liu, R.; Pan, L.; et al. Identification of exosomal miRNAs as diagnostic biomarkers for cholangiocarcinoma and gallbladder carcinoma. Signal Transduct. Target. Ther. 2020, 5, 77. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- De Boer, C.J.; van Krieken, J.H.; Janssen-van Rhijn, C.M.; Litvinov, S.V. Expression of Ep-CAM in normal, regenerating, metaplastic, and neoplastic liver. J. Pathol. 1999, 188, 201–206. [Google Scholar] [CrossRef]
- Al Ustwani, O.; Iancu, D.; Yacoub, R.; Iyer, R. Detection of circulating tumor cells in cancers of biliary origin. J. Gastrointest. Oncol. 2012, 3, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Reduzzi, C.; Vismara, M.; Silvestri, M.; Celio, L.; Niger, M.; Peverelli, G.; De Braud, F.; Daidone, M.G.; Cappelletti, V. A novel circulating tumor cell subpopulation for treatment monitoring and molecular characterization in biliary tract cancer. Int. J. Cancer 2020, 146, 3495–3503. [Google Scholar] [CrossRef]
- Yang, J.D.; Campion, M.B.; Liu, M.C.; Chaiteerakij, R.; Giama, N.H.; Mohammed, H.A.; Zhang, X.; Hu, C.; Campion, V.L.; Jen, J.; et al. Circulating tumor cells are associated with poor overall survival in patients with cholangiocarcinoma. Hepatology 2016, 63, 148–158. [Google Scholar] [CrossRef]
- Backen, A.C.; Lopes, A.; Wasan, H.; Palmer, D.H.; Duggan, M.; Cunningham, D.; Anthoney, A.; Corrie, P.G.; Madhusudan, S.; Maraveyas, A.; et al. Circulating biomarkers during treatment in patients with advanced biliary tract cancer receiving cediranib in the UK ABC-03 trial. Br. J. Cancer 2018, 119, 27–35. [Google Scholar] [CrossRef]
- Arnoletti, J.P.; Zhu, X.; Almodovar, A.J.; Veldhuis, P.P.; Sause, R.; Griffith, E.; Corpus, G.; Chang, J.C.; Fanaian, N.; Litherland, S.A. Portal Venous Blood Circulation Supports Immunosuppressive Environment and Pancreatic Cancer Circulating Tumor Cell Activation. Pancreas 2017, 46, 116–123. [Google Scholar] [CrossRef]
- Arnoletti, J.P.; Fanaian, N.; Reza, J.; Sause, R.; Almodovar, A.J.; Srivastava, M.; Patel, S.; Veldhuis, P.P.; Griffith, E.; Shao, Y.-P.; et al. Pancreatic and bile duct cancer circulating tumor cells (CTC) form immune-resistant multi-cell type clusters in the portal venous circulation. Cancer Biol. Ther. 2018, 19, 887–897. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Ridgway, L.D.; Wetzel, M.D.; Ngo, J.; Yin, W.; Kumar, D.; Goodman, J.C.; Groves, M.D.; Marchetti, D. The Identification and Characterization of Breast Cancer CTCs Competent for Brain Metastasis. Sci. Transl. Med. 2013, 5, 180ra48. [Google Scholar] [CrossRef] [Green Version]
- Hodgkinson, C.L.; Morrow, C.J.; Li, Y.; Metcalf, R.L.; Rothwell, D.G.; Trapani, F.; Polanski, R.; Burt, D.J.; Simpson, K.L.; Morris, K.; et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat. Med. 2014, 20, 897–903. [Google Scholar] [CrossRef]
- Ong, C.K.; Subimerb, C.; Pairojkul, C.; Wongkham, S.; Cutcutache, I.; Yu, W.; McPherson, J.R.; Allen, G.; Ng, C.C.Y.; Wong, B.H.; et al. Exome sequencing of liver fluke–associated cholangiocarcinoma. Nat. Genet. 2012, 44, 690–693. [Google Scholar] [CrossRef]
- Zou, S.; Li, J.; Zhou, H.; Frech, C.; Jiang, X.; Chu, J.S.C.; Zhao, X.; Li, Y.; Li, Q.; Wang, H.; et al. Mutational landscape of intrahepatic cholangiocarcinoma. Nat. Commun. 2014, 5, 5696. [Google Scholar] [CrossRef]
- Ross, J.S.; Wang, K.; Gay, L.; Al-Rohil, R.; Rand, J.V.; Jones, D.M.; Lee, H.J.; Sheehan, C.E.; Otto, G.A.; Palmer, G.; et al. New Routes to Targeted Therapy of Intrahepatic Cholangiocarcinomas Revealed by Next-Generation Sequencing. Oncologist 2014, 19, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Fujimoto, A.; Furuta, M.; Shiraishi, Y.; Gotoh, K.; Kawakami, Y.; Arihiro, K.; Nakamura, T.; Ueno, M.; Ariizumi, S.-I.; Nguyen, H.H.; et al. Whole-genome mutational landscape of liver cancers displaying biliary phenotype reveals hepatitis impact and molecular diversity. Nat. Commun. 2015, 6, 6120. [Google Scholar] [CrossRef]
- Nakamura, H.; Arai, Y.; Totoki, Y.; Shirota, T.; El Zawahry, A.; Kato, M.; Hama, N.; Hosoda, F.; Urushidate, T.; Ohashi, S.; et al. Genomic spectra of biliary tract cancer. Nat. Genet. 2015, 47, 1003–1010. [Google Scholar] [CrossRef]
- Sia, D.; Hoshida, Y.; Villanueva, A.; Roayaie, S.; Ferrer-Fabrega, J.; Tabak, B.; Peix, J.; Sole, M.; Tovar, V.; Alsinet, C.; et al. Integrative Molecular Analysis of Intrahepatic Cholangiocarcinoma Reveals 2 Classes That Have Different Outcomes. Gastroenterology 2013, 144, 829–840. [Google Scholar] [CrossRef] [Green Version]
- Gao, Q.; Zhao, Y.; Wang, X.; Guo, W.; Gao, S.; Wei, L.; Shi, J.; Shi, G.; Wang, Z.; Zhang, Y.; et al. Activating Mutations in PTPN3 Promote Cholangiocarcinoma Cell Proliferation and Migration and Are Associated with Tumor Recurrence in Patients. Gastroenterology 2014, 146, 1397–1407. [Google Scholar] [CrossRef]
- Zender, S.; Nickeleit, I.; Wuestefeld, T.; Sörensen, I.; Dauch, D.; Bozko, P.; El-Khatib, M.; Geffers, R.; Bektas, H.; Manns, M.P.; et al. A Critical Role for Notch Signaling in the Formation of Cholangiocellular Carcinomas. Cancer Cell 2013, 23, 784–795. [Google Scholar] [CrossRef] [Green Version]
- Villanueva, A.; Alsinet, C.; Yanger, K.; Hoshida, Y.; Zong, Y.; Toffanin, S.; Rodriguez–Carunchio, L.; Solé, M.; Thung, S.; Stanger, B.Z.; et al. Notch Signaling Is Activated in Human Hepatocellular Carcinoma and Induces Tumor Formation in Mice. Gastroenterology 2012, 143, 1660–1669.e7. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Y.; Pawlik, T.M.; Anders, R.; Selaru, F.M.; Streppel, M.M.; Lucas, D.J.; Niknafs, N.; Guthrie, V.B.; Maitra, A.; Argani, P.; et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat. Genet. 2013, 45, 1470–1473. [Google Scholar] [CrossRef]
- Chan-On, W.; Nairismägi, M.-L.; Ong, C.K.; Lim, W.K.; Dima, S.; Pairojkul, C.; Lim, K.H.; McPherson, J.R.; Cutcutache, I.; Heng, H.L.; et al. Exome sequencing identifies distinct mutational patterns in liver fluke–related and non-infection-related bile duct cancers. Nat. Genet. 2013, 45, 1474–1478. [Google Scholar] [CrossRef]
- Andersen, J.B.; Spee, B.; Blechacz, B.R.; Avital, I.; Komuta, M.; Barbour, A.; Conner, E.A.; Gillen, M.C.; Roskams, T.; Roberts, L.; et al. Genomic and Genetic Characterization of Cholangiocarcinoma Identifies Therapeutic Targets for Tyrosine Kinase Inhibitors. Gastroenterology 2012, 142, 1021–1031.e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nepal, C.; O’Rourke, C.J.; Oliveira, D.N.P.; Taranta, A.; Shema, S.; Gautam, P.; Calderaro, J.; Barbour, A.; Raggi, C.; Wennerberg, K.; et al. Genomic perturbations reveal distinct regulatory networks in intrahepatic cholangiocarcinoma. Hepatology 2018, 68, 949–963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arai, Y.; Totoki, Y.; Hosoda, F.; Shirota, T.; Hama, N.; Nakamura, H.; Ojima, H.; Furuta, K.; Shimada, K.; Okusaka, T.; et al. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology 2013, 59, 1427–1434. [Google Scholar] [CrossRef] [PubMed]
- Borger, D.R.; Zhu, A.X. IDHmutations: New genetic signatures in cholangiocarcinoma and therapeutic implications. Expert Rev. Anticancer Ther. 2012, 12, 543–546. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Dong, Q.; Zhang, C.; Kuan, P.-F.; Liu, Y.; Jeck, W.R.; Andersen, J.B.; Jiang, W.; Savich, G.L.; Tan, T.-X.; et al. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene 2013, 32, 3091–3100. [Google Scholar] [CrossRef] [Green Version]
- Drilon, A.; Laetsch, T.W.; Kummar, S.; Dubois, S.G.; Lassen, U.N.; Demetri, G.D.; Nathenson, M.; Doebele, R.C.; Farago, A.F.; Pappo, A.S.; et al. Efficacy of Larotrectinib inTRKFusion–Positive Cancers in Adults and Children. N. Engl. J. Med. 2018, 378, 731–739. [Google Scholar] [CrossRef]
- Doebele, R.C.; Drilon, A.; Paz-Ares, L.; Siena, S.; Shaw, A.T.; Farago, A.F.; Blakely, C.M.; Seto, T.; Cho, B.C.; Tosi, D.; et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: Integrated analysis of three phase 1–2 trials. Lancet Oncol. 2020, 21, 271–282. [Google Scholar] [CrossRef]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef] [Green Version]
- Ruys, A.T.; Koerkamp, B.G.; Wiggers, J.K.; Klümpen, H.-J.; Kate, F.J.T.; Van Gulik, T.M. Prognostic Biomarkers in Patients with Resected Cholangiocarcinoma: A Systematic Review and Meta-analysis. Ann. Surg. Oncol. 2013, 21, 487–500. [Google Scholar] [CrossRef]
- Ghidini, M.; Cascione, L.; Carotenuto, P.; Lampis, A.; Trevisani, F.; Previdi, M.C.; Hahne, J.C.; Said-Huntingford, I.; Raj, M.; Zerbi, A.; et al. Characterisation of the immune-related transcriptome in resected biliary tract cancers. Eur. J. Cancer 2017, 86, 158–165. [Google Scholar] [CrossRef] [Green Version]
- Sawada, R.; Ku, Y.; Akita, M.; Otani, K.; Fujikura, K.; Itoh, T.; Ajiki, T.; Fukumoto, T.; Kakeji, Y.; Zen, Y. Interleukin-33 overexpression reflects less aggressive tumour features in large-duct type cholangiocarcinomas. Histopathology 2018, 73, 259–272. [Google Scholar] [CrossRef]
- Ozawa, N.; Doi, S.; Tsujikawa, T.; Mabuchi, M.; Kajiyama, Y.; Sato, K.; Kikuchi, K.; Takahashi, M.; Kawamoto, M.; Yasuda, I. Intrahepatic cholangiocarcinoma producing granulocyte colony-stimulating factor and parathyroid hormone-related protein. Nihon Shokakibyo Gakkai Zasshi 2017, 114, 1285–1292. [Google Scholar]
- Suzumura, K.; Iimuro, Y.; Hirano, T.; Asano, Y.; Kuroda, N.; Okada, T.; Tanaka, S.; Nakasho, K.; Fujimoto, J. Granulocyte Colony-Stimulating Factor–Producing Cholangiocellular Carcinoma. Int. Surg. 2015, 100, 123–127. [Google Scholar] [CrossRef] [Green Version]
- Olaizola, P.; Lee-Law, P.; Arbelaiz, A.; Lapitz, A.; Perugorria, M.; Bujanda, L.; Banales, J. MicroRNAs and extracellular vesicles in cholangiopathies. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1293–1307. [Google Scholar] [CrossRef]
- Esparza-Baquer, A.; Labiano, I.; Bujanda, L.; Perugorria, M.J.; Banales, J.M. MicroRNAs in cholangiopathies: Potential diagnostic and therapeutic tools. Clin. Res. Hepatol. Gastroenterol. 2016, 40, 15–27. [Google Scholar] [CrossRef]
- Chusorn, P.; Namwat, N.; Loilome, W.; Techasen, A.; Pairojkul, C.; Khuntikeo, N.; Dechakhamphu, A.; Talabnin, C.; Chan-On, W.; Ong, C.K.; et al. Overexpression of microRNA-21 regulating PDCD4 during tumorigenesis of liver fluke-associated cholangiocarcinoma contributes to tumor growth and metastasis. Tumor Biol. 2013, 34, 1579–1588. [Google Scholar] [CrossRef]
- Wan, P.; Chi, X.; Du, Q.; Luo, J.; Cui, X.; Dong, K.; Bing, Y.; Heres, C.; Geller, D.A. miR-383 promotes cholangiocarcinoma cell proliferation, migration, and invasion through targeting IRF1. J. Cell. Biochem. 2018, 119, 9720–9729. [Google Scholar] [CrossRef]
- Collins, A.L.; Wojcik, S.; Liu, J.; Frankel, W.L.; Alder, H.; Yu, L.; Schmittgen, T.D.; Croce, C.M.; Bloomston, M. A differential microRNA profile distinguishes cholangiocarcinoma from pancreatic adenocarcinoma. Ann. Surg. Oncol. 2013, 21, 133–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigues, P.M.; Vogel, A.; Arrese, M.; Balderramo, D.C.; Valle, J.W.; Banales, J.M. Next-Generation Biomarkers for Cholangiocarcinoma. Cancers 2021, 13, 3222. https://doi.org/10.3390/cancers13133222
Rodrigues PM, Vogel A, Arrese M, Balderramo DC, Valle JW, Banales JM. Next-Generation Biomarkers for Cholangiocarcinoma. Cancers. 2021; 13(13):3222. https://doi.org/10.3390/cancers13133222
Chicago/Turabian StyleRodrigues, Pedro M., Arndt Vogel, Marco Arrese, Domingo C. Balderramo, Juan W. Valle, and Jesus M. Banales. 2021. "Next-Generation Biomarkers for Cholangiocarcinoma" Cancers 13, no. 13: 3222. https://doi.org/10.3390/cancers13133222






